Abstract
Galactose-inducible genes (GAL genes) in yeast Saccharomyces cerevisiae are efficiently transcribed only when the sequence-specific transcription activator Gal4p is activated. Activation of Gal4p requires the interaction between the Gal4p inhibitory protein Gal80p and the galactokinase paralog, Gal3p. It has been proposed that Gal3p binds to a Gal80p-Gal4p complex in the nucleus to activate Gal4p. Here, we present evidence that the Gal3p–Gal80p interaction occurs in the cytoplasm, and concurrently, Gal80p is removed from Gal4p at the GAL gene promoter. We also show that GAL gene expression can be activated by heterologous protein–protein interaction in the cytoplasm that is independent of galactose and Gal3p function. These results indicate that galactose-triggered Gal3p-Gal80p association in the cytoplasm activates Gal4p in the nucleus.
Cells use multiple layers of controls to direct spatial and temporal patterns of gene expression. Studies of the genetic switches that enable cells to activate and repress expression of specific groups of genes in response to environmental cues have enriched our understanding of the intricate cellular processes that regulate gene transcription. One well characterized genetic switch is the GAL switch that controls synthesis of galactose-metabolizing enzymes in the yeasts Saccharomyces cerevisiae (1, 2) and Kluyveromyces lactis (3).
The GAL gene regulatory circuit is composed of a transcription activator Gal4p, an inhibitory protein Gal80p, and a signal transducer protein Gal3p. Gal4p recognizes and binds to specific upstream activation sequences of the GAL structural genes (UASGAL) through its N-terminal DNA-binding domain. In the absence of galactose, the activity of Gal4p is inhibited by the binding of Gal80p to the C-terminal transcription activation domain of Gal4p (amino acids 768–881). The relief of Gal80p's inhibition of Gal4p in the presence of galactose depends on the function of Gal3p. Gal3p is a paralog of Gal1p (4), the galactokinase and the first enzyme of the galactose utilization pathway, but Gal3p does not have galactokinase activity (5, 6). Any one of several mutations dispersed in the Gal3p sequence (GAL3 constitutive mutant) causes the activation of GAL genes in the absence of galactose (7). Biochemical studies have shown that Gal3p directly interacts with Gal80p in a galactose- and ATP-dependent manner (3, 8), and that the Gal3p constitutive mutants interact with Gal80p in the absence of galactose (7). How the formation of the Gal3p-Gal80p complex transmits the galactose signal to the Gal80p-Gal4p complex in the nucleus to activate Gal4p has been unclear. A prevailing view has been that Gal3p interacts with the Gal80p-Gal4p complex in the nucleus and changes the conformation of the Gal80p-Gal4p complex to a form that is competent to activate transcription (9, 10). In previous work using two different methods, we were unable to detect Gal3p in the nucleus. In contrast, Gal80p was shown to be present in both the cytoplasm and the nucleus. Those results raised the possibility that the effective interaction between Gal3p and Gal80p may occur in the cytoplasm rather than the nucleus to modulate Gal80p–Gal4p interaction (11).
In this report, we present evidence that Gal3p can exert its induction function solely in the cytoplasm when it is tethered to the membranes. By using a chromatin immunoprecipitation analysis, we demonstrate that concurrent with the galactose signaling, Gal80p is removed from Gal4p at the GAL gene promoter. We also show that GAL gene expression can be activated by creating a binding site for Gal80p in the cytoplasm. These results indicate that galactose-triggered Gal3p-Gal80p association in the cytoplasm activates Gal4p in the nucleus, suggesting that the galactose signaling mechanisms are more dynamic than previously envisioned.
Materials and Methods
Plasmids and Yeast Strains.
Gal3p derivatives bearing the protein N-myristoylation signal (MGCTVSTQTIGDESDP, taken from the N-terminal domain of Gpa1p, the G protein α-subunit), an N-myristoylation signal variant (MACTVSTQTIGDESDP, the single Gly→Ala substitution abolishes the acylation and membrane targeting), or the mitochondria outer membrane signal anchor sequence (the first 29 amino acid residues of the Tom70p, a component of the translocase of the outer mitochondrial membrane complex) were generated by inserting oligonucleotides encoding each signal peptide sequence into GAL3 gene at the N terminus. It has been shown that the N-myristoylation signal used here is sufficient to target a heterologous protein to the plasma membrane (12). The first 29 amino acids of Tom70 protein is sufficient to target a heterologous protein to the outer membrane of mitochondria, leaving the bulk of protein extruding into the cytoplasm (13). The ADH2 promoter, FPR1, CNA1, HOM3, and CMD1 sequences were PCR-amplified by using yeast Sc723 genomic DNA as the template. The green fluorescent protein (GFP) and cyan fluorescence protein sequences were PCR-amplified by using pYGFP1 (14) and pDH3 (Yeast Resource Center, Univ. of Washington, Seattle) as template, respectively. Details of plasmid constructions and sequence information are available upon request (or at http://www.hmc.psu.edu/jhopper/plasmids-info.html).
All strains used in the study were derived from SJ21R (15), as described (7).
Microscopy.
Fluorescence was observed with a Nikon Optiphot-2 epifluorescence microscope equipped with a 100× objective. GFP was excited, and its emission fluorescence was detected by using Chroma filter no. 41017 (Chroma Technology, Brattleboro, VT). Images were acquired by using a SenSys KF1400 charge-coupled device camera (Photometrics, Tucson, AZ) controlled by QED software (Pittsburgh). The final figures were prepared by using Adobe PHOTOSHOP and Deneba CANVAS.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitations were performed as described (16, 17). Briefly, wild-type Sc723 cells (7) were grown to early exponential growth phase (absorbance at 600 nm ≈0.4) in 250 ml of yeast extract-peptone medium containing 2% glycerol, 3% lactic acid, and 0.05% glucose (-galactose, uninduced). For galactose-induced cultures, galactose was added to the uninduced cultures to a final concentration of 2%, and the cultures were incubated for an additional 20 min. Formaldehyde was added to a final concentration of 1%, the cultures were incubated for 20 min at room temperature, and glycine was added to a final concentration of 300 mM. All of the following steps were carried out at 4°C. Cells were washed twice with TBS (20 mM Tris⋅HCl/150 mM NaCl, pH 8.0), resuspended in 1 ml of lysis buffer (50 mM Tris⋅HCl, pH 8.0/10 mM EDTA/1% SDS/1 mM PMSF) and disrupted by vortexing with glass beads. The cross-linked chromatin was pelleted by centrifugation at 200,000 × g for 12 min, resuspended in 1 ml of lysis buffer, and sonicated to yield DNA fragments of 300 bps average size. Soluble chromatin was separated from insoluble materials by centrifugation at 13,000 × g for 15 min. Finally, the volume of soluble chromatin was adjusted to 10 ml with dilution buffer (16.7 mM Tris⋅HCl, pH 8.0/16.7 mM NaCl/1.2 mM EDTA/0.01% SDS/1.1% Triton X-100/1 mM PMSF); the resultant chromatin solution was stored in 1 ml aliquots at −80°C. For immunoprecipitation, 5 μl of rabbit anti-Gal4p serum, 5 μl of anti-Gal80p serum, or 5 μl of anti-Gal3p serum was added to 1 ml of chromatin solution. After overnight incubation on a rotator, 75 μl of protein A Sepharose beads were added, and the reactions were incubated for 2 h more. Precipitated protein-DNA complexes were eluted from the beads, cross-linked reversed, treated with proteinase K, and analyzed by quantitative radioactive PCR by using primer pairs GANG67/68 for the UASGAL region and GANG71/72 for an intergenic control region located 5 kb downstream from the GAL1 gene. The sequences of the primers are GANG67: CATGGCATTACCACCATATACATATCC; GANG68: GAAGGTTTGTGGGGCCAGGTTACTGC; GANG71: GTGCATTTGGCCTTCAATGAGC; GANG72: AAGTGATGTTCGACATACCTGTAAC. The template used for the various reactions was 1/30,000 of input DNA, 1/200 of anti-Gal4p antibody precipitated DNA, 1/50 of anti-Gal80p antibody precipitated DNA, and 1/25 of anti-Gal3p antibody-precipitated DNA. PCR conditions were as follows: 0.5 μM each primer, 0.25 mM each dNTP, 1.5 mM MgCl2, 0.06 mCi/ml [α-32P]dATP in a 20-μl reaction volume. The PCR cycle was as follows: 3 min at 94°C, followed by 25 cycles of 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C. PCR products were resolved on 8% polyacrylamide gels in 1× TBE buffer (89 mM Tris-base/89 mM boric acid/2mM EDTA). Quantitation of incorporated [α-32P]dATP in the PCR products was performed by using a PhosphorImager (Molecular Dynamics) analysis.
Results
Membrane-Associated Gal3p Retains Its Induction Function and Capacity.
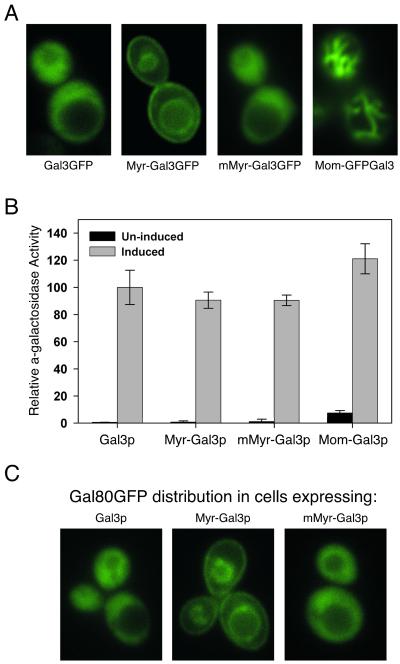
We have sequestered Gal3p in the cytoplasm precluding nuclear import of Gal3p and determined the effect of this sequestration on galactose induction of the GAL genes. Two different methods were used to anchor Gal3p in the cytoplasm. In the first method, we used a protein N-myristoylation signal to target Gal3p (Myr-Gal3p) to the cell plasma membrane and intracellular vesicle membranes. In the second method, Gal3p was targeted to the outer membrane of the mitochondria (Mom-Gal3p) by a signal anchor sequence. N-myristoylation (18, 19) was chosen because it is a cotranslational process that occurs when the nascent peptide is still attached to the ribosome. Thus, Myr-Gal3p was acylated and associated with the membrane before it was able to interact with Gal80p. Mitochondria targeting was chosen because it seems to be a very fast and efficient process (20, 21). After addition of these respective targeting signals, Myr-Gal3p was localized to the plasma membrane, and Mom-Gal3p was localized to the mitochondria. In contrast, wild-type Gal3p was shown to be located throughout the cytoplasm, as was Gal3p with a variant N-myristoylation signal having a single amino acid substitution that cannot be acylated (Fig. 1A).
Figure 1.
Sequestration of Gal3p to cellular membranes does not affect its induction function. (A) Localization of the Gal3p derivatives fused to GFP. Cells of yeast strain Sc781 (gal3Δ gal1Δ) carrying the indicated low-copy expression plasmid were grown to mid-log phase in medium containing 2% glycerol and 3% lactic acid as carbon source and induced with 2% galactose for 3 h before observation. GFP was inserted at the C termini of wild-type Gal3p, Myr-Gal3p, and mMyr-Gal3p. The vacuolar (yeast lysosomal) membrane also is readily seen in cells expressing Myr-Gal3GFP. For mitochondria-associated Gal3p, the GFP sequence is between the signal anchor sequence and the Gal3p sequence. Yeast mitochondria form the typical reticulum structure under the experiment conditions used. (B) MEL1 induction in cells carrying Gal3p derivatives. The results obtained after 6 h of galactose induction were normalized to those of cells carrying the wild-type Gal3p. The α-galactosidase induction assay was carried out as described (11). (C) Membrane-associated Gal3p sequesters Gal80p to membranes. Cells of yeast Sc786 strain (gal80Δ gal3Δ gal1Δ) carrying a low-copy plasmid expressing Gal80GFP together with wild-type Gal3p, or Myr-Gal3p, or mMyr-Gal3p, as indicated, were observed as in A.
We examined gene expression from two types of GAL gene promoters in cells carrying the membrane-bound and cytoplasm-sequestered Gal3p. We first used a sensitive and semiquantitative colony growth assay to assess the expression of an HIS3 reporter gene whose promoter bears four UASGAL sites. Cells of a gal3Δgal1Δ strain (Sc781) carrying Myr-Gal3p or Mom-Gal3p grew indistinguishably from cells harboring wild-type Gal3p on synthetic medium lacking His in our assays. Next, we quantified the expression of MEL1, a galactose responsive gene whose promoter bears a single UASGAL site. Despite the effective sequestration of Gal3p to the membrane, the MEL1 gene was expressed to wild-type levels upon galactose induction (Fig. 1B). The level of MEL1-encoded α-galactosidase in the extracts of cells carrying Myr-Gal3p was not significantly different from that detected in the extracts of cells carrying wild-type Gal3p (P > 0.1, Student's t test) or carrying a mMyr-Gal3p variant that abolished plasma association (P > 0.1). The mitochondria-associated Gal3p (Mom-Gal3p) gave rise to higher MEL1 expression than the wild-type Gal3p did (P < 0.05). We determined by Western analyses that membrane association of Gal3p did not increase steady-state levels of Gal3p. The cellular levels of Myr-Gal3p and the mMyr-Gal3p variant were slightly lower than that of wild-type Gal3p, whereas the Mom-Gal3p level was similar to that of wild-type Gal3p (data not shown).
Several lines of evidence indicated that the full induction of GAL genes observed in cells harboring membrane-bound Gal3p was due to the interaction between Gal3p and Gal80p at the cytoplasmic membranes. First, the targeting methods we used are very effective in sequestering Gal3p to the membranes. Based on cell fractionation and Western analyses, essentially all of Gal3p was associated with the membrane after the addition of the myristoylation signal. For the mitochondria-targeted Gal3p, we estimated that less than 10% of Mom-Gal3p was found in the soluble cytosolic fraction (data not shown). Second, Gal80p redistributed to the plasma membrane and the intracellular vesicle membrane in cells expressing Myr-Gal3p (Fig. 1C). Last, a small fraction of Gal3p that might have escaped the cytoplasmic sequestering methods used here would not have been sufficient to give rise to the levels of GAL gene expression observed in our experiments (Fig. 2). We constructed three plasmids providing different subcellular locations and expression levels of Gal3p, and we determined the expression levels of the MEL1 gene in cells carrying these plasmids. The results showed that a fivefold reduction of Gal3p resulted in a twofold reduction in MEL1 gene expression level, indicating that Gal3p is not in large excess in the cell and that quantitative interactions between Gal3p and Gal80p are required for the transcription of GAL genes. In comparison, a twofold reduction of Gal3p derivative in cells carrying Myr-Gal3p resulted in a 20% reduction in MEL1 gene expression level. Thus, membrane-associated Gal3p maintains not only Gal3p's induction function but also its induction capacity. Considering that GAL gene expression quantitatively relies upon the Gal3p–Gal80p interaction, we conclude that the active transcription of GAL genes in cells harboring Myr-Gal3p is due to the Gal3p–Gal80p interaction at the cytoplasmic membranes rather than to Gal3p molecules in nucleus.
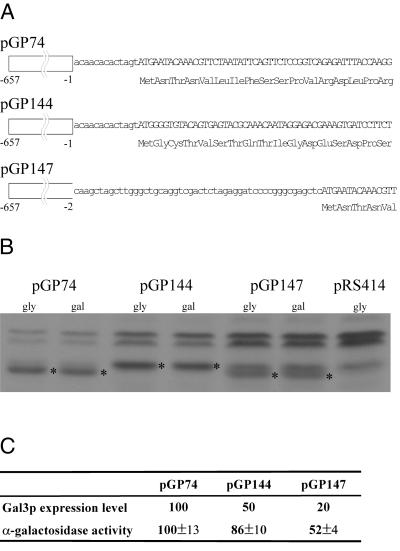
Figure 2.
Lowering the Gal3p expression level causes reduction in GAL gene expression. (A) Sequence differences between different Gal3p-encoding plasmids. pGP74 encodes a wild-type Gal3p expressed from an ADH2 promoter. pGP144 encodes the myristoylated Gal3p (Myr-Gal3p) expressed from an identical ADH2 promoter. pGP147 encodes a wild-type Gal3p expressed from a modified ADH2 promoter containing a 50-bp sequence insertion between the promoter sequence and the start codon of Gal3p. Numbers in the promoter region correspond to the distance from the start codon in the ADH2 gene. (B) Different Gal3p expression levels conferred by different Gal3p-encoding plasmids. Protein extracts from cells grown in the absence of galactose and from cells grown for 6 h in the presence of galactose were analyzed by Western blot. For the data shown here, the amount of protein extract analyzed was 16 μg, 40 μg, and 80 μg for cells carrying pGP74, pGP144, and pGP147 plasmids, respectively. The level of Gal3p expressed from pGP74 is similar to that of Gal3p expressed from its endogenous promoter under the galactose-induced condition. *, denotes detected Gal3p signal. The nonspecific bands detected by anti-Gal3p antiserum in this blot are indicated in the lane for cells carrying pRS414 (100 μg protein extract), the vector backbone for all three Gal3p-encoding plasmids used here. (C) MEL1 induction for cells carrying pGP74, pGP144, and pGP147. Cells carrying respective plasmids were induced with 0.5% galactose for 6 h, and cellular extracts were prepared. Aliquots of the cellular extracts were subjected to Western analyses as in B to provide the estimations for the Gal3p-expression levels. The α-galactosidase induction assay was carried out as described (11). Both Gal3p expression level and MEL1 induction results were normalized to those of cells carrying pGP74.
Gal80p Association with GAL Gene Promoter Is Reduced on Galactose Signaling.
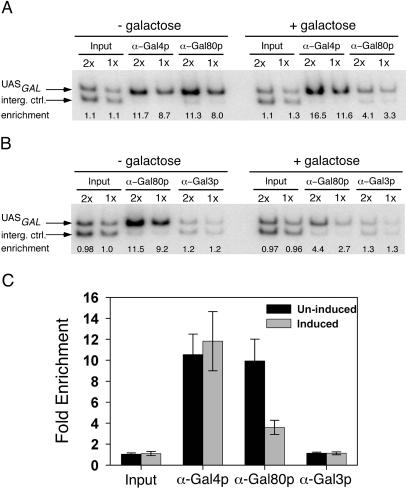
The results above indicate that Gal4p can be activated by the interaction between Gal80p and Gal3p solely in the cytoplasm. Because the association of Gal80p with Gal4p on the promoter of GAL gene is critical to the GAL gene transcriptional regulation mechanism, we examined whether the amount of Gal80p complexed with DNA-bound Gal4p is changed after galactose triggers the Gal3p–Gal80p interaction. We performed a formaldehyde-based in vivo cross-linking followed by chromatin immunoprecipitation to determine the extent of the association of Gal4p and Gal80p with the UASGAL region of the GAL1/GAL10 gene promoter. Because Gal80p is associated with the UASGAL through its binding to Gal4p only, comparison of the amount of the UASGAL DNA associated with Gal4p and Gal80p provided an assessment of the Gal4p–Gal80p interaction. We found that whereas Gal4p occupancy of the UASGAL site slightly increased upon galactose signaling, the Gal80p association with the UASGAL region decreased significantly. Repeated trials did not yield any evidence that Gal3p is associated with the GAL1/10 gene promoter (Fig. 3). These data indicate that concurrent with the galactose-induced interaction between Gal3p and Gal80p, the association between Gal80p and UASGAL-bound Gal4p decreases.
Figure 3.
Analysis of the association of Gal4p, Gal80p, and Gal3p with the UASGAL by chromatin immunoprecipitation. Yeast cells were grown to early-log phase in medium containing 2% glycerol, 3% lactic acid, and 0.05% glucose (-galactose, uninduced) and induced with 2% galactose for 20 min (+galactose, induced). The 344-bp PCR product (UASGAL) corresponds to the region extending from −600 to −257 relative to the +1 of the GAL1 gene. The 287-bp PCR product (interg. ctrl.) corresponds to an intergenic region located 5 kb downstream from the GAL1 promoter; it served as a control for background binding in the immunoprecipitation. The ratio between the 344-bp and 287-bp PCR products (fold enrichment) is calculated to normalize results and assess the promoter occupancy. (A) Gal4p and Gal80p occupancy at UASGAL sites in uninduced and induced cells. The results from twofold dilution of the template DNA used for the PCR reactions demonstrated that the reaction was within the linear range. Fold enrichments are indicated (enrichment). (B) Gal80p and Gal3p association with the UASGAL region evaluted before and after galactose induction. (C) Quantitation of the promoter occupancy by Gal4p, Gal80p, and Gal3p in uninduced and induced cells. Values are means (±SD) of at least five experiments carried out with two sets of independently prepared chromatin samples.
GAL Gene Expression Activated by Surrogate Protein–Protein Interactions in the Cytoplasm.
Considerable evidence has indicated that nuclear transport processes play important roles in transcriptional regulation (22). Because Gal80p appears to undergo rapid nucleo-cytoplasmic shuttling in vivo (11), it is likely that the galactose-triggered Gal3p–Gal80p interaction traps Gal80p in the cytoplasm, thereby causing the observed removal of Gal80p from the promoters of GAL genes and the activation of Gal4p. We note that the behavior of the Gal80p–Gal3p interaction in vivo is consistent with this view, as the membrane bound Myr-Gal3p is shown to sequester Gal80p to the membranes where Myr-Gal3p is located (Fig. 1C).
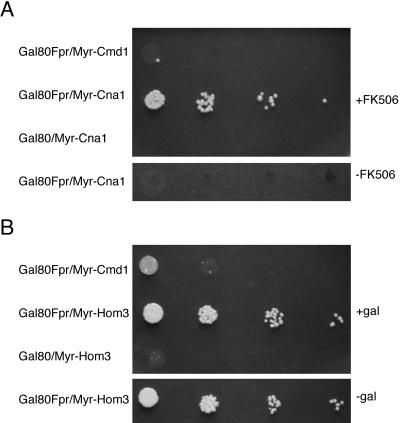
According to the scenario above, we might expect that substituting Gal3p with a surrogate-binding factor for Gal80p in the cytoplasm would activate GAL gene expression. To test this possibility, we took advantage of the interaction between Fpr1p and Cna1p that is elicited by a small membrane-permeable molecule FK506, a macrolide isolated from Streptomyces tsukubaensis (23–26). FK506 was used to trigger the interaction between a Gal80-Fpr1p fusion and plasma membrane-localized Myr-Cna1p. It has been shown that FK506 by itself does not activate transcription of the GAL genes (27). We assayed for the induction of the galactose-inducible HIS3 reporter gene. Cells of a gal80Δ gal3Δ gal1Δ strain (ScGP786) carrying both Gal80-Fpr1 and Myr-Cna1p plasmids exhibited evident growth only on the plate containing FK506 (Fig. 4A). It has been reported that Fpr1p also interacts with Hom3p, the aspartokinase, in the absence of FK506 in a yeast two-hybrid assay (28). Following a scheme that paralleled the one above, we found that the heterologous interaction between Gal80–Fpr1p and Myr–Hom3p was able to activate the HIS3 reporter gene expression (Fig. 4B). We also determined by Western analyses that the Gal80-Fpr1p fusion was present at a slightly higher level in cells containing both Gal80-Fpr1p and Myr-Hom3p, as compared with cells harboring the noninteracting controls, Gal80-Fpr1p and Myr-Cmd1p, indicating that activation of the HIS3 reporter gene was not caused by the degradation of Gal80-Fpr1p in cells expressing Myr-Hom3p. Because the surrogate Gal80p-binding factors are expected to only alter the subcellular distribution of Gal80p, these demonstrations of Gal3p- and galactose-independent activation of a galactose-inducible gene indicate that Gal4p activity is responsive to the reduced levels of Gal80p in the nucleus. Given the activation of Gal4p shown here, we suggest a Gal80p-trapping method might be useful as a genetic method to detect protein–protein interactions in the cytoplasm.
Figure 4.
Surrogate protein–protein interaction activates GAL gene expression. (A) FK506-dependent HIS3 reporter expression. Gal80-Fpr1 designates the Gal80-Fpr1p fusion. Myr-Cna1 designates the plasma membrane-targeted Cna1p. Myr-Cmd1 is a plasma membrane-targeted calmodulin and is used as a control. The nutrient agar plate used here contained 0.5% galactose, 3% glycerol, and 2% lactic acid as a carbon source. FK506 was used at 0.25 μg/ml where indicated. Identical results were obtained by using nutrient agar lacking galactose (data not shown). (B) Galactose-independent HIS3 reporter expression as a result of Gal80–Fpr1p and Myr–Hom3p interactions. The nutrient agar plate used here contained 3% glycerol and 2% lactic acid as carbon source. Galactose was used at 0.5% where indicated.
Discussion
Previous studies have shown that direct interaction between Gal3p and Gal80p plays a pivotal role in activating Gal4p-mediated GAL gene transcription upon galactose induction (3, 8). More recently, we provided evidence implicating the subcellular distribution dynamics of Gal3p and Gal80p in the mechanisms controlling Gal4p activity (11). The results presented in this report indicate that the tightly controlled and highly efficient galactose-responsive mechanism in yeast is achieved by modulating protein–protein interactions located in two different cellular compartments. Our results are consistent with a mechanism in which galactose triggers Gal3p-Gal80p complex formation in the cytoplasm to reduce the level of interaction of Gal80p with Gal4p in the nucleus (Fig. 5). This mechanism for activating Gal4p is not accounted for by the current prevailing model for GAL induction, which specifies that Gal3p interacts with the Gal80p-Gal4p complex in the nucleus and causes a postulated conformation change in the Gal80p and Gal4p complex (9, 10).
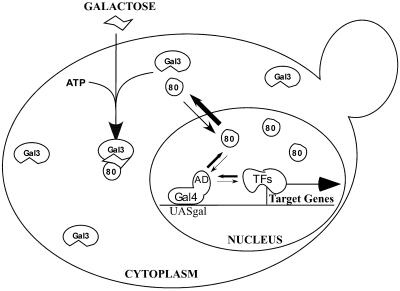
Figure 5.
Model for transcriptional regulation of GAL genes through modulation of protein–protein interactions in the nucleus and in the cytoplasm. Gal3p (Gal3) is located in the cytoplasm, whereas Gal80p (80) is located in both the cytoplasm and the nucleus and undergoes rapid nuclear-cytoplasmic shuttling. In the nucleus, a number of transcription factors (TFs) compete with Gal80p to bind to the activation domain (AD) of Gal4p (36–40). In the absence of galactose, binding of Gal80p to Gal4p effectively limits active transcription of the GAL genes. Cellular uptake of galactose triggers Gal3p–Gal80p interaction in the cytoplasm and results in redistribution of Gal80p from the nucleus to the cytoplasm. These events lead to a reduced probability of binding between Gal80p and Gal4p. Subsequently, recruitment of chromatin remodeling factors and PolII holoenzyme activate GAL gene transcription.
Gal3p Exerts Its Induction Function Solely in the Cytoplasm.
Here, we have shown that sequestration of Gal3p to membranes outside the nucleus does not impair galactose-triggered Gal4p-mediated GAL gene expression. For both plasma membrane-tethered and mitochondria outer membrane-inserted Gal3p, little, if any, Gal3p could enter the nucleus, yet GAL induction remained essentially the same as for wild-type Gal3p. These results would be difficult to explain by the existence of a small and undetectable fraction of Gal3p that escaped both types of membrane tethering in light of our results showing that GAL gene expression quantitatively depends upon the interaction between Gal3p and Gal80p.
The fact that membrane-bound Gal3p retains full induction capacity indicates that it is not necessary for Gal3p to enter the nucleus and act directly on the Gal80p-Gal4p complex to effect GAL gene expression. Consistently, we have been unable to detect an association of Gal3p with the UASGAL sites within the GAL gene promoters by a chromatin immunoprecipitation approach. It seems unlikely that the failure to detect this association is due to masking of Gal3p in the complex, as we have successfully detected the association of Gal4p and Gal80p with the GAL gene promoters in the same experiment. It also might be argued that Gal3p only needs to transiently associate with the Gal80p-Gal4p complex to induce the conformation change of the complex. However, our results showing that Gal80p redistributes to the membrane in cells containing Myr-Gal3p suggest that the in vivo Gal3p-Gal80p complex is relatively stable in the presence of galactose.
GAL Gene Activation Correlates with Decreased Binding Between Gal80p and Gal4p.
We have observed a significant decrease in Gal80p's association with the GAL gene promoter upon galactose signaling and GAL gene activation. The formal possibility that the reduced association observed was a result of lower cross-linking efficiency between Gal80p and the promoter-bound Gal4p in response to a putative conformation change of the complex does not seem very likely. Formaldehyde is a very reactive bipolar compound that efficiently cross-links amino and imino groups of amino acids and DNA, and, theoretically, only one condensation event is required to cross-link two entities in a complex. We suggest that the decreased binding is caused by either a reduced affinity between Gal80p and Gal4p or a lower level of Gal80p in the nucleus or both.
We also demonstrated that Gal4p activity is responsive to reduced levels of Gal80p in the nucleus by substituting Gal3p with a surrogate cytoplasmic Gal80p-binding factor. Based on those results, we suggest that Gal4p activation is due to a lower level of Gal80p in the nucleus as the interaction between Gal3p increases the dwell time of Gal80p in the cytoplasm. It should be pointed out, however, that the activation of the GAL genes by our engineered heterologous interactions is not a strong response compared with the response conferred by the galactose-triggered Gal3p–Gal80p interaction. There may be a number of reasons for the weaker response. The surrogate systems we used may not function as efficiently as the endogenous system because of differences in the stability of the surrogate protein–protein complex compared with the Gal80p–Gal3p complex. Also, the cellular uptake of the signal molecules (FK506 vs. galactose) could be different. Because there is evidence suggesting that the Gal80p–Gal3p interaction is capable of perturbing the Gal80p–Gal4p binding reaction (29), it is also possible that the surrogate systems do not fully recapitulate the elements of Gal3p induction function.
Implications of the Subcellular Distribution Dynamics of the GAL Gene Regulatory Factors.
It is now evident that cells exploit and integrate multiple layers of controls to achieve transcription regulation (30). Considerable evidence has implicated subcellular distributions of regulatory factors as intrinsic features in the signaling and transcriptional control pathways (22). The study we have presented here suggests that the activity of the transcription factor Gal4p is altered by a signal-dependent event initiated in the cytoplasm. This event could provide the cell with a mechanism to sense the local nutritional environment of the galactose-metabolizing pathway enzymes localized in the cytoplasm. Such a mechanism also might be used in other signaling systems regulating cytosolic pathways, as it may afford the cell the advantage of increased sensitivity of its response when the signal is limiting or fluctuating in the environment.
It has been recognized from several studies that controlling the subcellular distribution of transcription factors is not sufficient to regulate transcription activity (31, 32). Perhaps, this is because translocation of regulatory factors is largely a diffusion-driven process in most cell types (33–35). Thus, translocation alone may not be a sufficient mechanism to provide the properties demanded by many signal-responsive transcription switches. In future studies, it will be of interest to determine to what extent the translocation of Gal80p contributes to the regulation of GAL gene expression, as the results should advance our understanding of how translocation dynamics of the regulatory factors contributes to the transcriptional control mechanisms.
Our findings highlight the need to take into account the subcellular localization profiles and distribution dynamics of the Gal3, Gal80, and Gal4 proteins if we are to achieve a comprehensive understanding of the GAL regulatory system. We have determined by epitope tagging (both Gal3p and Gal80p were tagged with HA or GFP) and Western blotting that Gal3p is about fivefold more abundant than Gal80p in the cell, and that this ratio between Gal3p and Gal80p seems not to change upon galactose induction (data not shown). This ratio and the molar ratio of Gal80p to Gal4p must be viewed as integral parameters determining the mechanism controlling GAL gene expression, as the activation response of the GAL genes is expected to be conditioned by the linked equilibria between the Gal3p–Gal80p and the Gal80p–Gal4p binding reactions. Further investigations will address whether the Gal4p–Gal80p interaction dynamics in vivo is different from what has been measured in vitro, whether nucleocytoplasmic shuttling of Gal80p is a critical feature in the function of the GAL genetic switch, and whether the mechanism we elucidated here is applicable to other systems.
Acknowledgments
We thank S. D. Emr, C. Hill, A. K. Hopper, and D. R. Stanford for valuable discussions, P. Meluh for help with chromatin immunoprecipitation experiments, and Fujisawa Pharmaceutical for a generous supply of FK506. This work was supported by a National Institutes of Health grant (to J.E.H.).
Abbreviations
- UASGAL
upstream activation sequences of the GAL structural genes
- GFP
green fluorescence protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Oshima Y. In: The Molecular Biology of the Yeast Saccharomyces cerevisiae: Metabolism and Gene Expression. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 159–180. [Google Scholar]
- 2.Johnston M, Carlson M. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Jones E W, Pringle J R, Broach J R, editors. II. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 193–281. [Google Scholar]
- 3.Zenke F T, Engles R, Vollenbroich V, Meyer J, Hollenberg C P, Breunig K D. Science. 1996;272:1662–1665. doi: 10.1126/science.272.5268.1662. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe K H, Shields D C. Nature (London) 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 5.Bhat P J, Oh D, Hopper J E. Genetics. 1990;125:281–291. doi: 10.1093/genetics/125.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platt A, Ross H C, Hankin S, Reece R J. Proc Natl Acad Sci USA. 2000;97:3154–3159. doi: 10.1073/pnas.97.7.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blank T E, Woods M P, Lebo C M, Xin P, Hopper J E. Mol Cell Biol. 1997;17:2566–2575. doi: 10.1128/mcb.17.5.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki-Fujimoto T, Fukuma M, Yano K I, Sakurai H, Vonika A, Johnston S A, Fukasawa T. Mol Cell Biol. 1996;16:2504–2508. doi: 10.1128/mcb.16.5.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leuther K K, Johnston S A. Science. 1992;256:1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- 10.Platt A, Reece R J. EMBO J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng G, Hopper J E. Mol Cell Biol. 2000;20:5140–5148. doi: 10.1128/mcb.20.14.5140-5148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillen K M, Pausch M, Dohlman H G. J Cell Sci. 1998;111:3235–3244. doi: 10.1242/jcs.111.21.3235. [DOI] [PubMed] [Google Scholar]
- 13.McBride H M, Millar D G, Li J M, Shore G C. J Cell Biol. 1992;119:1451–1457. doi: 10.1083/jcb.119.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 15.Johnston S A, Hopper J E. Proc Natl Acad Sci USA. 1982;79:6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 17.Kuras L, Struhl K. Nature (London) 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 18.Johnson D R, Bhatnagar R S, Knoll L J, Gordon J I. Annu Rev Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- 19.Resh M D. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 20.Fujiki M, Verner K. J Biol Chem. 1993;268:1914–1920. [PubMed] [Google Scholar]
- 21.Funfschilling U, Rospert S. Mol Biol Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komeili A, O'Shea E K. Curr Opin Cell Biol. 2000;12:355–360. doi: 10.1016/s0955-0674(00)00100-9. [DOI] [PubMed] [Google Scholar]
- 23.Kino T, Hatanaka H, Hashimoto M, Nishiyama M, Goto T, Okuhara M, Kohsaka M, Aoki H, Imanaka H. J Antibiot. 1987;40:1249–1255. doi: 10.7164/antibiotics.40.1249. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Farmer J D, Jr, Lane W S, Friedman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 25.Cardenas M E, Muir R S, Breuder T, Heitman J. EMBO J. 1995;14:2772–2783. doi: 10.1002/j.1460-2075.1995.tb07277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffith J P, Kim J L, Kim E E, Sintchak M D, Thomson J A, Fitzgibbon M J, Fleming M A, Caron P R, Hsiao K, Navia M A. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 27.Biggar S R, Crabtree G R. EMBO J. 2001;20:3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alarcon C M, Heitman J. Mol Cell Biol. 1997;17:5968–5975. doi: 10.1128/mcb.17.10.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sil A K, Alam S, Xin P, Ma L, Morgan M, Lebo C M, Woods M P, Hopper J E. Mol Cell Biol. 1999;19:7828–7840. doi: 10.1128/mcb.19.11.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemon B, Tjian R. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, McKeon F. Nature (London) 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 32.Komeili A, O'Shea E K. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 33.Teruel M N, Meyer T. Cell. 2000;103:181–184. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 34.Ribbeck K, Gorlich D. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rout M P, Aitchison J D, Suprapto A, Hjertaas K, Zhao Y, Chait B T. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melcher K, Johnston S A. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirst M, Kobor M S, Kuriakose N, Greenblatt J, Sadowski I. Mol Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 38.Brown C E, Howe L, Sousa K, Alley S C, Carrozza M J, Tan S, Workman J L. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 39.Bhaumik S R, Green M R. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larschan E, Winston F. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]