Abstract
Acyl-CoA:monoacylglycerol acyltransferase (MGAT) catalyzes the synthesis of diacylglycerol, the precursor of physiologically important lipids such as triacylglycerol and phospholipids. In the intestine, MGAT plays a major role in the absorption of dietary fat because resynthesis of triacylglycerol is required for the assembly of lipoproteins that transport absorbed fat to other tissues. MGAT activity has also been reported in mammalian liver and white adipose tissue. However, MGAT has never been purified to homogeneity from mammalian tissues, and its gene has not been cloned. We identified a gene that encodes an MGAT (MGAT1) in mice. This gene has sequence homology with members of a recently identified diacylglycerol acyltransferase gene family. Expression of the MGAT1 cDNA in insect cells markedly increased MGAT activity in cell membranes. In addition, MGAT activity was proportional to the level of MGAT1 protein expressed, and the amount of diacylglycerol produced depended on the concentration of either of its substrates, oleoyl-CoA or monooleoylglycerol. In mice, MGAT1 expression and MGAT activity were detected in the stomach, kidney, white and brown adipose tissue, and liver. However, MGAT1 was not expressed in the small intestine, implying the existence of a second MGAT gene. The identification of the MGAT1 gene should greatly facilitate research on the identification of the intestinal MGAT gene and on the function of MGAT enzymes in mammalian glycerolipid metabolism.
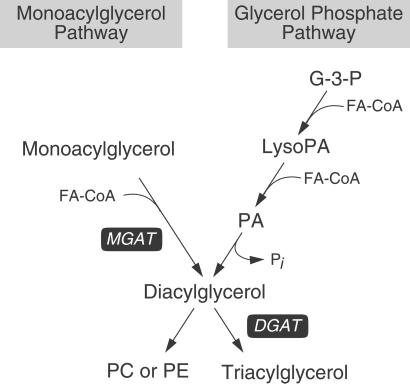
Diacylglycerol is the precursor of physiologically important lipids such as triacylglycerol and phospholipids, which store energy and form cellular membranes. Diacylglycerol is also a well established intracellular signaling molecule that activates protein kinase C (1). In eukaryotes, two major pathways exist for synthesizing diacylglycerol: the glycerol phosphate pathway and the monoacylglycerol pathway (Fig. 1). Both pathways generate diacylglycerol that can be used as a substrate by acyl-CoA:diacylglycerol acyltransferase (DGAT) for triacylglycerol synthesis (2, 3). In the glycerol phosphate pathway, which functions in most cells, diacylglycerol is derived by the dephosphorylation of phosphatidic acid produced by sequential acylations of glycerol phosphate. In the monoacylglycerol pathway, which has been reported predominantly in the intestine, diacylglycerol is formed directly from monoacylglycerol and fatty acyl-CoA in a reaction catalyzed by acyl-coA:monoacylglycerol acyltransferase (MGAT) (EC 2.3.1.22) (2, 3).
Figure 1.
The two major pathways for synthesizing diacylglycerol. In the monoacylglycerol pathway, MGAT produces diacylglycerol, the precursor of triacylglycerol and certain phospholipids, by covalently joining a fatty acyl moiety to monoacylglycerol. In the glycerol phosphate pathway, diacylglycerol is derived from the dephosphorylation of phosphatidic acid (PA), which is produced by sequential acylation of glycerol 3-phosphate (G-3-P). FA-CoA, fatty acyl-CoA; LysoPA, lysophosphatidic acid; Pi, inorganic phosphate; PC, phosphatidylcholine; PE, phosphatidylethanolamine.
MGAT is best known for its role in fat absorption in the intestine, where the fatty acids and sn-2-monoacylglycerol generated from the digestion of dietary fat (mainly triacylglycerol) are resynthesized into triacylglycerol in enterocytes for chylomicron synthesis and secretion. MGAT activity is also found at high levels in the livers of suckling rats (4) and in the white adipose tissue of migrating sparrows (5), where triacylglycerols are actively hydrolyzed to provide fatty acids for energy. MGAT preferentially acylates monoacylglycerols that contain a polyunsaturated fatty acyl moiety at the sn-2 position (6). Thus, MGAT may preserve essential fatty acids, all of which are polyunsaturated, by resynthesizing them into triacylglycerols. This function may be relevant in mammalian white adipose tissue, which possesses significant levels of MGAT activity (7). In addition, MGAT may play a role in signaling, because its product, diacylglycerol, and one of its substrates, 2-arachidonoylglycerol, activate signaling pathways (8, 9).
Like many enzymes that participate in neutral lipid synthesis, MGAT has proven difficult to purify to homogeneity, and an MGAT gene has not been identified. Several partial purifications of MGAT enzymes have been reported (10, 11), and a 43-kDa MGAT enzyme was purified recently from peanut cotyledons (12). Difficulties in the purification of MGAT may reflect its hydrophobicity or its involvement in an enzyme complex (13). Here we report the identification of a gene encoding a mammalian MGAT (MGAT1), which we found by its sequence homology to members of a recently identified DGAT2 gene family (14). We also report the biochemical characteristics of MGAT1 and its tissue distribution.
Materials and Methods
Cloning of MGAT1 cDNA.
Mouse expressed sequence tags encoding MGAT1 were identified by blast database searches through their sequence homology to DGAT2 from Mortierella rammaniana (GenBank accession no. AF391089). On the basis of these expressed sequence tags, primers were designed to amplify the MGAT1 coding sequence from mouse liver RNA by reverse transcription (SuperScript Choice System, GIBCO/BRL) and PCR (Takara Ex Taq, Panvera, Madison, WI). We previously deposited the sequences of mouse MGAT1 and its human homologue in GenBank (accession nos. AF384162 and AF384163) (14).
Insect Cell Expression Studies.
MGAT1 was tagged with an N-terminal FLAG epitope (MGDYKDDDDG, epitope underlined) and expressed in Spodoptera frugiperda (Sf9) insect cells as described (14). In some experiments, MGAT1 without FLAG was also expressed. The MGAT1 coding sequence (with or without FLAG) was subcloned into pVL1393 baculovirus transfer vector (PharMingen). Recombinant baculoviruses were generated by cotransfecting Sf9 insect cells with the transfer vector and BaculoGold DNA (PharMingen). High-titer viruses were obtained after two rounds of amplification. FLAG-tagged mouse DGAT1 (GenBank accession no. AF078752) and FLAG-tagged mouse DGAT2 (GenBank accession no. AF384160) were expressed as controls. Cells were typically infected with virus for 3 days, washed with PBS, and homogenized by 10 passages through a 27-gauge needle in 1 mM EDTA/200 mM sucrose/100 mM Tris⋅HCl, pH 7.4. Total membrane fractions (100,000 × g pellet) were resuspended in homogenization buffer and frozen at −80°C until use. Expression of MGAT1, DGAT2, and FLAG-tagged proteins was verified by immunoblotting membrane proteins (5 μg) with an antiserum against the C terminus (amino acids 295–316) of MGAT1, an antiserum against the C terminus (amino acids 372–378) of DGAT2, and an anti-Flag M2 antibody (Sigma), respectively.
In Vitro Acyltransferase Assays.
In general, acyltransferase activities were assayed under apparent Vmax conditions for 5 min in a final volume of 200 μl. Each reaction contained 100 μg of membrane proteins, 5 mM MgCl2, 1.25 mg/ml BSA, 200 mM sucrose, 100 mM Tris⋅HCl (pH 7.4), 25 μM acyl donor, and 200 μM acyl acceptor. Nonpolar acyl acceptors (diacylglycerol, monoacylglycerol, cholesterol, phosphatidic acid, and sphingosine) were dispersed as phosphatidylcholine liposomes (molar ratio ≈0.2), and polar acyl acceptors (glycerol 3-phosphate, dihydroxyacetone phosphate, lysophosphatidic acid, and lysophosphatidylcholine) were dissolved in water. Reactions were started by adding protein and terminated by adding 4 ml of chloroform/methanol (2:1 vol/vol). The extracted lipids were dried, separated by TLC with hexane/ethyl ether/acetic acid (80:20:1 vol/vol/vol), visualized with iodine vapor, and identified with lipid standards. For experiments with radiolabeled substrates, TLC plates were exposed to x-ray film or scraped to assess the incorporation of radioactivity into lipid products.
Individual assays were performed as follows. DGAT activity was measured as described (15). MGAT activity was determined by measuring the incorporation of the [14C]oleoyl moiety into diacylglycerol with 25 μM [14C]oleoyl-CoA (specific activity, ≈20,000 dpm/nmol) and 200 μM exogenously added sn-2-monooleoylglycerol. In some assays, sn-1-[14C]monooleoylglycerol or sn-2-[3H]monooleoylglycerol (specific activity, 18 and 1 μCi/μmol, respectively; 200 μM final concentration; American Radiolabeled Chemicals, St. Louis; 1 Ci = 37 GBq) was used as a tracer to measure MGAT activity in the presence of unlabeled oleoyl-CoA (25 μM) or other fatty acyl-CoA derivatives. The dependence of MGAT1 activity on monoacylglycerol and fatty acyl-CoA as substrates was determined by assaying MGAT1 with various concentrations of oleoyl-CoA or monooleoylglycerol in the presence of 400 μM monooleoylglycerol or 50 μM oleoyl-CoA, respectively. Diacylglycerol mass was quantified by densitometry after the lipid products were separated by TLC and visualized by immersing the plate in a solution of 10% cupric sulfate and 8% phosphoric acid and heating at 180°C for 30 min. Stereoisomers of monoacylglycerol (sn-1-, sn-2-monooleoylglycerol, and sn-3-monostearoylglycerol) and fatty acyl-CoAs [malonyl-CoA, n-octanoyl-CoA (C8:0), lauroyl-CoA (C12:0), palmitoyl-CoA (C16:0), stearoyl-CoA (C18:0), arachidoyl-CoA (C20:0), oleoyl-CoA (C18:1), linoleoyl-CoA (C18:2), and arachidonoyl-CoA (C20:4)] were from Sigma. MGAT activity in tissues was measured in particulate fractions prepared from pooled tissues of three 15-week-old male mice.
Mammalian Cell Expression Studies.
For mammalian cell expression, FLAG-tagged MGAT1 was subcloned into a pcDNA3 vector and transfected into COS-7 or Chinese hamster ovary cells with Fugene 6 (Roche Diagnostics). FLAG-tagged DGAT1, DGAT2, and ACAT2 (acyl-CoA:cholesterol acyltransferase 2, accession no. AF078751) were expressed as controls. Membrane fractions were prepared as described for insect cells. Expression of FLAG-tagged proteins (in 20 μg of membrane proteins) was verified by immunoblotting with the anti-FLAG antibody. For immunocytochemistry, cells were grown and transfected on glass cover slips. Two days after transfection, cells were fixed in acetone/methanol (1:1) for 2 min and incubated in PBS containing 3% BSA and 0.2% Triton X-100 for 1 h at room temperature. Samples were then incubated sequentially with 4 μg/ml anti-FLAG antibody for 1 h and 10 μg/ml FITC-conjugated goat anti-mouse IgG (CalBiochem) for 30 min. Antibodies were diluted in PBS containing 3% BSA and 0.02% Triton X-100. MGAT activities in membranes of transfected cells were assayed as described above.
MGAT1 Tissue Expression Pattern in Mice.
To determine the tissue distribution of MGAT1 expression in mouse, a total RNA blot (SeeGene, Seoul, South Korea), a blot of total RNA from indicated tissues, a poly(A)+ blot (CLONTECH), and a blot of poly(A)+ RNA from indicated tissues were hybridized with 32P-labeled probes generated by random priming (Amersham Pharmacia) with MGAT1 cDNA as the template. These blots were stripped and hybridized with DGAT2 probes as a control.
Results
Identification of an MGAT1 Gene.
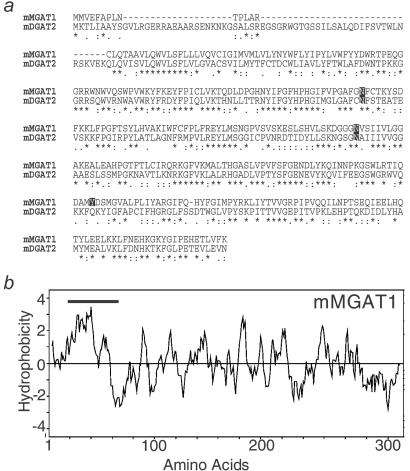
Mouse MGAT1 cDNA was originally identified as a DGAT candidate (DC) gene (mouse DC2 in ref. 14) through its homology to genes encoding DGAT2 (14, 16). The ORF of the MGAT1 cDNA predicts a 335-aa protein that is 40% identical to mouse DGAT2 (Fig. 2a) and has a predicted molecular mass of 38.8 kDa. Like DGAT2, MGAT1 contains sequences similar to a domain of phosphate acyltransferases (14). MGAT1 also possesses two putative N-linked glycosylation sites and a potential tyrosine phosphorylation site. The hydrophobicity plot for MGAT1 is similar to that for DGAT2 and predicts at least one transmembrane domain (amino acids 21–43) in the amino terminus (Fig. 2b). Sequences for the human MGAT1 homologue have been reported as human DC2, a member of the DGAT2 gene family (14). The mouse MGAT1 gene is located on chromosome 1 (GenBank accession no. AC079223), and its human homologue is on chromosome 2 (GenBank accession no. NT005126).
Figure 2.
Mouse MGAT1 protein sequence analysis. (a) Alignment of predicted mouse MGAT1 (mMGAT1) amino acid sequences with mouse DGAT2 (mDGAT2). Amino acid residues identical for both MGAT1 and DGAT2 are indicated with an asterisk; conservation of strong groups is indicated with a colon; conservation of weak groups is indicated with a period. Two putative N-linked glycosylation sites and one potential tyrosine phosphorylation site are also indicated. (b) Hydrophobicity plot of mMGAT1 as assessed by Kyte–Doolittle analysis (20). The bold line indicates a predicted transmembrane domain (http://www.cbs.dtu.dk/services/TMHMM/).
Mouse MGAT1 Expressed in Insect Cells.
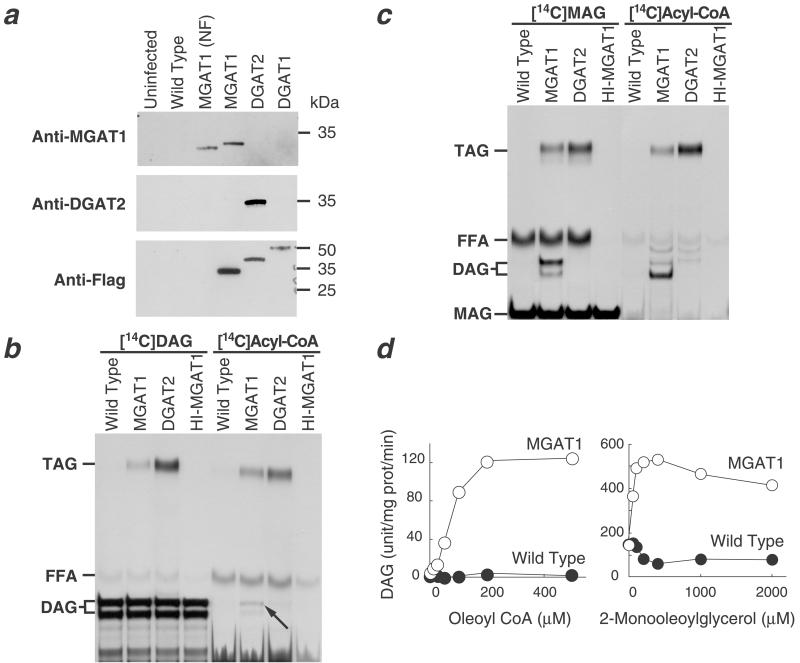
To examine the biochemical activity of MGAT1 protein, we expressed FLAG-tagged and nontagged versions of the cDNA in insect cells. The non-FLAG-tagged version migrated on SDS/PAGE with an apparent molecular mass of ≈33 kDa. As expected, the FLAG-tagged version migrated slightly more slowly because of the FLAG epitope (Fig. 3a). Because MGAT1 shares sequence homology with DGAT2, we first examined whether membranes expressing MGAT1 have DGAT activity. With either [14C]dioleoylglycerol or [14C]oleoyl-CoA as the radiolabeled substrate, MGAT1-expressing membranes incorporated more radioactivity into triacylglycerols than membranes expressing wild-type viral proteins or heat-inactivated MGAT1 (Fig. 3b), indicating that these membranes have DGAT activity. However, this DGAT activity was significantly less than that in control membranes expressing DGAT2, even though MGAT1 protein was expressed at a higher level.
Figure 3.
Expression of mouse MGAT1 cDNA in Sf9 insect cells. (a) Immunoblots of insect cell membranes. Expression of MGAT1 and DGAT2 was verified by immunoblotting with specific antibodies or an anti-FLAG antibody. Membrane proteins (5 μg) were analyzed from uninfected Sf9 cells, cells infected with wild-type virus, non-FLAG-tagged MGAT1 (MGAT1-NF), FLAG-tagged-MGAT1 (MGAT1), FLAG-tagged-DGAT2 (DGAT2), or FLAG-tagged-DGAT1 (DGAT1) recombinant baculoviruses. (b) Weak DGAT activity conferred by expression of MGAT1. DGAT activity was detected by incorporation of [14C]dioleoylglycerol ([14C]DAG) or [14C]oleoyl-CoA ([14C]Acyl-CoA) into triacylglycerol. The arrow indicates the incorporation of [14C]oleoyl-CoA into diacylglycerol by membranes expressing MGAT1. In this TLC system, the three stereoisomers of diacylglycerol migrate as a doublet, in which the upper species is sn-1,3-diacylglycerol and the lower species is a combination of sn-1,2- and sn-2,3-diacylglycerol. FFA, free fatty acid; HI-MGAT1, heat-inactivated MGAT1; TAG, triacylglycerol. (c) Strong MGAT activity conferred by expression of MGAT1. MGAT activity was detected by using either sn-1-[14C]monooleoylglycerol ([14C]MAG) or [14C]oleoyl-CoA ([14C]acyl-CoA) as the radioactive substrate. (d) Dependence of MGAT activity in membranes expressing MGAT1 on substrate concentrations. MGAT activity was assessed by measuring diacylglycerol mass in membranes after the assay. Values are the mean ± SD of four measurements.
Because [14C]oleoyl-CoA was incorporated into diacylglycerol in membranes expressing MGAT1 (Fig. 3b, arrow), we suspected that MGAT1 possesses MGAT activity. To test this possibility, membranes expressing MGAT1 were assayed with either [14C]monooleoylglycerol or [14C]oleoyl-CoA as the labeled substrate. In both cases, membranes expressing MGAT1 catalyzed the incorporation of the label into diacylglycerol (Fig. 3c), establishing that the MGAT1 protein possesses MGAT activity. This MGAT activity was confirmed by its dependence on MGAT substrates; when unlabeled MGAT substrate (monooleoylglycerol or oleoyl-CoA) was provided over a range of concentrations while the other substrate was held constant, the mass of diacylglycerol synthesized depended on the concentration of substrate (Fig. 3d). Further, the acyltransferase activity of MGAT1 seemed to be specific for monoacylglycerol (and possibly diacylglycerol) as the acyl group acceptor. No acyltransferase activity was found in MGAT1-expressing membranes when glycerol 3-phosphate, dihydroxyacetone phosphate, lysophosphatidate, lysophosphatidylcholine, sphingosine, or cholesterol were used as the [14C]oleoyl-CoA acceptor (not shown). [14C]Monooleoylglycerol was incorporated into triacylglycerol by insect cell membranes expressing DGAT2 (Fig. 3c). This finding suggests that DGAT2 may possess an MGAT activity that generates diacylglycerol, which can then serve as a substrate for its robust DGAT activity.
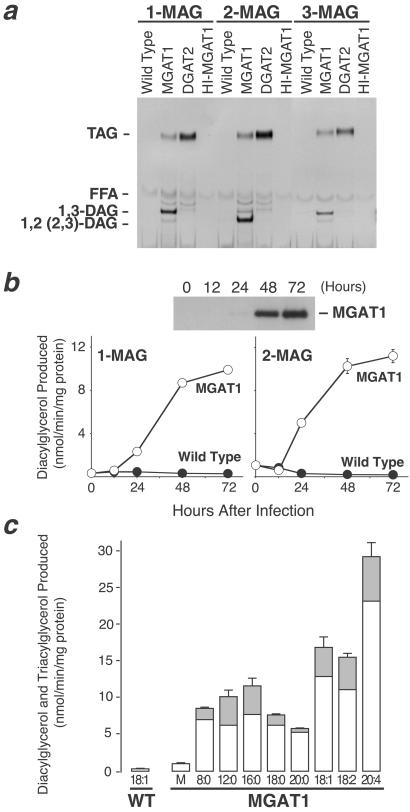
Next, we determined whether MGAT1 can acylate each of the three stereoisomers of monoacylglycerol. In these assays, [14C]oleoyl-CoA was used as the acyl donor and either sn-1-monooleoylglycerol, sn-2-monooleoylglycerol, or sn-3-monostearoylglycerol as the acyl acceptor. When sn-1-monooleoylglycerol or sn-3-monostearoylglycerol was used, the major product was sn-1,3-diacylglycerol. When sn-2-monooleoylglycerol was used, the major product was sn-1,2(2,3)-diacylglycerol (Fig. 4a). These results indicate that MGAT1 can acylate each of the stereoisomers of monoacylglycerol and that it does so mainly at the sn-1 or sn-3 position. We also found that the specific activities of MGAT1 with either sn-1-monooleoylglycerol or sn-2-monooleoylglycerol as the substrate were similar and that both increased proportionally with MGAT1 protein levels (Fig. 4b). These findings suggest that MGAT1 expressed in insect cells can use sn-1-monooleoylglycerol and sn-2-monooleoylglycerol equally well as substrates in vitro.
Figure 4.
Substrate specificities of MGAT1. (a) Ability of MGAT1 expressed in insect cells to acylate each of the three monoacylglycerol stereoisomers [sn-1-monooleoylglycerol (1-MAG), sn-2-monooleoylglycerol (2-MAG), or sn-3-monostearoylglycerol (3-MAG)]. FFA, free fatty acid. (b) Proportional increases in MGAT1 protein and MGAT activity in insect cell membranes. The specific activities of MGAT1 with sn-1-monooleoylglycerol (1-MAG) and sn-2-monooleoylglycerol (2-MAG) substrates are similar. Values are the mean ± SD of three measurements. (c) Preference of MGAT1 for unsaturated fatty acyl-CoAs. sn-2-[3H]monooleoylglycerol was used as the acyl acceptor for different fatty acyl-CoAs (50 μM each). M, malonyl-CoA; 8:0, n-octanoyl-CoA; 12:0, lauroyl-CoA; 16:0, palmitoyl-CoA; 18:0, stearoyl-CoA; 20:0, arachidoyl-CoA; 18:1, oleoyl-CoA; 18:2, linoleoyl-CoA, and 20:4, arachidonoyl-CoA. White and gray bars represent radioactivity recovered from incorporation of the label into diacylglycerol and triacylglycerol, respectively. Because sn-2-[3H]monooleoylglycerol was used the labeled substrate, incorporation of radioactivity into diacylglycerol and triacylglycerol represents total MGAT activity. Values are the mean ± SD of four measurements and are representative of two independent experiments.
We also determined whether MGAT1 preferred specific fatty acyl-CoAs as substrates in assays using sn-2-[3H]monooleoylglycerol as the acyl acceptor. MGAT1 used a variety of fatty acyl-CoAs, and MGAT activities were highest with CoA derivatives containing unsaturated fatty acids (Fig. 4c). The highest activity was found with arachidonoyl-CoA (C20:4).
Mouse MGAT1 Expressed in Mammalian Cells.
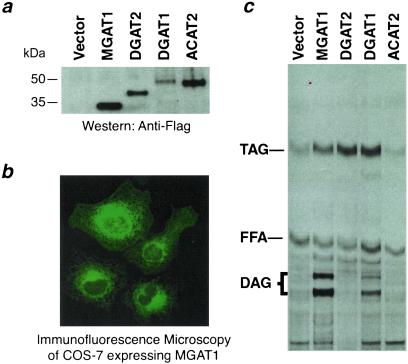
We also expressed mouse MGAT1 cDNA and control cDNAs transiently in monkey kidney COS-7 cells (Fig. 5a). In cells expressing MGAT1, immunofluorescence microscopy demonstrated a perinuclear and reticular staining pattern, consistent with distribution of the protein in the endoplasmic reticulum (Fig. 5b); however, we cannot exclude that a minor portion of MGAT1 may be located in other organelles such as the Golgi apparatus. COS-7 cell membranes expressing MGAT1 incorporated a significant amount of radioactivity into diacylglycerol when [14C]oleoyl-CoA and sn-2-monooleoylglycerol were provided as substrates (Fig. 5c), indicating that these membranes possess MGAT activity. Similar levels of MGAT activity were found when sn-1-monooleoylglycerol was used as the acyl acceptor (not shown). In contrast, MGAT activity was not detected in control membranes, except in those expressing DGAT1, which seemed to possess a low level of MGAT activity. Similar results were found when these cDNAs were expressed in Chinese hamster ovary cells (not shown).
Figure 5.
Expression of MGAT1 in mammalian cells. (a) Immunoblotting of FLAG-tagged proteins to verify expression of MGAT1 and control proteins. Cells were transfected with vector only or with FLAG-tagged versions of MGAT1, DGAT2, DGAT1, or ACAT2. (b) Immunocytochemistry of COS-7 cells expressing FLAG-tagged MGAT1. An anti-FLAG antibody and FITC-conjugated secondary antibody were used. (c) Acyltransferase activities in COS-7 cell membranes. [14C]oleoyl-CoA was used for enzyme assays.
Tissue Distribution of Mouse MGAT1 Expression.
MGAT1 mRNA expression was highest in the stomach and kidney; lower levels of expression were present in white and brown adipose tissue, uterus, and liver (Fig. 6 a and b). MGAT1 mRNA expression was not detected in the small intestine. Because MGAT activity had not previously been reported in mice, we assessed MGAT activity in membranes from mouse tissues. As demonstrated for many other species, the highest activity was found in the small intestine (Fig. 6c). MGAT activity was also detected at significant levels in the stomach, kidney, adipose tissue, and liver, where MGAT1 is expressed.
Figure 6.
Tissue expression pattern of mouse MGAT1. (a) MGAT1 mRNA expression was detected by Northern analysis with 20 μg of total RNA. The same blots were probed for DGAT2 as a control for the presence of mRNA. (b) MGAT1 mRNA expression in liver was detected by Northern analysis with 2 μg (Left) or 5 μg (Right) poly(A)+ RNA. Sm, small; Sk, skeletal; WAT, white adipose tissue; BAT, brown adipose tissue. (c) MGAT activity in mouse tissues. Values are the mean ± SD of four measurements.
Discussion
MGAT is a microsomal enzyme that is important in the biosynthesis of cellular triacylglycerols because it generates their precursor diacylglycerols, in particular, in specific tissues where degradation and resynthesis of triacylglycerols are prominent. In this study, we identified a gene that encodes an MGAT enzyme, MGAT1, in mice. Evidence that MGAT1 is an MGAT is provided by the finding that MGAT1 expressed in insect cell membranes catalyzed diacylglycerol synthesis with either [14C]monoacylglycerol or [14C]oleoyl-CoA as a substrate. Further, MGAT1 expressed in membranes increased diacylglycerol mass, and this increase depended on the concentration of the substrate. Moreover, MGAT1 also exhibited MGAT activity in mammalian cell lines. The activity of MGAT1 was specific for catalyzing diacylglycerol synthesis because MGAT1 incorporated fatty acyl-CoA into diacylglycerol but not into other lipids.
MGAT1 expressed in insect cell membranes also resulted in more labeled substrate incorporation into triacylglycerol than did control membranes (wild-type and heat-inactivated MGAT1), suggesting that MGAT1 also possesses weak DGAT activity. This possibility seems likely given the homology of MGAT1 to DGAT2. On the other hand, it is possible that MGAT1 activity produces labeled diacylglycerol, which can then be used as substrate for the endogenous DGAT activity present in insect cells to generate triacylglycerol. Our experiments also suggest that DGAT1 and DGAT2 may possess some weak MGAT activities. Studies performed with purified proteins will be needed to elucidate whether these weak activities are intrinsic to these enzymes. We reported sequences encoding a human homologue of MGAT1 (hDC2 in ref. 14). Whether the human MGAT1 homologue functions as an MGAT awaits the cloning and expression of a full-length MGAT1 transcript from human tissue cDNA.
In our study, mouse MGAT1 expressed in insect cells and mammalian cells used both sn-2-monoacylglycerol and sn-1-monoacylglycerol efficiently as substrates. However, in rat tissues, sn-2-monoacylglycerol was the preferred substrate for both small intestine and liver MGAT activity (6, 11), although its stereoisomer sn-1(3)-monoacylglycerol can also be used as a substrate (17, 18). It is unclear whether mouse MGAT1 possesses broader substrate specificity than MGAT enzymes in rat tissues, or whether our results are unique to our expression and assay systems. Mouse MGAT1 also preferred unsaturated fatty acyl-CoAs as substrates, consistent with a proposed role of MGAT in preserving essential fatty acids in specific tissues through reesterification cycles (6). The preference of MGAT1 for unsaturated fatty acyl-CoAs contrasts with a reported preference of human intestinal MGAT for saturated fatty acyl-CoAs (19).
In mice, MGAT1 mRNA was highly expressed in stomach and kidney. MGAT activity in these two tissues has not previously been characterized, although MGAT activity has been reported in rat kidney (4). Therefore, the physiological function of MGAT1 in these tissues is unknown. In contrast, MGAT1 was not expressed in the small intestine, a tissue in which MGAT activity has often been demonstrated. Findings were similar for the human MGAT1 homologue, which was expressed in kidney and liver but not in the small intestine as assessed by Northern analysis (C.-L.E.Y. and R.V.F., unpublished data). These findings imply that a second MGAT gene exists. Because the functions of several related genes of the DGAT2 gene family remain uncharacterized, these genes are candidates for an intestinal MGAT.
The identification of a mouse MGAT gene provides a molecular probe for studying the role of MGAT enzymes in glycerolipid synthesis and lipid metabolism. The identification of MGAT1 will make it easier to identify MGAT genes in different organisms and an MGAT gene that functions in the intestine. The cloning of MGAT genes will facilitate the development of pharmacological inhibitors of MGAT enzymes. Because of MGAT's prominent role in intestinal fat absorption, inhibitors of MGAT in the intestine may be attractive pharmaceutical targets for modulating fat absorption.
Acknowledgments
We thank Bethany Taylor for manuscript preparation, Gary Howard and Stephen Ordway for editorial assistance, and Rosalind Coleman and Mimi Zeiger for helpful comments on the manuscript. We also thank Kathy Lardizabal and Toni Voelker for contributions to our work on DGAT2-related genes. This work was supported by National Institutes of Health Grant DK56084 (to R.V.F.), Canadian Institute for Health Research postdoctoral fellowship (to S.S.), and the J. David Gladstone Institutes.
Abbreviations
- MGAT
acyl-CoA:monoacylglycerol acyltransferase
- DGAT
acyl-CoA:diacylglycerol acyltransferase
- ACAT
acyl-coA:cholesterol acyltransferase
References
- 1.Ron D, Kazanietz M G. FASEB J. 1999;13:1658–1676. [PubMed] [Google Scholar]
- 2.Lehner R, Kuksis A. Prog Lipid Res. 1996;35:169–201. doi: 10.1016/0163-7827(96)00005-7. [DOI] [PubMed] [Google Scholar]
- 3.Bell R M, Coleman R A. Annu Rev Biochem. 1980;49:459–487. doi: 10.1146/annurev.bi.49.070180.002331. [DOI] [PubMed] [Google Scholar]
- 4.Coleman R A, Haynes E B. J Biol Chem. 1984;259:8934–8938. [PubMed] [Google Scholar]
- 5.Mostafa N, Bhat B G, Coleman R A. Lipids. 1994;29:785–791. doi: 10.1007/BF02536701. [DOI] [PubMed] [Google Scholar]
- 6.Xia T, Mostafa N, Bhat B G, Florant G L, Coleman R A. Am J Physiol. 1993;265:R414–R419. doi: 10.1152/ajpregu.1993.265.2.R414. [DOI] [PubMed] [Google Scholar]
- 7.Jamdar S C, Cao W F. Arch Biochem Biophys. 1992;296:419–425. doi: 10.1016/0003-9861(92)90592-k. [DOI] [PubMed] [Google Scholar]
- 8.Nishizuka Y. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 9.Panikashvili D, Simeonidou C, Ben-Shabat S, Hanus L, Breuer A, Mechoulam R, Shohami E. Nature (London) 2001;413:527–531. doi: 10.1038/35097089. [DOI] [PubMed] [Google Scholar]
- 10.Manganaro F, Kuksis A. Can J Biochem Cell Biol. 1985;63:341–347. doi: 10.1139/o85-050. [DOI] [PubMed] [Google Scholar]
- 11.Bhat B G, Bardes E S-G, Coleman R A. Arch Biochem Biophys. 1993;300:663–669. doi: 10.1006/abbi.1993.1092. [DOI] [PubMed] [Google Scholar]
- 12.Tumaney A W, Shekar S, Rajasekharan R. J Biol Chem. 2001;276:10847–10852. doi: 10.1074/jbc.m100005200. [DOI] [PubMed] [Google Scholar]
- 13.Lehner R, Kuksis A. J Biol Chem. 1995;270:13630–13636. doi: 10.1074/jbc.270.23.13630. [DOI] [PubMed] [Google Scholar]
- 14.Cases S, Stone S J, Zhou P, Yen E, Tow B, Lardizabal K D, Voelker T, Farese R V., Jr J Biol Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 15.Cases S, Smith S J, Zheng Y-W, Myers H M, Lear S R, Sande E, Novak S, Collins C, Welch C B, Lusis A J, et al. Proc Natl Acad Sci USA. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lardizabal K D, Mai J T, Wagner N W, Wyrick A, Voelker T, Hawkins D J. J Biol Chem. 2001;276:38862–38869. doi: 10.1074/jbc.M106168200. [DOI] [PubMed] [Google Scholar]
- 17.Coleman R A, Haynes E B. J Biol Chem. 1986;261:224–228. [PubMed] [Google Scholar]
- 18.Lehner R, Kuksis A, Itabashi Y. Lipids. 1993;28:29–34. doi: 10.1007/BF02536356. [DOI] [PubMed] [Google Scholar]
- 19.Bierbach H. Digestion. 1983;28:138–147. doi: 10.1159/000198977. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]