Abstract
Benign prostatic hyperplasia (BPH) is a disease of unknown etiology that significantly affects the quality of life in aging men. Histologic BPH may present itself either as symptomatic or asymptomatic in nature. To elucidate the molecular differences underlying BPH, gene expression profiles from the prostate transition zone tissue have been analyzed by using microarrays. A set of 511 differentially expressed genes distinguished symptomatic and asymptomatic BPH. This genetic signature separates BPH from normal tissue but does not seem to change with age. These data could provide novel approaches for alleviating symptoms and hyperplasia in BPH.
Benign prostatic hyperplasia (BPH) is one of the most common diseases affecting aging men. Currently, it is estimated that the disease progressively involves three fourths of the male population over 75 years of age (1). Clinical manifestations range widely from minimally bothersome symptoms to urinary retention and renal failure (2). Other factors such as age-induced detrusor dysfunction, neural alterations in the bladder and prostate, and, at least in some patients, chronic inflammation caused by bacterial endotoxin in the prostate also are thought to contribute to the symptoms (3). Although it is generally held that BPH is influenced by androgens, recent data suggest that the action of androgens alone may not explain the hyperplastic development of the prostate gland (4, 5). A number of mitogenic growth factors have been implicated in the pathophysiology of BPH (6), but, despite decades of intensive research efforts, the etiology and pathophysiology remain unclear (7–9). Furthermore, the underlying molecular differences producing symptoms in some but not all patients with histologic BPH are largely unknown. Finally, why BPH is generally restricted to the transition zone of the prostate and why it remains benign are still enigmatic issues.
Gene-expression profiling facilitated by the development of DNA microarrays (10, 11) represents a major advance in global gene-expression analysis. In a single assay, the quantitative expression of each gene in response to a change in the cellular state can be measured in parallel. In recent years, several investigators have applied this technology in a variety of ways such as classification of disease samples, gene function during development and differentiation, target identification and validation, pathway dissection, and cellular responses to physiological perturbation or pharmacological treatment (12).
Previous attempts to understand the pathophysiology of BPH have been hampered by the lack of appropriate cell lines, the lack of a suitable animal model, and the implication of a multitude of factors with diverse biological functions. In the present paper, we have taken a different approach to gain new insights into the disease process; we have used human tissue samples and oligonucleotide microarray technology to determine global changes in gene expression without bias to any particular factor as the overriding cause of BPH. Here, we present an analysis of the observed expression profiles and their relationship to phenotypic properties of the various disease groups.
Materials and Methods
Sample Selection and Description.
Five different groups of prostate tissue samples were identified for this study. Because BPH is a disease associated with aging, two groups of “normal” individuals were identified. First, normal prostate tissues were isolated from five young organ donors. These individuals were all 20 years or younger. This group was designated as Normal 1 (N1). The second set of five normal donors ranged from 30 to 50 years in age. This group was designated Normal 2 (N2). Symptomatic BPH samples were obtained from eight patients who underwent open prostatectomies to relieve obstruction. Asymptomatic BPH samples were obtained from organ donors or from histological BPH lesions in five patients who underwent cystoprostatectomy because of bladder cancer. The fifth group consisted of eight samples that were obtained by radical prostectomy from individuals with prostate cancer but who had no symptoms of BPH. This group was designated as BPH with cancer.
The weight of the prostates from the group of young organ donors (N1 group of organ donors less than 30 years of age) ranged from 28 to 50 grams, with an average weight of 38 grams. Most of these prostates were histologically unremarkable, with no changes of benign prostatic hyperplasia. No evidence of prostatitis was seen in these prostates.
The second group (Group 2) of prostates was from organ donors with the age of the donors ranging from 20 to 50 years of age. The weight of the prostate ranged from 30 to 70 grams with an average weight of 48 grams. Two of the five cases showed focal areas of benign prostatic hyperplasia (epithelial predominant). No evidence of prostatitis was seen in these patients.
Group 3 consisted of patients with symptomatic benign prostatic hyperplasia. The specimens evaluated consisted of four retropubic prostatectomies and four transurethral prostate resections. The hyperplasia seen in all of these cases was predominantly epithelial. The retropubic prostatectomies had weights ranging from 100 to 193 grams, with an average weight of 130 grams. Two of these cases had mild prostatitis, one had moderate prostatitis, and one had marked prostatitis. The transurethral prostate resections had an average weight of 55 grams. Two of these transurethral prostate resection specimens showed mild to moderate prostatitis.
The group of patients with prostate adenocarcinoma and associated benign prostatic hyperplasia (Case set 4) consisted of a total of 5 cases. Three cases had Gleason scores of 6 (moderately differentiated prostatic adenocarcinoma) and two cases had Gleason scores of 7 (poorly differentiated tumor). Two of the cases evaluated had a tumor node metastasis (TNM) stage of T3A with extracapsular penetration by the prostatic adenocarcinoma. The remaining three cases had a TNM stage T2 lesion (tumor identified within one of both lobes of the prostate). The extent of involvement of the prostate by the tumor ranged from 5 to 15%. The total prostate volume involved by the tumor averaged 7.5% (range 5–15%). The average weight of the prostate was 60 grams (range 34–86 grams). The hyperplasia was predominantly epithelial. No significant prostatitis was seen.
Group 5 consisted of either older donors (age greater than 50 years of age) or patients with prostates harvested from a cystoprostatectomy specimen (prostate resected as part of a resection of urothelial carcinoma). The weight of the prostate ranged from 30 to 65 grams, with an average of 47 grams. The hyperplasia was epithelial predominant. Two of these prostates had changes of mild prostatitis.
All of the prostates evaluated showed an epithelial-predominant prostatic hyperplasia. No definite stromal hyperplasia was seen in any of the cases evaluated.
Sample Acquisition.
The selected tissue samples were acquired from the University of Pittsburgh Medical Center under stringent Institutional Review Board guidelines with appropriate informed consent. Samples (>500 mg) were excised and snap frozen in liquid nitrogen within 30 min of excision and stored at −80°C until extraction of RNA. All samples were submitted for pathology evaluation. In every case, the tissue was excised from the junction between the ejaculatory duct and the prostatic urethra in the transition zone of the prostate. In particular, BPH tissue from patients with early stage prostate cancer was carefully excised away from the cancer lesion macroscopically, and their histological diagnosis was confirmed microscopically.
Sample Preparation and Data Analysis.
Sample preparation, hybridization to the 42 K Affymetrix HuGeneFL array, and raw data collection was done exactly as described by Tackels-Horne et al. (13). The raw data were analyzed with Affymetrix software, GENE CHIP V.3.0 and EXPERIMENTAL DATA MINING TOOL V.1.0. S-Plus was used to perform the ANOVA, principal component analysis (PCA), and hierarchical clustering analyses. For the PCA, we used the correlation matrix on nontransformed expression values. For the clustering analysis, we used average linkage clustering with correlation of nontransformed expression values as the distance metric. The data were viewed by using TREEVIEW (14).
Peptide Synthesis and Antibody Production.
A synthetic peptide, CPGQEREGTPPIEERKVE (amino acid residues 44–60), of human JM27 was used for the production of polyclonal antibodies in rabbits. Before using, the antibodies were affinity-purified by using the synthetic peptide.
Immunohistochemistry.
Immunohistochemistry was performed on formalin-fixed paraffin-embedded sections of tissue obtained from the same sample that was used for the microarray analysis. Immunohistochemical examination was performed with the standard avidin-biotin technique, and a protease pretreatment step was included.
Results and Discussion
Gene Selection and Principal Component Analysis.
As an experimental strategy, we first identified a set of genes (individual probe elements on the microarray) that are differentially expressed among the four groups of normal and disease samples. Each of the 42,843 genes on the microarray was fitted in an ANOVA model, and P values corresponding to each of six possible pair-wise comparisons among the four sample groups then were determined for each gene (for details of individual groups, see Table 1). The selection criteria required a gene to have P < 0.001 for two or more of the pair-wise comparisons. These criteria resulted in a nonredundant set of 511 genes.
Table 1.
Details of the samples used to compare gene expression analysis
| Sample group | Age (yrs) | No. of samples |
|---|---|---|
| Normal (N) | 13–50 | 10 |
| BPH without symptoms (O) | 51–65 | 5 |
| BPH with symptoms (S) | 42–77 | 8 |
| BPH with cancer (C) | 60–70 | 8 |
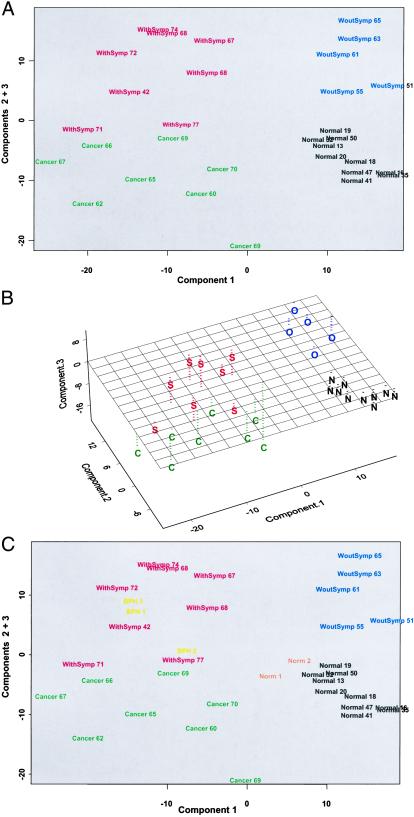
To determine whether this 511 gene set serves as a basis to discriminate among the various groups of samples, we performed a PCA on this gene set. The samples were plotted by using the scores for the first three principal components. As shown in Fig. 1A, each of the four sample groups can be clearly distinguished from one another in this analysis. Component 1 (36% of the variability) discriminated between Normal and asymptomatic BPH vs. BPH cancer and symptomatic BPH. Component 2 (10% of the variability) distinguished Normal from asymptomatic BPH, and Component 3 (8% of the variability) distinguished BPH cancer from symptomatic BPH.
Figure 1.
PCA. (A) A two-dimensional plot of the data showing the separation of the four sample groups. (B) A three-dimensional plot of the same data. (C) A two-dimensional plot of the data including the additional normal (Norm 1 & 2) and symptomatic BPH samples (BPH 1, 2, and 3). The four sample groups are normal (N), asymptomatic BPH (O), symptomatic BPH (S), and BPH with cancer (C) and include the age of the individual patient. S-PLUS was used to perform the ANOVA, PCA, and hierarchical clustering analyses. For the PCA, the correlation matrix on nontransformed expression values were used.
A two-dimensional representation of the PCA plot wherein Component 1 is plotted against Components 2 + 3 is presented in Fig. 1B. Each of the four sample groups is clustered in a different quadrant of this figure. Notably, within each sample group, there was no noticeable age-related clustering of samples. Further, although there was an overlap of ages between the samples from various groups, age does not seem to be a confounding factor for the analysis based on this gene set. We had initially identified two subgroups of normal subjects. A younger set included individuals ranging in age from 13–20 years, whereas the older set ranged from 31–51 years (Table 1). At a molecular level, however, the two subsets were indistinguishable and hence were grouped together. In all subsequent analysis, this combined group is referred to as the Normal group.
The intra-group variability (i.e., the tightness of clustering) differed between the four groups (Fig. 1A). Normal samples exhibited the least intra-group variability, followed by asymptomatic BPH and symptomatic BPH. BPH with cancer samples exhibited the most sample-to-sample variability. On the other hand, the inter-group variation segregated the asymptomatic BPH group with the normal group more than the symptomatic BPH group (Fig. 1B).
Asymptomatic samples were obtained from men with no record of being treated for BPH but had histological evidence when examined retrospectively. Because asymptomatic samples clearly exhibit the BPH phenotype at the microscopic level, one would expect the two BPH groups to exhibit more similarity than disparity. Furthermore, the BPH with cancer group was distinct from the asymptomatic BPH group but more similar to the symptomatic BPH group (Fig. 1B). Because transition-zone tissue was obtained from men with prostate cancer with no clinical symptoms of BPH, gene-expression similarity to either the normal or the asymptomatic BPH groups would be expected. On the contrary, the BPH with cancer group could be readily distinguished from every other group.
To extend these findings, additional samples from two normal and three symptomatic BPH men were studied in a subsequent PCA by using the same set of 511 genes. As shown in Fig. 1C, the two additional normal samples clustered with the previous normal samples and the three additional symptomatic BPH samples clustered with the other symptomatic BPH samples. PCAs applying a less stringent (P < 0.01) or a more stringent (P < 0.0001) gene selection criteria (1,854 genes and 110 genes, respectively) yielded essentially similar results (data not shown).
Interrogating the differential gene-expression data by using PCA distinguishes each of the disease groups from normal tissue. Furthermore, within the three disease groups, PCA clearly separates them from each other. In fact, recent data suggest that the occurrence of symptoms related to clinical BPH are not caused by differences in the histological composition of the prostate (15), lending further credence to these findings.
Hierarchical Clustering of Genes Expressed in Normal and BPH Tissues.
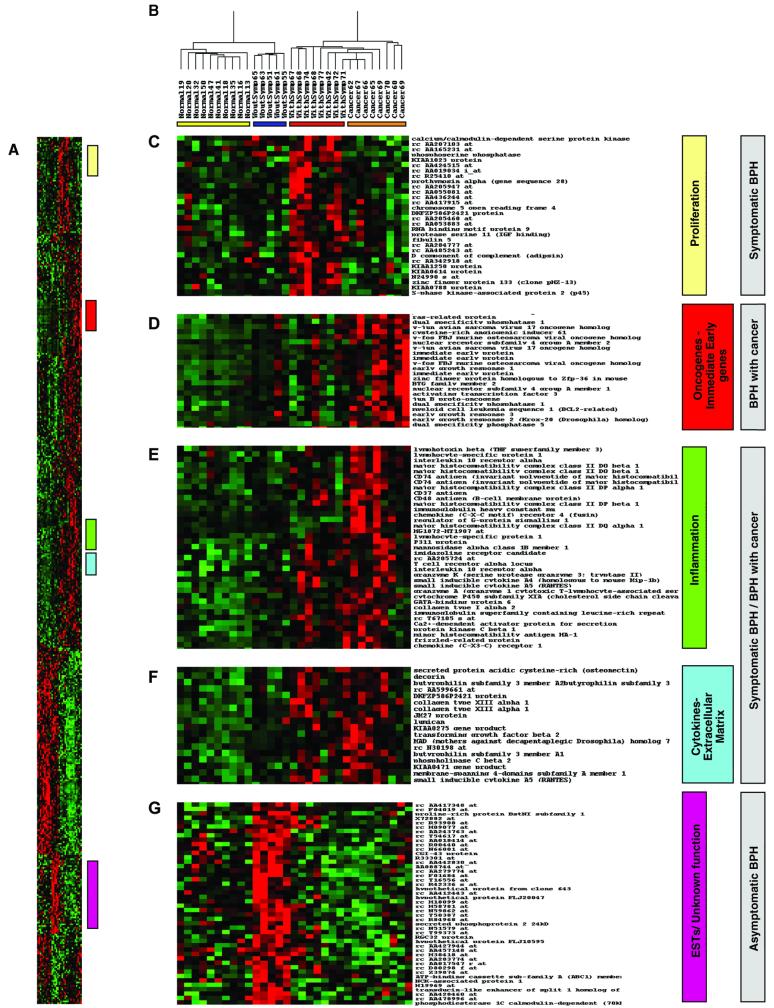
Applying two-dimensional hierarchical clustering to the expression data from the set of 511 genes measured across all 31 samples used for the PCA demonstrates an unambiguous separation of normal and disease groups as well as the separation of three subsets of BPH tissue samples (Fig. 2A). A dendrogram showing a major division in the distribution of samples is illustrated in Fig. 2B. The first node includes all normal and asymptomatic BPH samples, whereas the second node includes all symptomatic BPH and BPH with cancer individuals. It is remarkable that within each branch of the two nodes, the individuals in each group cluster together on a sub-branch.
Figure 2.
Two-dimensional hierarchical clustering. (A) Expression pattern of 511 genes obtained by PCA in 31 experimental samples. Rows represent individual genes, and columns represent individual samples. Each cell in the matrix represents the expression level of a single transcript in a single sample, with red and green indicating transcript levels above and below the median for that gene across all samples, respectively. Color saturation is proportional to the magnitude of the difference from the mean. Expression values are represented as Z-scores [(expression-gene mean) gene SD]. (B) Dendrogram showing overall similarity in expression profiles in the respective samples. Four clear subdivisions are present, each representing a distinct sample group. A colored bar under each group denotes this separation. (C–G) Colored bars alongside the matrix denote clusters of genes with similar cellular functions that are associated with the specific disease phenotypes (gray boxes).
As shown in Fig. 2 C–G, various subsets of clusters contain genes associated with related functions, as well as expressed sequence tag (EST) and genes of unknown functions. A set of genes comprising mostly ESTs and several genes associated with cell proliferation (for example, calcium/calmodulin-dependent serine kinase, phosphoserine phosphatase, S-phase kinase-associated protein 2 or p45) were significantly up-regulated in the symptomatic BPH group compared with any other group (Fig. 2C). Similarly, a subset of genes including oncogenes and immediate early genes (for example, ras-related protein, v-jun, v-fos, immediate early protein, and early growth response gene) was highly up-regulated in the BPH cancer group alone (Fig. 2D). Two subsets of genes including several inflammatory mediators (for example, lymphotoxin beta, immunoglobulins, and chemokine receptors; Fig. 2E), cytokines, and extracellular matrix-associated molecules (for example, RANTES, osteonectin, lumican; Fig. 2F) appear to distinguish symptomatic BPH and BPH with cancer as a separate group distinct from the normal or asymptomatic BPH groups. Finally, a cluster of genes including mostly ESTs and genes with potential ORFs of unknown function clearly distinguishes the asymptomatic BPH group from all of the others (Fig. 2G).
The strong correlation between inflammation and symptomatic BPH is striking. Asymptomatic histological inflammation and latent infection are common phenomena in BPH (16–18). Furthermore, Mahapokai et al. (19) have recently reported that in a hormonally induced BPH dog model, hyperplasia was followed by cell-mediated and humoral immune responses. The expression profiling data demonstrate the strong correlation between inflammation and symptomatic BPH, suggesting new therapeutic approaches such as anti-inflammatory agents could be developed to alleviate BPH symptoms (20).
A group of oncogenes and immediate early genes predominantly influence the clustering of the BPH cancer patients into a separate group distinct from symptomatic and asymptomatic BPH (Fig. 2D). Considering that the BPH cancer samples were taken from the histologically confirmed noncancerous transition zone in men with no symptoms of BPH, it was expected that these samples would cluster with the asymptomatic BPH samples. In fact, they appear as a distinct group, more similar to the symptomatic BPH group.
Several members of a cluster common to symptomatic BPH and BPH with cancer represent genes involved in various types of cancers. For example, butyrophilin, a gene involved in cell proliferation (21), RAB2L, a member of the RAS oncogene family involved in signal transduction (22), protocadherin, a member of a large family of genes involved in cell–cell interaction and adhesion (23), osteonectin, an antiadhesive protein known to be involved in cell–matrix interactions, migration, and angiogenesis, and JM 27, are all components of this common cluster (Fig. 2F).
In particular, osteonectin promotes the invasive ability of bone-metastasizing prostate (and breast) cancer cells but not that of non-bone-metastasizing tumor cells (24, 25). Osteonectin does not stimulate the growth of prostate cancer cells in vitro or in vivo (24). The fact that molecules such as osteonectin promoting invasion but not proliferation are being expressed in symptomatic BPH challenges previous concepts of the nature of BPH.
In the symptomatic BPH group, a cluster comprising mostly ESTs and several genes associated with cell proliferation clearly distinguished this group from every other group. The expression of proliferation-associated genes is not surprising in a hyperplastic state. Similarly, asymptomatic BPH also is identified with a cluster of genes comprised largely of ESTs and genes of unknown function. Elucidating the functions of these ESTs could provide new clues to the etiology of BPH and may yield novel markers and/or therapeutic targets for BPH.
Immunohistochemistry and Tissue Distribution of a Prostate-Specific Gene.
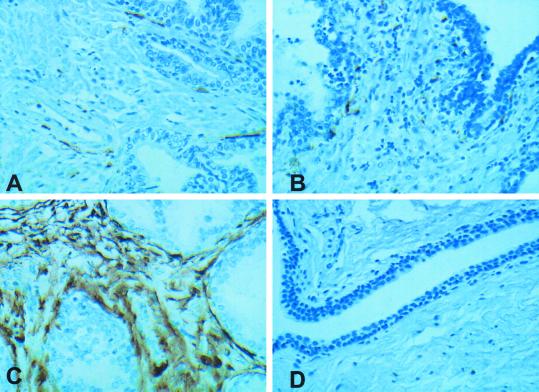
JM27, a gene up-regulated in prostate cancer (26), is also differentially up-regulated in symptomatic BPH (present data). To examine the expression and subcellular location of the encoded protein, an immunohistochemical analysis was performed on the same samples used for gene-expression studies. Compared with normal prostate (Fig. 3A) or asymptomatic BPH (Fig. 3B), JM27 protein was overexpressed in symptomatic BPH (Fig. 3C). Antibody specificity is confirmed by no reaction with preimmune serum (Fig. 3D). Within the prostate tissue, expression of JM27 protein was confined to stromal cells with no expression in the epithelial (glandular) cells. By using tissue arrays from several normal and diseased prostate samples, we have extended and confirmed these observations (data not shown). In every case, the JM27 protein was significantly up-regulated only in symptomatic BPH and was confined to the stromal compartment (R.D. and R.H.G., unpublished work). Taken together, the data provide good evidence for the differential expression of JM27 both at the transcript as well as the protein level.
Figure 3.
Immunohistochemical localization of JM27. Normal prostate (A), asymptomatic (B) and symptomatic BPH (C) tissue samples stained with anti-peptide JM27 antibody, respectively. (D) Symptomatic BPH tissue stained with preimmune serum as a control. All images were taken at ×400 magnification.
The microarray expression of JM27 from more than 570 samples covering a broad range of normal human tissues showed remarkable tissue-specificity (Fig. 4). JM27 is expressed only in the prostate and in certain female reproductive (for example, uterine) tissue samples. These data suggest that JM27 is a potential candidate marker and/or therapeutic target for symptomatic BPH. JM27 is homologous to a family of MAGE/GAGE-like proteins containing RGD motifs frequently found in cell-adhesion proteins. GAGE/MAGE antigens were previously reported to be targets for tumor-specific cytotoxic lymphocytes in melanoma (27), suggesting that overexpression of JM27 may be involved in the progression of BPH, prostate cancer (26), and other tumors of the female reproductive tract (28).
Figure 4.
Tissue-specific expression of JM27. The expression values consisting of present (blue), marginal (yellow), and absent (red) calls for the JM27 gene fragment (AA463726) are plotted across prostate normal, symptomatic BPH, asymptomatic BPH, BPH with cancer, and 20 different normal human tissue sample sets. For each sample set, vertical bars are displayed at the median value and the upper 75 and 25 percentile range (bound by an orange box). The extreme vertical bars are located three SDs from the median. The x axis shows graduated markers indicating expression intensity. The number of samples in each set is noted within parentheses, and the percentage of samples where the gene is scored present is indicated as an orange bar at the right of the sample set name.
Finally, these data suggest several commonalities between symptomatic BPH, BPH with cancer, and prostate cancer. In fact, BPH in itself seems to be closely related to prostate cancer, because both diseases involve overgrowth of epithelial cells. It has been suggested that tumor development proceeds by a process analogous to Darwinian evolution, in which a succession of genetic changes, each conferring some growth advantage, leads to the progressive conversion of normal human cells into cancer cells (29). Unlike prostate cancer however, BPH is rarely associated with genetic abnormalities and is an overgrowth of a more normal epithelium (8). Comparing global changes in gene expression in BPH and prostate cancer will provide further insights into these disease processes.
Acknowledgments
We thank members of the Gene Logic GeneChip array production group for their expertise, and Dr. Tatsuji Chuman, Dr. Doug Dolginow, and Dr. Jeff Cossman for their support. We also thank Dr. Subbu Yerramilli, Dr. Glenn Hoke, Dr. Uwe Scherf, and Dr. Joel B. Nelson for their thoughtful comments on the manuscript.
Abbreviations
- BPH
benign prostatic hyperplasia
- PCA
principal component analysis
- EST
expressed sequence tag
References
- 1.Caprino L. Minerva Urol Nefrol. 2000;52:87–92. [PubMed] [Google Scholar]
- 2.Medina J J, Parra R O, Moore R G. Med Clin N Am. 1999;83:1213–1229. doi: 10.1016/s0025-7125(05)70159-0. [DOI] [PubMed] [Google Scholar]
- 3.Roper W G. Med Hypotheses. 1998;50:61–65. doi: 10.1016/s0306-9877(98)90179-7. [DOI] [PubMed] [Google Scholar]
- 4.Berthon P, Waller A S, Villette J M, Loridon L, Cussenot O, Maitland N J. Int J Cancer. 1997;73:910–916. doi: 10.1002/(sici)1097-0215(19971210)73:6<910::aid-ijc25>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Bartsch G, Rittmaster R S, Klocker H. Eur Urol. 2000;37:376–380. doi: 10.1159/000020181. [DOI] [PubMed] [Google Scholar]
- 6.Wong Y C, Wang Y Z. Int Rev Cytol. 2000;199:65–116. doi: 10.1016/s0074-7696(00)99002-8. [DOI] [PubMed] [Google Scholar]
- 7.Ziada A, Rosenblum M, Crawford E D. Urology. 1999;53, Suppl. 3a:1–6. doi: 10.1016/s0090-4295(98)00532-9. [DOI] [PubMed] [Google Scholar]
- 8.De Marzo A M, Coffey D S, Nelson W G. Urology. 1999;53, Suppl. 3a:29–39. doi: 10.1016/s0090-4295(98)00536-6. [DOI] [PubMed] [Google Scholar]
- 9.Cabelin M A, Te A E, Kaplan S A. Curr Opin Urol. 2000;10:301–306. doi: 10.1097/00042307-200007000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 11.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;13:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 12.Debouck C, Goodfellow P N. Nature (London) 1999;21:658–660. doi: 10.1038/4475. [DOI] [PubMed] [Google Scholar]
- 13.Tackels-Horne D, David M, Williams A J, Wilson D J, Eskandari T, Vogt L M, Boland J F, Scherf U, Vockley J G. Cancer. 2001;92:395–405. doi: 10.1002/1097-0142(20010715)92:2<395::aid-cncr1335>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 14.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paez-Borda A, Lujan-Galan M, Martin-Oses E, de la Cal M A, Garcia M, Martin-Benito J, Barba C, Bergenguer-Sanchez A. Acta Urol Esp. 1999;23:518–522. [PubMed] [Google Scholar]
- 16.Anim J T, Udo C, John B. Acta Histochem. 1998;100:439–449. doi: 10.1016/S0065-1281(98)80040-8. [DOI] [PubMed] [Google Scholar]
- 17.Nickel J C, Downey J, Young I, Boag S. Br J Urol. 2000;84:976–981. doi: 10.1046/j.1464-410x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- 18.Elsasser-Beile U, Przytulski B, Gierschner D, Grussenmeyer T, Katzenwadel A, Leiber C, Deckart A, Wetterauer U. Prostate. 2000;45:1–7. doi: 10.1002/1097-0045(20000915)45:1<1::aid-pros1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 19.Mahapokai W, van den Ingh T S, van Mil F, van Garderen E, Schalken J A, Mol J A, van Sluijs F J. Vet Immunol Immunopathol. 2001;78:297–303. doi: 10.1016/s0165-2427(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 20.Malkowicz S B, McKenna W G, Vaughn D J, Wan X S, Propert K J, Rockwell K, Marks S H, Wein A J, Kennedy A R. Prostate. 2001;48:16–28. doi: 10.1002/pros.1077. [DOI] [PubMed] [Google Scholar]
- 21.Seto M H, Liu H L, Zajchowski D A, Whitlow M. Proteins. 1999;35:235–249. [PubMed] [Google Scholar]
- 22.Isomura M, Okui K, Fujiwara T, Shin S, Nakamura Y. Cytogenet Cell Genet. 1996;74:263–265. doi: 10.1159/000134431. [DOI] [PubMed] [Google Scholar]
- 23.Nollet F, Kools P, van Roy F. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 24.Jacob K, Webber M, Benayahu D, Kleinman H K. Cancer Res. 1999;59:4453–4457. [PubMed] [Google Scholar]
- 25.Thomas R, True L D, Bassuk J A, Lange P H, Vessella R L. Clin Cancer Res. 2000;6:1140–1149. [PubMed] [Google Scholar]
- 26.Bull J H, Ellison G, Patel A, Muir G, Walker M, Underwood M, Khan F, Paskins L. Br J Cancer. 2001;84:1512–1519. doi: 10.1054/bjoc.2001.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkman U, Vasmatzis G, Lee B, Pastan I. Cancer Res. 1999;59:1445–1448. [PubMed] [Google Scholar]
- 28.Brinkman U, Vasmatzis G, Lee B, Yerushalmi N, Essand M, Pastan I. Proc Natl Acad Sci USA. 1998;95:10757–10762. doi: 10.1073/pnas.95.18.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]