Abstract
Human cytomegalovirus (HCMV) exploits a range of strategies to evade and modulate the immune response. Its capacity to down-regulate MHC I expression was anticipated to render infected cells vulnerable to natural killer (NK) attack. Kinetic analysis revealed that during productive infection, HCMV strain AD169 first enhanced and then inhibited lysis of primary skin fibroblasts by a CD94/NKG2A+NKG2D+ILT2+ NK line. The inhibition of cytotoxicity against strain AD169-infected fibroblasts was abolished by prior treatment of targets or effectors with anti-MHC I and anti-CD94 monoclonal antibodies, respectively, implying a CD94/HLA-E-dependent mechanism. An HCMV strain AD169, UL40 deletion mutant could not inhibit CD94/NKG2A+ NK killing against skin fibroblasts. The contribution of UL40 to evasion of primary NK cells then was tested in a system where targets and effectors were MHC-matched. Primary NK cells activated with IFNα as well as cultured primary NK cell lines showed increased killing against ΔUL40-infected fibroblasts compared with AD169-infected targets. This effect was abrogated by depletion of CD94+ cells. These findings demonstrate that HCMV encodes a mechanism of evasion specifically targeted against a proportion of CD94+ NK cells and show that this system functions during a productive infection.
Keywords: NK cells‖inhibition
Human cytomegalovirus (HCMV) is the prototypical β-herpesvirus and an important pathogen, being the major viral cause of congenital malformation and associated with severe morbidity in immunocompromised individuals. Primary infection is followed by lifelong persistence. The cellular immune response is crucial in controlling infection and providing resistance to disease (1). HCMV encodes an impressive arsenal of genes that act to counter MHC I-restricted cytotoxic T lymphocyte recognition by down-regulation of MHC I at various stages of the replicative cycle (reviewed in ref. 2).
Although down-regulation of MHC I expression is associated with protection against antigen-specific cytotoxic T lymphocytes, it also may enhance natural killer (NK) recognition of HCMV-infected cells. The importance of NK cells for control of CMV has been shown in vivo: in mice, an NK resistance locus, cmv1, has been identified (3) and mapped to the gene for Ly-49H, an NK-activating receptor (4, 5); in humans, an absence of NK cells increases susceptibility to several herpesvirus infections including HCMV (6). A number of human NK-activating and -inhibitory receptors have now been identified (reviewed in refs. 7–9). Members of the killer inhibitory receptor (KIR) family and the leukocyte Ig-like receptors (LIR-1/ILT-2 and LIR-2/ILT-4) bind HLA class I molecules. The HCMV MHC I homolog gpUL18 exhibits high affinity for LIR-1/ILT-2 (10, 11); however, the function of gpUL18 during virus infection is yet to be resolved. A surface C-type lectin receptor CD94/NKG2A/B also has been shown to interact specifically with the nonclassical MHC I molecule, HLA-E, to deliver an inhibitory signal to NK cells (12–14). HLA-E exhibits limited polymorphism and binds a restricted set of peptides derived from the leader sequence of classical MHC I molecules and HLA-G (15–17). Class I leader peptides are loaded onto HLA-E by a transporter associated with antigen processing (TAP)-dependent pathway (16, 18). Consequently, conventional HLA-E surface expression is blocked by HCMV gpUS6, an inhibitor of TAP function (19, 20). However, an HLA-E-binding peptide is contained within the leader sequence of HCMV gpUL40. We recently demonstrated that adenoviral expression of UL40 in cell lines induced TAP-independent up-regulation of HLA-E cell-surface expression (21) and elicited protection against lysis by NK cells (21, 22). Recent data has suggested that this effect may be enhanced by IFNγ (23), but no studies to date have investigated UL40 in the context of an active HCMV infection.
In vitro cytotoxicity assays have reported variously both inhibition and enhancement of NK killing of HCMV- infected cells (24–28). In this study, data are presented demonstrating that productive HCMV infection can induce protection to NK attack. In particular, the role of UL40 in specifically eliciting protection against CD94/NKG2A+ NK cells was examined systematically during productive virus infection.
Materials and Methods
Cell Lines.
The NK cell line, DEL, was derived from a patient suffering a lymphoid dysplasia associated with a peripheral demyelinating neuropathy. To generate the DEL cell line, patient peripheral blood mononuclear cell (PBMCs) were cultured in RPMI medium 1640 supplemented with L-glutamine, penicillin/streptomycin (GIBCO/BRL), 10% heat-inactivated human AB serum (RPMI-AB), and 1,000 units/ml recombinant IL-2 (Proleukin, Chiron). The derivative line, DEL, expressed CD94 and CD57 (Fig. 1A) and bound HLA-E tetramer (Fig. 1B), and its surface phenotype was NKG2A+, CD16+, CD56+, ILT2+, but KIR− (Fig. 1B). Reverse transcription–PCR also showed mRNA for DAP12, NKG2A, -D, -F, and -G but not NKG2C or -E (data not shown). The NK cell line, NKL, was maintained as described (29). The primary NK cell lines (SW-, DD-, and BM-NK) were generated by culturing in RPMI-AB and 1,000 units/ml recombinant IL-2 after depletion of adherent cells and T cells using the mAb OKT3 (American Type Culture Collection) and Dynabeads (Dynal, Bromborough, U.K.) per manufacturer instructions. Primary NK cell lines were regularly given feeders consisting of irradiated, allogeneic PBMCs and immortalized B lymphoblastoid cell lines. Primary skin fibroblast lines DD-SF, SW-SF, and BM-SF were generated from skin biopsies as described in ref. 30. All fibroblast lines were maintained in DMEM/10% FCS; immortalized B lymphoblastoid cell lines (EW-BCL and SM-BCL) were maintained in RPMI medium 1640/10% FCS.
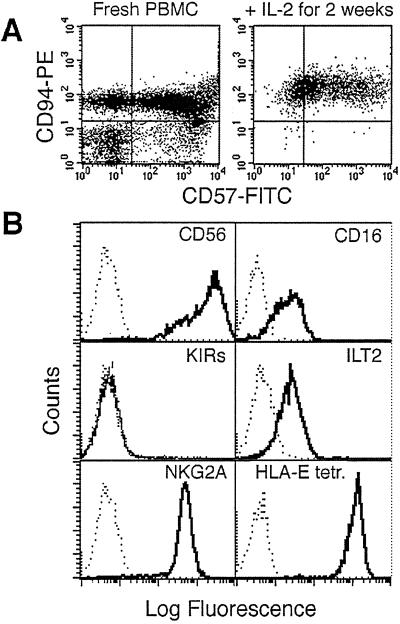
Figure 1.
Characterization of DEL, a CD94/NKG2A+ NK line. (A) Fluorescence-activated cell sorter analysis of CD94 and CD57 expression directly on the patient's PBMCs (≈75% CD94+, Left) and after stimulation with IL-2 to generate DEL (99.4% CD94+, Right). (B) Fluorescence-activated cell sorter histograms showing the expression of specific cell surface markers on DEL cells and binding of the HLA-E tetramer.
Antibodies and Flow Cytometry.
Flow cytometric analysis was carried out by using standard protocols on a DEL NK cell line depleted of T cells by using a FACSCalibur with CELLQUEST software (Becton Dickinson). Antibodies used included anti-CD56-PC5 (N901), anti-CD94-PE (HP-3B1, Immunotech, Luminy, France, and Becton Dickinson), anti-CD16-FITC (NKP15), anti-CD57 (HNK-1) (Becton Dickinson), anti-KIR2DL1/KIR2DS1 (EB6), and anti-KIR2DL2/3/KIR2DS2 (GL183) (Beckman Coulter). A number of antibodies were generously donated: anti-ILT-2/LIR-1 (GHI/75) by D. Mason (Oxford University, Oxford), anti-NKG2A (Z199) by A. Moretta (University of Genova, Genova, Italy), anti-KIR3DL1/KIR3DL2/KIR2DS4 (5.133) by M. Colonna (Basel Institute, Basel) and anti-KIR3DL1 (DX9), and anti-MHC I (DX17) and anti-CD94 (DX22) by L. Lanier (University of California, San Francisco). Unlabeled antibodies were visualized by anti-mouse IgG-FITC (Sigma). HLA-E tetramer was refolded with peptide VMAPRTLFL (12).
Viruses.
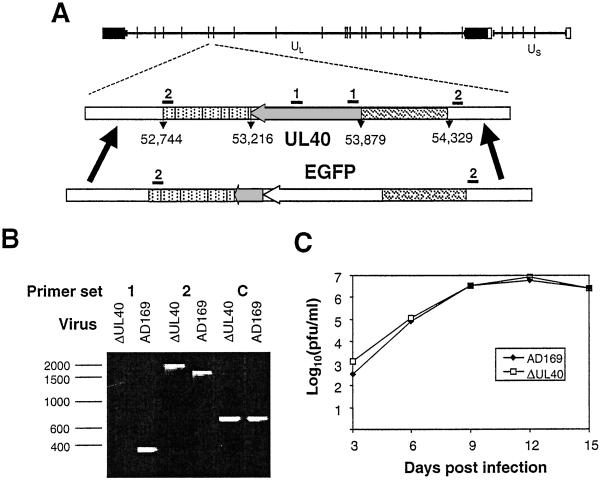
An HCMV strain AD169 UL40 deletion mutant (ΔUL40) was generated by insertion of the enhanced green fluorescent protein (EGFP) gene (Fig. 2A). The UL40 promoter (nucleotides 54,329–53,879, terminating one nucleotide upstream of the translational start site) was amplified by PCR and fused to the EGFP ORF in pEGFP-N1 (CLONTECH). A second PCR-generated fragment encompassing 191 bp from the 3′ end of UL40 extending through the polyadenylation signal sequence (nucleotides 53,403–52,744) then was fused to the UL40 promoter/EGFP cassette in plasmid pAL320. Nucleotide designations refer to the HCMV strain AD169 genome sequence (GenBank accession no. X17403). pAL320 linearized by restriction endonuclease digestion was cotransfected with purified HCMV genomic DNA into human fibroblasts by using Effectene (Qiagen, Chatsworth, CA). Recombinant viruses were identified by using a fluorescence microscope (Leica DMIRBE) and plaque-purified. ΔUL40 was characterized by PCR with primers specific for the deleted element and flanking the expression cassette DNA (Fig. 2B). The PCR primers were: set 1, 5′-CACTCGTATCGGCTTCAC-3′ and 5′-CGAAAGTGGTCCTGACAG-3′; set 2, 5′-AAGCATCATCGGGGTAACGGG-3′ and 5′-GATCTAGACGTCGTCTTGAAACACCG-3′; and set C, 5′-GACTCGAGAGTTTCTCTTCCGCGTCT-3′ and 5′-CGGGATCCTGATTGTTTGGCTGCTGA-3′.
Figure 2.
Generation of an HCMV strain AD169, UL40 deletion mutant, ΔUL40. (A) A schematic representation of the HCMV strain AD169 genome shows the inverted repeats and HindIII sites. EGFP was inserted into the viral genome under the control of the UL40 promoter while deleting most of the UL40 ORF. The relative positions of PCR primer sets 1 and 2 are shown. (B) PCR analysis of viral DNA showing the absence of a PCR product with the ΔUL40 template with primer set 1, consistent with a UL40 deletion. With primer set 2, the increased mobility of the DNA fragment generated by using the HCMVΔUL40 DNA, relative to the parental virus, corresponds to an appropriate EGFP insertion. Primer set C is a positive control amplifying a remote region of the genome within the β2.7 gene. (C) Growth curves showing no difference between parent strain, AD169, and ΔUL40. Human fetal foreskin fibroblasts were infected with either strain AD169 or ΔUL40 [multiplicity of infection (MOI) = 0.1] for 90 min at 37°C. At 3, 6, 9,12, and 15 days postinfection (pi), the tissue culture supernatant was harvested. The virus titer at each time point then was determined by a plaque assay on human fetal foreskin fibroblasts. The experiment was repeated with duplicate samples. pfu, plaque-forming units.
NK Cytotoxicity Assays.
Cytotoxicity was measured in a 51Cr-release assays carried out as described (12) with some minor changes. Primary NK lines were depleted of CD3+ T cells 48 h before use, resulting in >90% CD3−, CD56+ cultures. All assays were carried out in RPMI medium 1640 supplemented with 2% human AB serum. For antibody-blocking studies, effectors and targets were incubated before the assay with mAbs DX22 and DX17 (10 μg/ml final concentration), respectively, for 15 min at 37°C. Between 103 (in primary NK assays to accommodate large effector/target ratios) and 2.5 × 103 (DEL NK assays) targets were used per well. The final means and standard deviations were determined from triplicate or quadruplicate cultures. Primary NK assays were carried out as described in ref. 24. In brief, PBMCs were cultured overnight in the presence or absence of 500 units/ml IFNα (Roferon, Hoffman–La Roche) in RPMI-AB. Before the assay, CD94+ cells were stained with anti-CD94-PE mAb and depleted with Dynabeads. Assays were carried out over 4 h on fibroblasts infected for 72 h with the indicated HCMV strains/mutants (MOI = 10).
Results
HCMV Infection Inhibits CD94/NKG2A+ NK Killing.
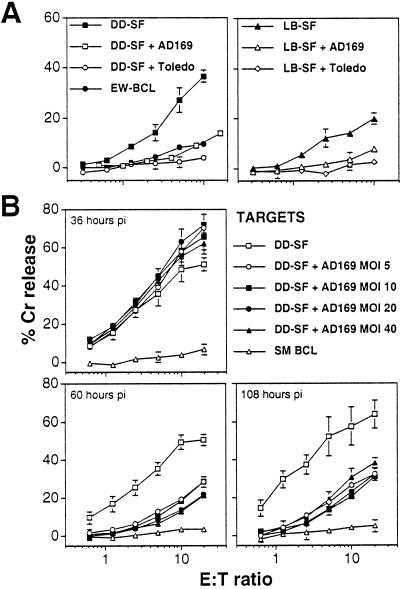
To examine the effect of HCMV infection on CD94/NKG2A+ NK-cell killing, a CD94/NKG2A+ NK cell line, DEL, was derived that exhibited classical NK cell-mediated cytotoxicity by killing K562 cells (data not shown). Fig. 1 summarizes the surface phenotype of this line, showing high expression of CD94, NKG2A, and CD56, lower levels of ILT-2/LIR-1 and CD16, and no expression of KIRs. Efficient HCMV replication in vitro is restricted almost exclusively to primary human fibroblasts. DD-SF and LB-SF skin fibroblasts express significant levels of classical MHC I yet remained susceptible to NK DEL attack (Fig. 3A). Infection of these fibroblasts with both HCMV strains AD169 and Toledo elicited efficient protection against DEL-mediated NK cytotoxicity (Fig. 3A), but HCMV strain Toledo infections seemed to provide consistently more efficient protection against NK cell-mediated lysis than infections with strain AD169 (Fig. 3A). Importantly, this inhibition of killing also was observed against another well defined CD94/NKG2A+ NK line, NKL (ref. 29; Table 1).
Figure 3.
HCMV-induced protection against NK killing by DEL and its kinetics. (A) Infection with HCMV strains, AD169 or Toledo, inhibited CD94/NKG2A+ DEL NK-mediated cytolysis as measured in a chromium-release assay in uninfected target cells, DD-SF (Left) or LB-SF (Right) primary skin fibroblasts. Fibroblasts were infected with HCMV (MOI = 10) for 72 h. (B) Enhancement of cytotoxicity in DD-SF fibroblasts infected with HCMV strain AD169 for 36h but inhibition of cytotoxicity after 60 and 108 h. Varying effector/target (E:T) ratios are shown, and standard deviations are indicated by error bars.
Table 1.
Summary of cytotoxicity in primary NK and NKL assays
| Subject/line | E/T ratio | Undepleted effectors
|
CD94-depleted effectors
|
||||
|---|---|---|---|---|---|---|---|
| AD169*
|
ΔUL40* | AD169* | ΔUL40* | ||||
| Mouse IgG | anti-CD94† | anti-MHC I† | |||||
| I.PBMC | |||||||
| DD expt 1 | 40:1‡ | 39 ± 2§ | nd | nd | 53 ± 4§ | nd | nd |
| DD expt 2 | 200:1 | 35 ± 7§ | nd | nd | 48 ± 3§¶ | 35 ± 0.3 | 33 ± 2¶ |
| DD expt 3 | 200:1 | 20 ± 2§ | 24 ± 1 | 18 ± 2 | 28 ± 3§¶ | 13 ± 2 | 20 ± 2¶ |
| SW expt 1 | 100:1 | 8 ± 2§ | nd | nd | 12 ± 1§¶ | 2 ± 2 | 7 ± 1¶ |
| SW expt 2 | 200:1 | 8 ± 3§ | nd | nd | 20 ± 3§¶ | 6 ± 1 | 7 ± 1¶ |
| SW expt 3 | 200:1 | 3 ± 0.1§ | 4 ± 1 | 8 ± 2 | 11 ± 2§¶ | 2 ± 0.1 | 4 ± 1¶ |
| BM expt 1 | 200:1 | 0 ± 3§ | 6 ± 3 | 3 ± 3 | 17 ± 2§ | 3 ± 0.4 | 11 ± 4 |
| NK Lines | |||||||
| DD-NK1 | 20:1 | 27 ± 2§ | 34 ± 3 | 43 ± 4 | 43 ± 7§ | ||
| DD-NK2 | 20:1 | 11 ± 2§ | 12 ± 1 | 13 ± 0.1 | 24 ± 1§ | ||
| SW-NK1 | 40:1 | 8 ± 6§ | 16 ± 4 | 19 ± 4 | 22 ± 6§ | ||
| SW-NK2 | 40:1 | 9 ± 6 | 24 ± 8 | 14 ± 5 | 17 ± 1 | ||
| BM-NK1 | 40:1 | 1 ± 1§ | 3 ± 0.1 | 4 ± 0.3 | 8 ± 1§ | ||
| BM-NK2 | 80:1 | 2 ± 0.3§ | 5 ± 1 | 7 ± 4 | 5 ± 1§ | ||
| NKL | 20:1 | 11 ± 2§ | 26 ± 2 | 29 ± 1 | 21 ± 1§ | ||
I.PBMC, IFNα-treated PBMCs; E/T, effector/target ratio; nd, not done; all numbers show percentage of Cr release ± SD.
Autologous fibroblast targets (except for NKL) infected with the indicated HCMV strains for 72 h.
AD169-infected fibroblasts were treated with the indicated mAbs.
DD expt 1 was carried out by using CD3-depleted PBMCs, resulting in a lower effector/target ratio.
§, P < 0.05 (AD169 vs. ΔUL40) and ¶, P < 0.05 (undepleted vs. CD94-depleted) using t test assuming unequal variance.
Resistance to NK Killing Is Associated with the Late Phase of Infection.
HCMV-encoded gene functions implicated in the evasion of MHC I-restricted cytotoxic T lymphocytes act at various stages of the replication cycle. The rate of infection can be influenced also by the amount of input virus, measured by the MOI. HCMV inhibition of NK DEL cells therefore was characterized further by performing time-course and dose-response experiments. HCMV infection induced a small but significant increase in sensitivity to NK DEL-mediated cytolysis at 36 h pi (Fig. 3B). However, HCMV-induced inhibition of NK cell killing was observed consistently after 60 h of infection. Resistance to NK cell attack was maintained to 108 h pi and thus is not a transient effect. Efficient protection against NK attack was observed at an MOI of 5 plaque-forming units per cell; however, the kinetics and magnitude of protection were not affected substantially by increasing the input MOI up to 40 plaque-forming units per cell (Fig. 3B).
Antibody Blocking of the CD94/HLA-E Interaction Abrogates HCMV-Induced NK Inhibition.
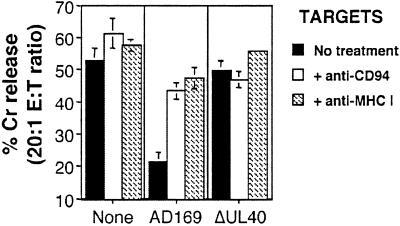
Antibody-blocking experiments demonstrated that inhibition of DEL killing by HCMV strain AD169 was reversed efficiently in both DD-SF and LB-SF fibroblasts by treatment of either the effectors with anti-CD94 or the targets with anti-MHC I antibodies (Fig. 4). Nonspecific mouse Ig had no effect on the observed inhibition of NK DEL (data not shown). HCMV strain AD169 evasion of NK DEL cell killing thus is likely to depend on an interaction between CD94/NKG2A NK receptors and MHC I expressed on infected cells. Considering expression of all classical HLA class I are down-regulated within 24 h of HCMV infection, whereas the nonclassical HLA-E is up-regulated (21, 31), it is likely that HLA-E is the target MHC I for this inhibition. This up-regulation of HLA-E would be consistent with an UL40-dependent mechanism, because UL40 has been shown to induce HLA-E cell-surface expression (21).
Figure 4.
Anti-CD94 and anti-MHC I reverses the AD169-induced inhibition of the NK cell line DEL. Infection (MOI = 10 for 72 h) with HCMV strain AD169 inhibited DEL killing as compared with uninfected DD-SF fibroblasts. Treatment of effectors with anti-CD94 or targets with anti-MHC I resulted in the total recovery of DEL killing against strain AD169-infected cells. Results are from an effector/target (E:T) ratio of 20:1, and standard deviations are indicated by error bars.
A Role for UL40 in HCMV-Induced Protection.
The potential role of UL40 expression in mediating the observed resistance to CD94/NKG2A+ NK cytolysis during productive HCMV infection was examined. To test directly whether UL40 expression is required during productive infection to evade CD94/NKG2A+ NK killing, an HCMV UL40 deletion mutant (ΔUL40) was generated as described in the Fig. 2 legend. ΔUL40 is based on HCMV strain AD169 but contains EGFP under the control of the UL40 promoter while simultaneously deleting the first 475 bp of the UL40 ORF. The 37-aa signal sequence encoding the HLA-E-binding peptide thereby was deleted while preserving an uncharacterized overlapping ORF (UL39) transcribed off the opposite strand. The nature and purity of ΔUL40 was confirmed by Southern blot (data not shown) and PCR (Fig. 2B) analyses. ΔUL40 replicated as efficiently as the parent virus and with similar growth kinetics (Fig. 2C); thus UL40 was dispensable for in vitro virus replication, and any effects of ΔUL40 on NK killing could not be caused by differences in the progression of infection.
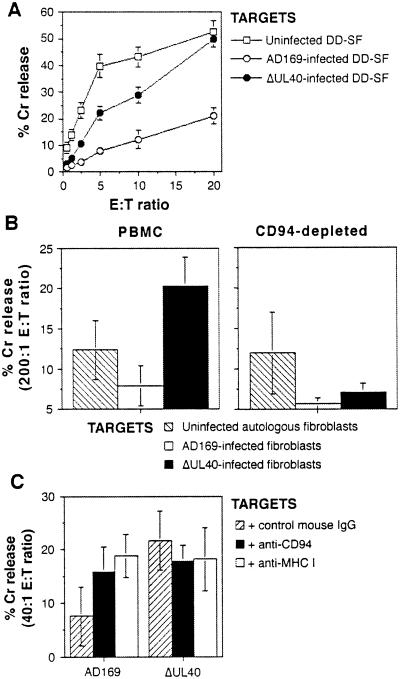
When the ΔUL40 virus was used to infect fibroblasts, the virus lost the capacity to induce protection against killing by NK DEL cells (Fig. 5A). The anti-CD94 and anti-MHC I blocking antibodies also had no significant effect on lysis of ΔUL40-infected cells (Fig. 4). The physiological significance of this effect then was tested by assays on primary NK cells. PBMCs exhibit little NK activity in the absence of stimulation. Therefore, PBMCs first were stimulated with IFNα to induce cytotoxicity (24). IFNα-activated PBMCs exhibited greater cytotoxicity against ΔUL40-infected (compared with strain AD169-infected) autologous fibroblasts (Fig. 5B). To identify the effector population involved in this increased killing, depletions were carried out immediately before performing the assay. Increased cytotoxicity against ΔUL40-infected targets was abrogated by depletion of cells expressing CD94 (Fig. 5B). These experiments were carried out in three different healthy subjects (SW, DD, and BM), and a summary of the results is shown in Table 1. In some subjects, there was significant killing of ΔUL40-infected fibroblasts (Table 1) and K562 targets (data not shown) even after CD94 depletion, which likely was caused by residual CD94+ NK cells (depletions were >88%) and CD94− NK cells that represented ≈20–50% of CD3−, CD56+ cells in these three donors (data not shown).
Figure 5.
Effects of the ΔUL40 deletion mutant on NK cytotoxicity. (A) Infection (MOI = 10 for 72 h) with the ΔUL40 mutant resulted in increased DEL killing as compared with strain AD169-infected DD-SF fibroblasts. (B) Infection of autologous fibroblast targets with the ΔUL40 mutant but not HCMV strain AD169 resulted in significant killing by IFNα-activated PBMCs (Left). Depletion of CD94+ PBMCs resulted in the loss of killing against ΔUL40-infected targets (Right). Results are from an effector/target (E:T) ratio of 200:1. Results from one representative experiment of seven are shown. (C) Cytotoxicity assays using primary NK cell lines against autologous, HCMV-infected targets showed killing against ΔUL40-infected but not AD169-infected fibroblasts. Treatment of cultures with anti-CD94 or anti-MHC I recovered killing of AD169-infected targets. Primary lines were cultured as described in Materials and Methods and depleted of CD3+ T cells before assay. Results from one representative experiment of six, at an effector/target ratio of 40:1, are shown. Standard deviations are indicated in all experiments by error bars.
Primary NK Cell Lines and Inhibition by UL40.
Primary NK lines also were generated and used in NK cytotoxicity assays against autologous, AD169-infected or ΔUL40-infected fibroblasts. A significant increase in killing was observed against ΔUL40-infected targets compared with AD169-infected fibroblasts from SW-NK1 (Fig. 5C) and five other NK lines (Table 1). Anti-CD94 and anti-MHC I mAbs reversed the AD169-induced inhibition of NK line killing by >50% in 8 of 12 cases (3/6 for anti-CD94 and 5/6 for anti-MHC I; Fig. 5C; Table 1). All lines showed >35% killing of K562 targets (data not shown).
Discussion
In recent years, a broad range of NK-inhibitory and -activating receptors has been identified, and a number of HCMV genes have been shown to encode proteins with the potential of interacting with these receptors or their ligands to induce NK inhibition. However, the physiological action of these molecules during a productive HCMV infection has been hard to prove. We have shown, using an HCMV strain AD169 deletion mutant, that the UL40 gene can act during a productive HCMV infection, which occurs against certain defined CD94+ NK cell lines as well as primary CD94+ NK cells.
Using the NK line, DEL, we defined the kinetics of HCMV strain AD169-induced NK inhibition. HCMV-induced inhibition of NK DEL cell killing was associated with the late phase of virus infection, being reproducibly observed from 60 h pi. In contrast, HCMV strain AD169 infection stimulated DEL-mediated killing at 36 h pi; down-regulation of cell surface MHC I expression occurs from 24 h pi and may increase sensitivity to NK cell attack. Because UL40 transcription can be detected as early as 8 h pi (21), this early-phase window of enhanced susceptibility to CD94/NKG2A+ NK cells was unexpected. It is possible that gpUL40 translation is delayed or a threshold level of expression must be reached to induce overt protection. Alternatively, because NK killing is controlled by a balance between activating and inhibitory signals, inhibition induced by UL40 may not be the dominant effect at 24–48 h pi. The delayed kinetics of the response may partly explain some apparently conflicting results in the literature. Studies reporting that HCMV infection enhances susceptibility to NK attack that were performed during the early phase of infection (24–48 h pi; refs. 24 and 25) and those reporting induction of protection at later time points (4–8 days pi; refs. 26 and 27) therefore are consistent with our observations.
DEL is an NK cell line developed for this study that exhibits a clonal phenotype after culture with IL-2 (Fig. 1A) and is characteristic of the CD56bright, CD16mid NK subset [positive for CD25 (IL2Rp55), CD57, CD69, MHC class II, CD45RO, and CD71 and negative for CD3, CD4, CD5, and CD8 (data not shown)]. This line ignores the presence of MHC I on the surface of primary skin fibroblasts and lyses them. However, NK inhibition by HCMV infection is still strong enough to inhibit this aggressive killing. DEL may be particularly sensitive to UL40-mediated evasion of NK cytolysis, because it expresses a high level of the CD94/NKG2A NK receptor compared with NK lines derived from normal individuals (data not shown), which is reflected by the high levels of binding to HLA-E-tetrameric complexes (Fig. 1B). However, similar UL40-mediated protection of HCMV-infected fibroblasts has also been observed with another CD94/NKG2A+ NK line, NKL (Table 1). Furthermore, our assays using activated primary NK cells and cultured NK cell lines suggest that UL40 is a major gene responsible for inhibition of at least a proportion of NK cells expressing CD94/NKG2A (Fig. 5; Table 1). The primary NK lines also highlight the variable nature of the NK responses with differences in antibody blocking and the killing of lines, between individuals as well as within the same subject (Table 1). The data imply that without NK-inhibitory genes HCMV infection indeed does induce increased sensitivity to NK cells, resulting in killing of even MHC-matched autologous fibroblast targets. It is likely also that a number of HCMV-encoded NK-inhibitory mechanisms exist that are directed at NK cells of different phenotype and specificity.
The sensitivity of DEL to the UL40 mechanism of NK inhibition and its resistance to classical MHC I-induced inhibition also may be caused by the presence or absence of other activating or inhibitory receptors. DEL expressed only low levels of the inhibitory receptor, ILT-2/LIR-1, and no detectable expression of a range of characterized KIRs (KIR2DL1, KIR2DL2/3, KIR3DL1, and KIR3DL2; Fig. 1B). HCMV gpUL18 has high affinity for ILT-2/LIR-1 (10, 11) and potentially could induce NK inhibition. Reverse transcription–PCR carried out to phenotype the expression of other NKG2 receptor family members revealed relatively high levels of mRNAs for NKG2A and NKG2D, a lower level for NKG2F, NKG2H, and DAP12, but no NKG2C and E mRNAs (data not shown). An inhibitory role for soluble HCMV gpUL16 has been suggested, because it binds MIC B and ULBPs (32, 33) and therefore could block binding between MIC B and its activating receptor NKG2D/DAP10 (34). Both the UL18 and UL16 genes are present in HCMV strain AD169 and ΔUL40; however, neither of these potentially inhibitory HCMV genes were dominant over UL40 in our assays. UL16 and UL18 may well further influence the interactions between HCMV and NK cells, but more study with multiple UL16, UL18, and UL40 mutant viruses would be needed to clarify these interactions. The expression of other, activating, natural cytotoxicity receptors (NCRs) such as NKp30 (35), NKp44 (36), and NKp46 (37) were not investigated in this study but may have a further impact on the cytotoxic behavior of NK cells (7, 38).
From these results, it is obvious that the interaction between HCMV-infected cells and NK cells is complex and is influenced by the phase of infection, the nature of activating and inhibitory receptors expressed by NK cells, the influence of soluble factors, and the virus isolate. Reports have implied that attenuated HCMV laboratory strains are more vulnerable to NK attack than less-adapted viruses or clinical isolates (27, 39). In our hands, HCMV strain Toledo behaves similarly to fresh clinical isolates in being more resistant to DEL- and NKL-mediated cytolysis. Furthermore, HCMV strain Toledo and the clinical strain 742 induced protection against DEL-mediated cytolysis that could only be recovered partially by blocking antibodies to CD94 and MHC I (data not shown). The genome of the Toledo strain resembles more closely a clinical isolate, containing an additional 13–15 kb of sequence spontaneously lost by the laboratory strains AD169 and Towne (40). This deleted element is already known to encode two chemokines and a tumor necrosis factor receptor homologue (41, 42). Interestingly, both strains AD169 and Towne have been used as live vaccines and are associated with reduced pathogenicity in vivo. In conclusion, we propose that UL40-mediated up-regulation of HLA-E operates to provide efficient protection against at least a proportion of CD94/NKG2A+ NK cells during HCMV infection. Future work may reveal additional CD94-dependent and independent mechanisms of NK evasion encoded by other HCMV strains and clinical isolates.
Acknowledgments
We are grateful to Christian Willberg for supplying HLA-E tetramers and D. Mason, A. Moretta, M. Colonna, and L. Lanier for the gift of mAbs. Thanks to T. Hoy and J. Fisher for cell sorting, which was carried out by the Flow Cytometry Core Facility, University of Wales College of Medicine. Thanks also to M. Wills for helpful discussions. This work was supported by grant funding from the Wellcome Trust. V.M.B. was a Royal Society University Research Fellow and C.R. was supported by La Fondation pour la Recherche Medicale and the Medical Research Council.
Abbreviations
- HCMV
human cytomegalovirus
- NK
natural killer
- KIR
killer inhibitory receptor
- LIR
leukocyte Ig-like receptor
- PBMC
peripheral blood mononuclear cell
- EGFP
enhanced green fluorescent protein
- MOI
multiplicity of infection
- pi
postinfection
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Walter E A, Greenberg P D, Gilbert M J, Finch R J, Watanabe K S, Thomas E D, Riddell S R. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 2.Tortorella D, Gewurz B E, Furman M H, Schust D J, Ploegh H L. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 3.Scalzo A A, Fitzgerald N A, Simmons A, La Vista A B, Shellam G R. J Exp Med. 1990;171:1469–1483. doi: 10.1084/jem.171.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown M G, Dokun A O, Heusel J W, Smith H R, Beckman D L, Blattenberger E A, Dubbelde C E, Stone L R, Scalzo A A, Yokoyama W M. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 5.Lee S H, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal S M. Nat Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 6.Biron C A, Byron K S, Sullivan J L. N Engl J Med. 1989;320:1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 7.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari M C, Biassoni R, Moretta L. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 8.Moretta L, Biassoni C, Bottino C, Mingari M C, Moretta L. Immunol Today. 2000;21:420–422. doi: 10.1016/s0167-5699(00)01673-x. [DOI] [PubMed] [Google Scholar]
- 9.Lanier L L. Annu Rev Immunol. 1998;16:359–363. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 10.Chapman T L, Heikeman A P, Bjorkman P J. Immunity. 1999;11:603–613. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 11.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu M L. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 12.Braud V M, Allan D S, O'Callaghan C A, Soderstrom K, D'Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H, Lanier L L, McMichael A J. Nature (London) 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 13.Borrego F, Ulbrecht M, Weiss E H, Coligan J E, Brooks A G. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty D E. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braud V, Jones E Y, McMichael A J. Eur J Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- 16.Lee N, Goodlett D R, Ishitani A, Marquardt H, Geraghty D E. J Immunol. 1998;160:4951–4960. [PubMed] [Google Scholar]
- 17.Llano M, Lee N, Navarro F, Garcia P, Albar J P, Geraghty D E, Lopez-Botet M. Eur J Immunol. 1998;28:2854–2863. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Braud V M, Allan D S, Wilson D, McMichael A J. Curr Biol. 1998;8:1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 19.Lehner P J, Kartunnen J T, Wilkinson G W, Creswell P. Proc Natl Acad Sci USA. 1997;94:6904–6909. doi: 10.1073/pnas.94.13.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt E W, Gupta S S, Lehner P J. EMBO J. 2001;20:387–396. doi: 10.1093/emboj/20.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomasec P, Braud V M, Rickards C, Powell M B, McSharry B P, Gadola S, Cerundolo V, Borysiewicz L K, McMichael A J, Wilkinson G W. Science. 2000;287:1031–1033. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 22.Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart J W, Pla M, Weiss E H. J Immunol. 2000;164:5019–5022. doi: 10.4049/jimmunol.164.10.5019. [DOI] [PubMed] [Google Scholar]
- 23.Carboni C, Mousavi-Jazi M, Wakiguchi H, Carbone E, Karre K, Soderstrom K. Eur J Immunol. 2001;31:2926–2935. doi: 10.1002/1521-4141(2001010)31:10<2926::aid-immu2926>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Borysiewicz L K, Rodgers B, Morris S, Graham S, Sissons J G. J Immunol. 1985;134:2695–2701. [PubMed] [Google Scholar]
- 25.Leong C C, Chapman T L, Bjorkman P J, Formankova D, Mocarski E S, Phillips J H, Lanier L L. J Exp Med. 1998;187:1681–1687. doi: 10.1084/jem.187.10.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fletcher J M, Prentice H G, Grundy J E. J Immunol. 1998;161:2365–2374. [PubMed] [Google Scholar]
- 27.Cerboni C, Mousavi-Jazi M, Linde A, Soderstrom K, Brytting M, Wahren B, Karre K, Carbone E. J Immunol. 2000;164:4775–4782. doi: 10.4049/jimmunol.164.9.4775. [DOI] [PubMed] [Google Scholar]
- 28.Huard B, Fruh K. Eur J Immunol. 2000;30:509–515. doi: 10.1002/1521-4141(200002)30:2<509::AID-IMMU509>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 29.Robertson M J, Cochran K J, Cameron C, Le J M, Tantravahi R, Ritz J. Exp Hematol (Charlottesville, Va) 1996;24:406–415. [PubMed] [Google Scholar]
- 30.Borysiewicz L K, Morris S, Page J D, Sissons J G. Eur J Immunol. 1983;13:804–809. doi: 10.1002/eji.1830131005. [DOI] [PubMed] [Google Scholar]
- 31.Zhu H, Cong J-P, Mamtora G, Gingeras T, Shenk T. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosman D, Mullberg J, Sutherland C L, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny N J. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 33.Kubin M, Cassiano L, Chalupny J, Chin W, Cosman D, Fanslow W, Mullberg J, Rousseau A-M, Ulrich D, Armitage R. Eur J Immunol. 2001;31:1428–1437. doi: 10.1002/1521-4141(200105)31:5<1428::AID-IMMU1428>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Song Y, Bakker A B, Bauer S, Spies T, Lanier L L, Phillips J H. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 35.Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, Marcenaro E, Accame L, Malaspina A, Biassoni R, Bottino C, Moretta A, Moretta A. J Exp Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, Augugliaro R, Moretta L, Moretta A. J Exp Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, Biassoni R, Moretta A. J Exp Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sivori S, Pende D, Bottino C, Marcenaro E, Pessino A, Biassoni R, Moretta L, Moretta A. Eur J Immunol. 1999;29:1656–1666. doi: 10.1002/(SICI)1521-4141(199905)29:05<1656::AID-IMMU1656>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 39.Waner J L, Nierenberg J A. J Med Virol. 1985;16:233–244. doi: 10.1002/jmv.1890160304. [DOI] [PubMed] [Google Scholar]
- 40.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benedict C A, Butrovich K D, Lurain N S, Corbeil J, Rooney I, Schneider P, Tschopp J, Ware C F. J Immunol. 1999;162:6967–6970. [PubMed] [Google Scholar]
- 42.Penfold M E, Dairaghi D J, Duke G M, Saederup N, Mocarski E S, Kemble G W, Schall T J. Proc Natl Acad Sci USA. 1999;96:9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]