Abstract
I-TevI, the phage T4 td intron-encoded endonuclease, recognizes a lengthy DNA target and initiates intron mobility by introducing a double-strand break in the homing site. The enzyme uses both sequence and distance determinants to cleave the DNA 23–25 bp upstream of the intron insertion site. I-TevI consists of an N-terminal catalytic domain and a C-terminal DNA-binding domain separated by a long, flexible linker. The DNA-binding domain consists of three subdomains: a zinc finger, a minor-groove binding α-helix, and a helix–turn–helix. In this study, a mutational analysis was undertaken to assess the roles of these subdomains in substrate binding and cleavage. Surprisingly, the zinc finger is not required for DNA binding or catalysis. Rather, the zinc finger is a component of the linker and directs the catalytic domain to cleave the homing site at a fixed distance from the intron insertion site. When the cleavage site (CS) is shifted outside a given range, wild-type I-TevI defaults to the fixed distance, whereas zinc-finger mutants have lost the distance determinant and search out the displaced cleavage sequences. Although counterintuitive, a protein containing a 19-aa deletion of the zinc finger can extend further than can wild-type I-TevI to cleave a distant CS sequence, and a Cys-to-Ala mutant of the ligands for zinc, nominally a longer protein, can retract to cleave at a closer CS sequence. Models are presented for the novel function of the zinc finger, as a molecular constraint, whereby intramolecular protein–protein interactions position the catalytic domain by “catalytic clamp” and/or “linker-organizer” mechanisms.
Homing endonucleases, associated with introns and inteins in all three biological kingdoms, promote the insertion (homing) of their genetic elements into unoccupied allelic sites (1–6). These enzymes are grouped into four families based on conserved sequence elements: GIY-YIG, LAGLIDADG, H-N-H, and His-Cys (2, 7). I-TevI is a 28-kDa homing endonuclease of the GIY-YIG family, encoded by the td intron of phage T4. The enzyme is remarkable in that it is site specific, yet recognizes a target site of ≈37 bp in a sequence-tolerant fashion (8). Its primary binding site is centered on the intron insertion site (IS), yet the cleavage site (CS) is 23–25 bp upstream from the IS. I-TevI binds its DNA substrate as a monomer, making extensive minor groove and phosphate backbone contacts, and inducing a distortion at the CS (8, 9). The enzyme consists of two functionally distinct domains, an N-terminal catalytic domain and a C-terminal DNA-binding domain, connected by a long linker (10, 11). Varying the distance between the CS and the IS has led to the suggestion that the linker is flexible and can extend and retract for the catalytic domain to find its CS. However, when the CS is out of range, the linker will default to cleavage at the wild-type distance (10).
The DNA-binding domain contacts a 20-bp region of the homing site and binds with the same affinity as the full-length protein (11). The crystal structure of a complex of the DNA-binding domain, residues 130–245, with a 20-bp DNA duplex has recently been determined (12). Surprisingly, rather than being a compact folded structure, the domain includes three subdomains connected by extended regions (Fig. 1A). The three subdomains are a zinc finger (a in Fig. 1 A and B) that makes few DNA contacts, an α-helix that binds in the minor groove (d in Fig. 1 A and B), and a helix–turn–helix subdomain (f in Fig. 1 A and B) that sits on the major groove. The protein and the DNA make contacts along their entire lengths, as the protein wraps completely around the DNA (Fig. 1C). Most protein–DNA contacts involve the phosphate backbone. Interestingly, the few base contacts that do occur are in the extended regions linking the individual subdomains (b, c, and e in Fig. 1 A–C). This structure suggests that the binding domain of I-TevI evolved by assembly of multiple DNA-binding and recognition subdomains, providing redundant contacts, which is consistent with the observed sequence tolerance of the enzyme.
Figure 1.
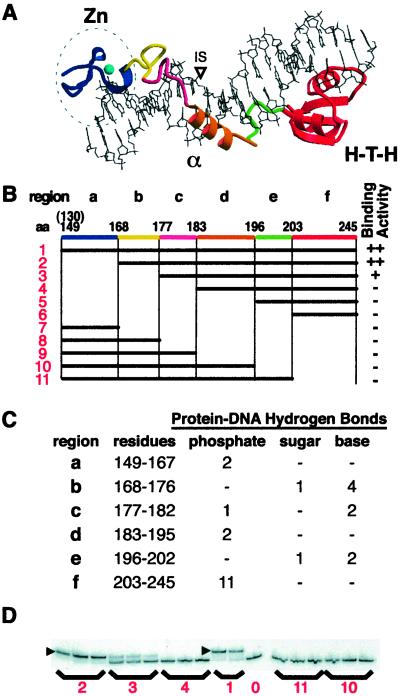
Binding activity of deletion derivatives. (A) The crystal structure of the I-TevI DNA-binding domain bound to substrate (12). Three subdomains are: Zn, zinc finger; α, α-helix; H-T-H, helix–turn–helix. The colored elements correspond to segments a–f of equivalent color in B. (B) Deletion derivatives. The horizontal black bars show the portion of the binding domain that is expressed in each construct, numbered 1–11. Protein 1 starts at residue 130 but the visible crystal structure begins at residue 149. Region a, zinc finger; b and c, extended region I; d, α-helix; e, extended region II; and f, H-T-H. Qualitative binding activities with cell lysates are shown on the right. ++, strong binding; +, weak binding; −, no detectable binding. (C) Protein hydrogen bond contacts to DNA organized by protein region and DNA contact. (D) EMSAs for representative constructs. ▸ = shifted complex. Numbers correspond to constructs in B. Lane 0 = DNA only.
To initiate a functional analysis of the three subdomains and their extended joining elements, a mutational study was undertaken. Although the data are consistent with important DNA recognition elements residing in amino acids C terminal to residue 168, they indicate that the zinc finger is dispensable for binding and cleavage by I-TevI. Rather, the zinc finger appears to have a novel function, to act as a distance determinant in the linker.
Materials and Methods
Expression Plasmids for I-TevI Deletion Derivatives.
The overexpression plasmid for the entire DNA-binding domain (130C; construct 1 in Figs. 1 and 2) (11), a derivative of pET-3a, was used as the template for the long-way-around PCR to generate 24 variants. For subdomain deletion derivatives, primers were chosen to amplify the desired portions of the vector and the DNA encoding the binding domain. Because both primers carried a unique KpnI restriction site, the amplified DNA could be readily ligated and transformed for expression. The KpnI site resulted in the insertion of Gly-Thr at the deletion site, regardless of location. Constructs 1 and 1a (Fig. 2) were prepared previously and do not contain the Gly-Thr insertion (11).
Figure 2.
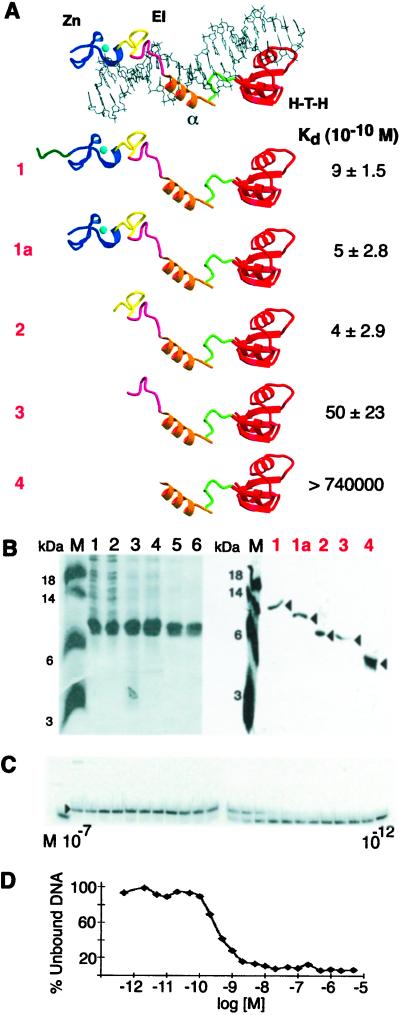
Kd determination for I-TevI subdomain derivatives. (A) Summary of deletion derivatives and Kds. Protein–DNA complex labeled as in Fig. 1A. EI, elongated region I. Data from 3–5 experiments were used to derive the Kds. (B) Purification of I-TevI deletion derivatives. (Left) Purification of construct 2. M, molecular mass markers; lane 1, crude extract; lane 2, soluble lysate; lane 3, 0.2% polyethyleneimine supernatant; lane 4, 30–60% saturated (NH4)2SO4 pellet and heparin column load; lanes 5 and 6, heparin column fractions. (Right) Purified protein samples that were used for the Kd determinations of constructs 1, 1a, 2, 3, and 4. (C) EMSA with construct 1a. I-TevI derivatives were bound to 100-bp DNA (3.4 × 10−12 M) containing the homing site. The binding reaction mixtures contained the protein (1a) at concentrations ranging from 1.0 × 10−7 M to 1.0 × 10−12 M. Protein concentration ranges were as follows for the remaining proteins: 1, 2.5 × 10−7 M to 2.5 × 10−11 M; 2, 2.0 × 10−6 M to 2.0 × 10−13 M; and 3, 5.0 × 10−6 M to 5.0 × 10−13 M. Protein 4 showed no evidence of binding to homing site DNA at 7.4 × 10−5 M. (D) Bjerrum plot of the binding experiment depicted in C.
Full-length I-TevI mutants were generated for in vitro transcription/translation. The wild-type I-TevI encoding plasmid pSP6–716 was prepared previously as follows. The EcoRI–HindIII fragment from pUC9, which contained the AccI–PvuII fragment of the td intron encoding I-TevI in its SmaI site, was inserted into pSP65 under the control of the SP6 promoter (13, 14). ΔZn was prepared with the same primers used to make the subdomain deletion derivatives, using pSP6–716 as the template and ligating at the KpnI site. Two derivatives, ΔC151 and ΔC151/153, also under the control of the SP6 promoter, were prepared in an earlier study (14).
Gene Editor in vitro site-directed mutagenesis (Promega) was used to generate Cys-to-Ala mutations of the zinc finger. The cysteines were mutated in pairs using the following two primers: 5′-CCCATAAGTTTGCTAAGGCCGGTGTTCGCATAC-3′ to give C151/153A, and 5′-CTGCTTATACTGCTAGTAAAGCCAGAAATCGTTC-3′ to give C164/167A. Use of both mutagenic primers in one step provided CZnA, with all four cysteines mutated to alanines.
I-TevI Subdomain Cleared Lysates and Purification.
The proteins were expressed under the control of the T7 promoter in BL21(DE3)pLysS. After growth at 37°C in TBYE (1% tryptone, 0.5% sodium chloride, 0.5% yeast extract, pH 7.0) supplemented with ampicillin (200 μg/ml) to OD600 0.6, the cultures were induced with 1 mM isopropyl β-d-thiogalactoside and shaken for 3 h at 30°C. Cells were harvested by centrifugation, and the pellets were resuspended in protein lysis buffer (50 mM Tris, pH 8.0/2 mM EDTA/1 mM DTT). Cells were lysed by brief sonication, and after centrifugation the cleared lysates were used directly.
I-TevI subdomain derivatives were purified essentially as described (11). Briefly, 10% polyethyleneimine was added to soluble cell lysates to 0.2% to remove nucleic acids and some contaminating proteins. The proteins were precipitated by the addition of ammonium sulfate to 55% saturation and then purified by heparin agarose chromatography.
Electrophoretic Mobility-Shift Assays (EMSAs).
Binding reactions (20 μl) containing 32P-labeled DNA and the appropriate protein were performed with 50 mM 2-mercaptoethanol, 2 mM EDTA, 50 mM Tris (pH 8.0), 20 μg/ml poly dI/dC, and 10 μg/ml BSA. The 100-bp DNA substrate was generated by PCR. Proteins were diluted (cleared lysates or purified protein) on ice with 50 mM Tris (pH 8.0), 2 mM EDTA, and then added to reaction mixtures, which were incubated for 10 min at 25°C. The reactions were quenched with 4 μl stop-load buffer (0.05 μM EDTA/5% SDS/25% glycerol/0.025% bromophenol blue) and electrophoresed at 30 mA for 1 h on a native 8% polyacrylamide gel at 4°C. The dried gel was visualized on a PhosphorImager (Storm, Molecular Dynamics), and the data were quantitated with imagequant (Molecular Dynamics).
In Vitro Expression and I-TevI Cleavage Assays.
All full-length proteins were expressed in vitro. RNA was transcribed from HindIII-linearized templates using SP6 RNA polymerase. Wheat germ extract (Promega) was used to translate the RNAs with [35S]methionine according to the recommended protocol. Aliquots of the translated proteins were fractionated on a 20% SDS/polyacrylamide gel to allow for normalization of relative concentrations as measured by a PhosphorImager.
Cleavage reactions (20 μl) were performed at 37°C on 250 ng of ScaI-linearized plasmid DNA containing either a wild-type or mutant td homing site (Fig. 3 C and D) (10) in I-TevI cleavage buffer (50 mM Tris⋅HCl, pH 8.0/10 mM MgCl2/100 mM NaCl). Time courses were carried out by using a reaction mix. Two reaction volumes were removed from the mixture to serve as negative controls (DNA only, and unprogrammed wheat germ extract). In vitro-translated protein was added to the reaction mix, and 20-μl aliquots were removed and quenched with 0.25 vol of stop-load buffer as described (10) at 5-, 10-, 20-, and 30-min time points. The quenched reactions were then incubated with 1 μl RNase A (500 μg/ml) for 15 min, and afterward with 1 μl Proteinase K (20 mg/ml) for 1 h, both at 37°C. Samples were separated on 0.7% (wt/vol) agarose gels in TAE buffer (40 mM Tris-acetate/1 mM EDTA) for 1.5 h at 120 V. Negatives of the ethidium bromide-stained gels were scanned with masterscan software, and the extent of cleavage was determined by analyzing the density of product bands relative to full-length DNA (Scanalytics, Billerica, MA).
Figure 3.
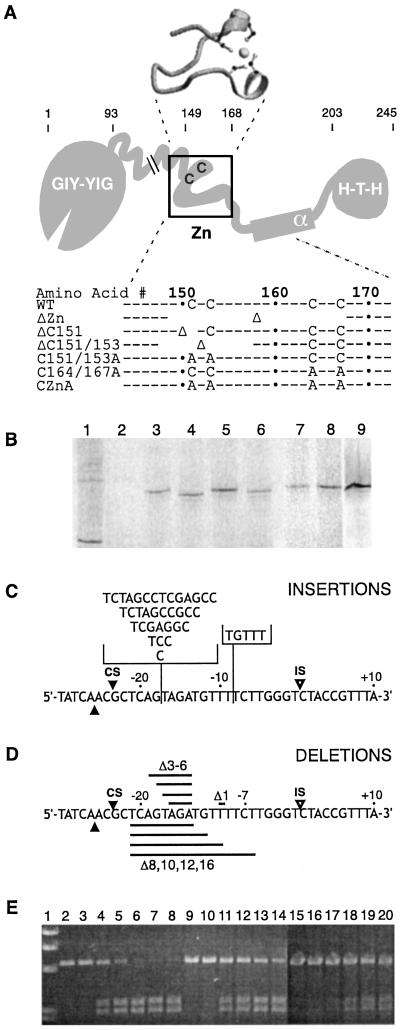
Endonuclease and substrate variants. (A) Schematic representation of full-length I-TevI variants. The structure of the zinc finger as seen in the cocrystal structure (12) is shown above the schematic. C and A indicate relevant cysteines and alanines; Δ indicates deletion; spaces indicate the extent of deletion; and ⋅ indicates every 10th amino acid. The regions of I-TevI are: GIY-YIG, catalytic domain; Zn, zinc finger; α, α-helix; H-T-H, helix–turn–helix. (B) In vitro translation of I-TevI zinc-finger deletion variants. Wheat germ extract and 35S-labeled methionine were used for translations with RNA runoff transcript (10). Lane 1, bovine mosaic virus translation product as a positive control; lane 2, unprogrammed extract as a negative control; lane 3, wild-type I-TevI; lane 4, ΔZn; lane 5, ΔC151; lane 6, ΔC151/153; lane 7, C151/153A; lane 8, C164/167A; and lane 9, CZnA. (C) Schematic of the homing-site insertion variants. The locations for insertions of base pairs (+1, +3, +5, +7, +10, +14) are indicated on the top strand with the vertical line. The horizontal brackets contain the inserted sequences in each case. IS and open arrowheads indicate the IS while CS and closed arrowheads indicate the CSs on each strand. (D) Schematic of the homing-site deletion variants. The deletions of base pairs (−1, −3, −4, −5, −6, −8, −10, −12, −16) are indicated on the bottom strand by the horizontal black bands. (E) Representative cleavage data with cleavage products resolved on an agarose gel. Lane 1, molecular weight markers; lanes 2–8, wild-type I-TevI on wild-type DNA; lanes 9–14, CZnA mutant on +3 DNA; and lanes 15–20, CZnA mutant on +7 DNA. Lanes 2, 9, and 15 contain DNA only; lanes 3, 10, and 16 contain DNA with unprogrammed lysates. Incubation times were 1, 2, 4, 7, and 10 min for lanes 4–8, and 5, 10, 20 and 30 min for lanes 11–14 and 17–20, respectively.
CS Mapping.
The CS mapping was performed as described (15). Double-stranded DNA was denatured and annealed to a primer upstream or downstream of the td homing site (primers W1763/OP16 and W1764/OP4) (10). The priming reaction was mixed with [α-32P]dATP and Sequenase 2.0 (United States Biochemical) and split in half. Extension reactions were carried out by using 2.5 μl of 2.5 mM deoxynucleotides to generate cleavage substrate, which was prewarmed (1 min at 37°C) and incubated for 5–20 min at 37°C with the appropriate volume of protein or unprogrammed wheat germ extract. Cleavage reactions were phenol-extracted, ethanol-precipitated, and then resuspended in Sequenase stop buffer. The other half of the priming reaction was incubated with dideoxynucleotides to generate the sequence ladders. Reactions were separated on 6% (wt/vol) polyacrylamide/8.25 M urea gels in TBE buffer (89 mM Tris/89 mM boric acid/2 mM EDTA).
Results
Delineation of Binding Activity with Subdomain Deletion Derivatives.
Deletion derivatives of the I-TevI DNA-binding domain were generated to evaluate the relative contributions of each subdomain to DNA binding. A series of deletions was based on contacts found in the cocrystal of the binding domain complexed with its homing-site DNA (12) (Fig. 1 B and C).
Protein expression and solubility were determined by SDS/PAGE of soluble lysates from cells expressing plasmid-encoded deletion derivatives. Although proper folding of the truncated proteins has not been definitively verified, they were sufficiently soluble and stable to be detected by SDS/PAGE, as is typical of well-folded proteins (Fig. 2B). The ability of the proteins to bind homing-site DNA was assessed by EMSAs using cleared lysates (Figs. 1D and 2C). N-terminal deletions would presumably leave intact most of the phosphate contacts while removing base contacts (Fig. 1C) (12). Qualitatively, the deletion of amino acids 130–167 (Fig. 1B, construct 2), which removes the zinc finger (a in Fig. 1B), did not have an effect on binding. However, removal of amino acids 130–176 (Fig. 1B, construct 3), which deletes a region of extended structure and at least five protein-to-DNA hydrogen bonds (b in Fig. 1C), diminished binding 10-fold. The removal of amino acids 130–182 (Fig. 1B, construct 4), a stretch containing more extended structure and three more hydrogen bonds along with hydrophobic contacts (c in Fig. 1C), abolished binding completely, as did any subsequent deletions from the N-terminal end. Therefore, the region 168–182 (b and c in Fig. 1B) and beyond appears to be important for binding.
The C-terminal deletions remove residues that contribute most of the structure's phosphate contacts and many hydrophobic surface interactions, while leaving intact residues that contribute most of the base interactions (Fig. 1C) (12). The removal of residues 203–245, the H-T-H subdomain (Fig. 1B, construct 11), abolished binding. None of the subsequent deletions from the C-terminal end bound I-TevI substrate (Fig. 1B, constructs 7–10), although the proteins appear well expressed and soluble. This finding suggests that the C-terminal H-T-H subdomain (f in Fig. 1B) may also be essential for binding activity, although a role in the proper folding of the protein remains a possibility.
The apparent dissociation constants (Kds) were determined for key derivatives (Fig. 2). These included the 130C derivative, containing residues 130–245 (11) (Figs. 1B and 2A, construct 1), a protein containing only the residues visible in the crystal structure, 148–245 (Fig. 2A, construct 1a), and the derivatives that displayed the stepwise decrease in ability to bind I-TevI substrate in Fig. 1B (constructs 2–4). These derivatives were purified to apparent homogeneity in a fashion analogous to that used for purification of the 130C derivative (Fig. 2 A and B, construct 1) (11).
Dissociation constants (Kds) were determined by EMSA (16) with 32P-labeled DNA at sub-Kd levels (100-fold < Kd; Fig. 2C). An example of the binding data from the EMSA used to determine apparent Kds is shown in Fig. 2D, with the fraction of free DNA plotted against the log of protein concentration (Bjerrum plot). These data indicate that the protein–DNA contacts in extended region I (E1 in Fig. 2A) are likely important, because their removal dramatically increased the Kd from nanomolar (10−10 to 10−9 M) to high micromolar concentrations at best (>10−5 M) (Fig. 2A, constructs 1–3 vs. construct 4). In contrast, removal of the zinc-finger subdomain did not increase the Kd of the derivatives (Fig. 2, construct 1a vs. construct 2), consistent with its having little or no role in affinity of the DNA-binding domain for its substrate.
Probing the Role of the Zinc Finger in I-TevI Cleavage Activity.
Because the zinc finger has no role in binding of DNA by the C-terminal domain, we next investigated its role in the cleavage activity of full-length protein by using two classes of mutants (Fig. 3). The first class consisted of full-length protein in which all or parts of the zinc finger were deleted. The constructs include a 19-aa deletion of the entire zinc-finger subdomain (ΔZn, amino acids 149–167), a 2-aa deletion including Cys-151 (ΔC151, amino acids 150–151), and a 10-aa deletion comprising half of the zinc finger, including Cys-151 and Cys-153 (ΔC151/153, amino acids 148–157). The second class comprised proteins containing Cys-to-Ala substitutions of the four cysteines that make up the zinc coordination site. The Cys-to-Ala mutagenesis was carried out in pairs to generate three variants Cys-151–Ala and Cys-153–Ala (C151/153A), Cys-164–Ala and Cys-167–Ala (C164/167A), and all four cysteines of the zinc finger, C151, C153, C164 and C167, to alanines (CZnA). Both classes of mutation are predicted to interfere with the formation of the zinc finger by removing or replacing the cysteines that chelate the zinc ion (Fig. 3A), although this hypothesis has not been formally tested. Because full-length wild-type I-TevI is toxic to Escherichia coli, the mutant proteins were expressed in vitro with wheat germ extract (17). The protein derivatives were generated with [35S]methionine to determine relative quantities of the translated proteins that migrated as single labeled bands of slightly differing sizes, as predicted, after SDS/PAGE (Fig. 3B).
The zinc-finger mutants were tested for cleavage of the homing site, to probe the possible requirement of the zinc-finger subdomain for catalysis (Figs. 3 C and D and 4, substrate 0). Cleavage of a linear substrate into two bands was monitored over time (Fig. 3E). All of the zinc-finger mutants were able to cleave the wild-type homing site (Fig. 4), although at lower efficiency than wild-type I-TevI. The cleavage activities of the I-TevI zinc-finger derivatives on wild-type homing site were expressed as percentages of wild-type I-TevI activity: wild-type I-TevI (100%), ΔZn (24%), ΔC151 (19%), ΔC151/153 (14%), C151/153A (19%), C164/167A (18%), and CZnA (22%). For these estimates, short time courses (0.5–10 min) were performed in the linear range (data not shown). This 4- to 7-fold reduction in cleavage activity may be caused by suboptimal protein–DNA interactions or reduced stability, rather than by a specific reduction in catalytic efficiency. CS mapping confirmed the specificity of catalysis, as reported below.
Figure 4.
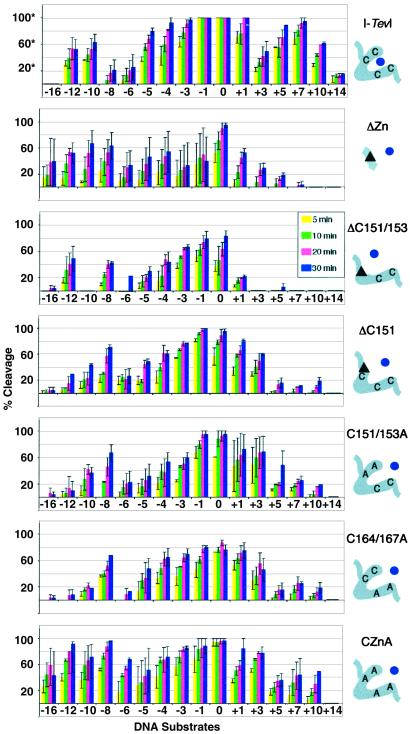
Summary of cleavage data of zinc-finger mutants. Each graph represents the compiled data for one protein on the different DNA homing sites. The homing sites are ordered from deletions to wild type to insertions from left to right [−16, −12, −10, −8, −6, −5, −4, −3, −1, 0 (wild type), +1, +3, +5, +7, +10, and +14]. The average percent cleavage at four time points is presented for each protein/DNA pair (5, 10, 20, and 30 min). *, The % cleavage for wild-type I-TevI was derived at a 3-fold lower protein concentration than was used for the mutants. The data for which 100% cleavage is seen with no error bars indicate that the DNA was completely cleaved under the conditions of this experiment; cleavage activity is in reality higher. Accurate relative activities of the mutants compared with wild-type I-TevI on wild-type homing-site DNA are given in the text. The mutant proteins were all run at equivalent concentrations and can be compared with one another directly. Error bars represent the SD in the data from 2–5 cleavage assays. The schematic on the right corresponds to the boxed portion of I-TevI in Fig. 3A. The zinc ion (dot) is shown either coordinated (wild-type I-TevI) or not (mutants). The triangles correspond to deletions.
Probing the Role of the Zinc Finger in Linker Flexibility.
To determine whether the I-TevI zinc finger may be involved in the flexibility of the linker between the catalytic and DNA-binding domains, we assessed the cleavage activities of the zinc-finger mutants on a set of DNA insertion and deletion derivatives of the homing site (Fig. 3 C and D). This library was previously constructed to show that the linker of intact I-TevI allows the catalytic domain to extend and retract to find the native cleavage sequence, as well as to show that the enzyme has a distance measure (10).
Observations from this extensive data set are as follows. First, the zinc-finger deletion derivatives are generally less active on DNA substrates that have insertions between the CS and the IS than on substrates with deletions between the CS and IS (Fig. 4, compare 0 to +14 with 0 to −16). These data indicate a reduction in the ability of the catalytic domain to stretch out to find the native CS, while maintaining a good degree of its ability to pull back.
Second, a decrease in product formation is seen in all cases when the normal CS is rotated by approximately half a helical turn from wild-type CS by deletions, and less consistently by insertions between the IS and the CS. Sensitivity to the face of the helix was observed previously for wild-type I-TevI (10); this property has been maintained by the zinc-finger mutants. However, the decrease for wild-type I-TevI is seen on −8 and +3 substrates, whereas the decrease for the mutant proteins is seen on −6 and +5 substrates. The latter is closer to what would be expected for B-form DNA, suggesting that the zinc-finger mutants may not distort the DNA in the same way as wild-type I-TevI (9).
Third, derivatives with some of the cysteines intact (ΔC151, ΔC151/153, C151/153A, and C164/167A) appear to have a narrower range of activity on the different substrates than do the completely compromised derivatives (ΔZn and CZnA). To test whether the free cysteines may be oxidized to cystines, a change that would compromise the overall structure of the mutant protein, cleavage assays were performed with and without 10 mM DTT. Comparable results were seen between the experiments (data not shown), indicating that formation of spurious disulfide bonds is not involved.
Finally, there is little correlation between the amount of protein deleted and the cleavage ability on deletion or insertion substrates. For example, ΔC151 (a 2-aa deletion) is active on a broader range of substrates than is ΔC151/153 (a 10-residue deletion), although ΔZn (a 19-residue deletion) cleaves all of the deletion substrates, but with greater variability in activity (note the large error bars for ΔZn in Fig. 4). Likewise, ΔZn is more active on the insertion derivatives than is ΔC151/153, despite the reduction in protein length. Moreover, the potentially extended zinc-finger point mutants do not have a greater tendency to cleave insertions of the homing site than to cleave deletions. We must therefore conclude that I-TevI does not simply use linker length as a means to find its remote CS.
CS Mapping to Evaluate Sequence and Distance Preferences.
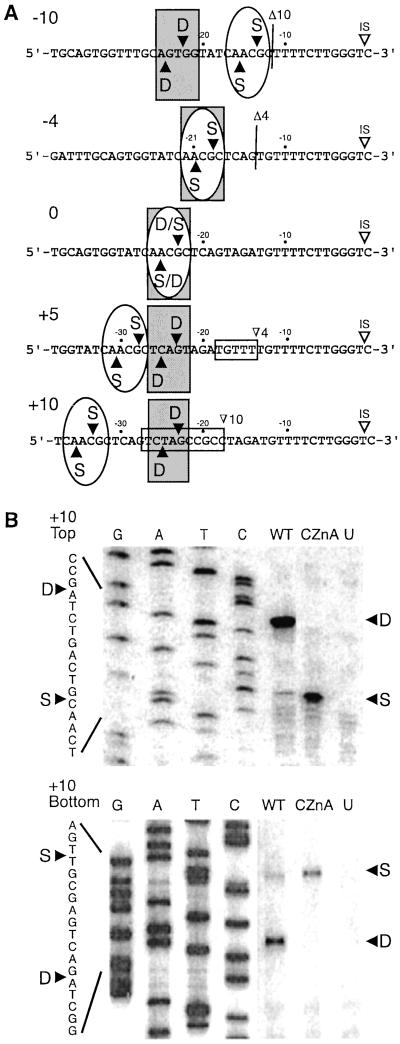
The specific CSs were mapped for I-TevI, ΔZn, and CZnA on wild-type homing-site DNA and selected homing-site mutants. Both ΔZn and CZnA cleave at the same positions as does wild-type I-TevI on wild-type substrate, at 23 and 25 bp from the IS (Fig. 5A). CSs were also mapped for I-TevI, ΔZn, and CZnA on substrates with a 4-bp deletion (−4), a 10-bp deletion (−10), a 5-bp insertion (+5), and a 10-bp insertion (+10). All three I-TevI derivatives pulled back and cleaved the −4 substrate at the wild-type cleavage sequence. Whereas wild-type I-TevI cleaves −10 substrate predominantly only 1 bp from the normal cleavage distance (22 and 24 bp from the IS) (10), ΔZn and CZnA pull back to cleave predominantly at the cleavage sequence, which is 13 and 15 bp from the IS. Furthermore, wild-type I-TevI cleaves the +5 and +10 substrates predominantly at the usual distance of 23 and 25 bp from the IS, while the mutants again cleave predominantly at the cleavage sequence, albeit with reduced activity. In these cases the mutant proteins need to extend the catalytic domain by five and 10 additional bp to reach the correct sequence, but they do not default to distance, as does wild-type I-TevI. The striking difference between wild-type I-TevI and the mutants is illustrated by the comparison of wild type to CZnA on +10 substrate (Fig. 5B). Thus, the dominant CS for wild-type I-TevI is at distance, while the dominant CS for ΔZn and CZnA is at the cleavage sequence (Table 1). Most remarkably, ΔZn, with a 19-aa deletion, prefers to stretch out to cleave at sequence rather than to default to distance, suggesting that this and other zinc-finger mutants have lost a distance determinant. In the absence of this distance constraint, the sequence preference of the catalytic domain again becomes manifest.
Figure 5.
CS mapping for I-TevI variants. (A) CSs indicated on each of the DNA sequences. Vertical lines indicate the site of the 10- and 4-bp deletions. The horizontal boxes indicate the 5- and 10-bp insertions. Gray rectangles indicate cleavage by wild-type I-TevI, and ovals indicate cleavage by ΔZn and CZnA. (B) Top and bottom strand CS mapping for wild-type I-TevI and CZnA on the +10 homing site variant (Fig. 3). G, A, T, and C indicate the bases represented in the sequencing ladders. WT, cleavage by wild-type I-TevI for 5 min; CZnA, cleavage by CZnA for 20 min; U, unprogrammed extract; S, cleavage at the wild-type cleavage sequence; D, cleavage at the wild-type cleavage distance from the IS. Other labels are as in Fig. 3.
Table 1.
Preferred cleavage sites for selected I-TevI zinc-finger mutants
| HS | Wild-type I-TevI | ΔZn | CZnA |
|---|---|---|---|
| −10 | S < D 1:16* | S > D 2:1* | S = D 1:1* |
| −4 | S | S | S |
| 0 | S and D | S and D | S and D |
| +5 | S < D 1:7* | S = D 1:1* | S > D 3:1* |
| +10 | S < D 1:9* | S > D 2:1* | S > D 16:1* |
Cleavage at sequence (S) vs. distance (D).
Discussion
The Zinc Finger of I-TevI Is a Component of the Flexible Linker Rather Than of the DNA-Binding Domain.
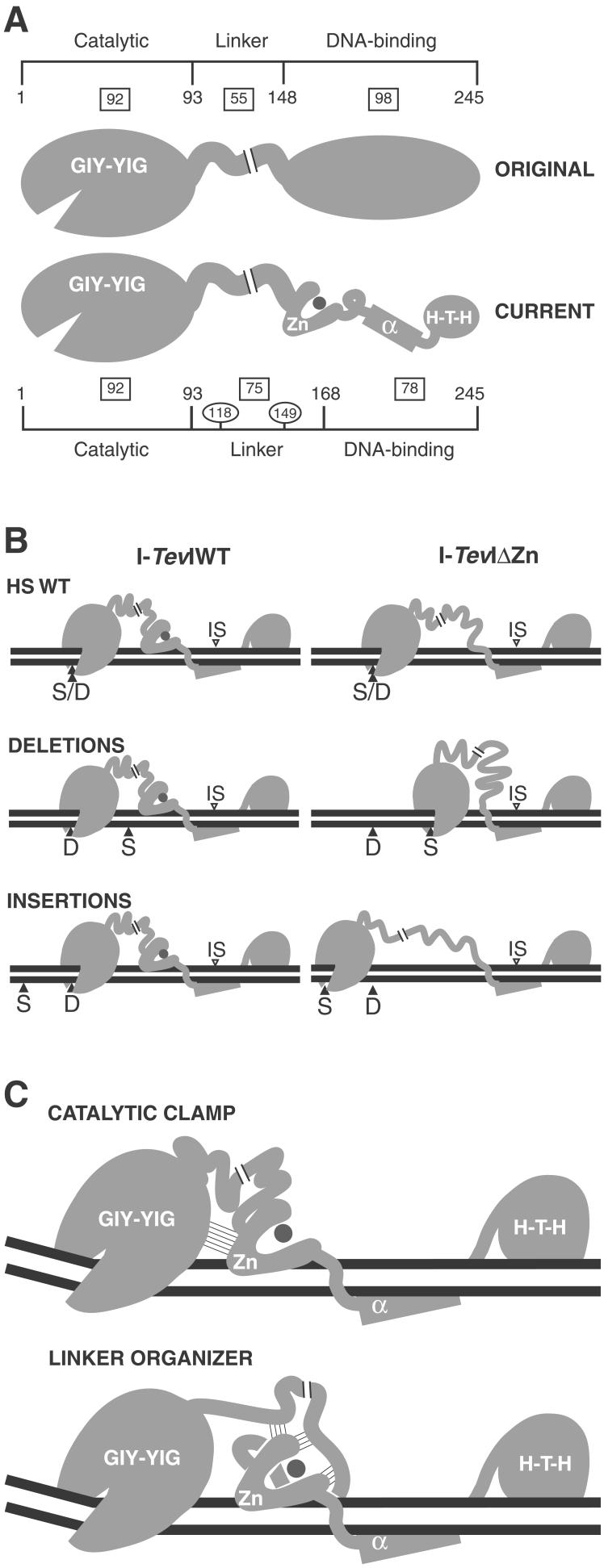
The recently solved structure of the modular DNA-binding domain of intron endonuclease I-TevI complexed with its homing-site duplex (12) has stimulated an analysis of the roles of its individual subdomains. The binding ability of nested sets of deletion derivatives suggests that the H-T-H module at the C terminus, the central minor groove-binding helix, and their joining region are required for DNA binding. However, most of the joining segment between the α-helix and the zinc finger plays a nonessential role, and the zinc finger itself is dispensable (Figs. 1 A and B and 2A). Most of the binding energy for the interaction between I-TevI and its substrate resides in contacts made by the DNA-binding domain, because the domain and full-length derivatives of the enzyme have comparable binding affinity (11). Our results indicate that the zinc finger in I-TevI has no role in the binding affinity of either the C-terminal domain or the full-length protein, and that rather than being a component of the DNA-binding domain it forms part of the flexible linker (Fig. 6A).
Figure 6.
Models for the role of the zinc finger in I-TevI. The regions of I-TevI were: GIY-YIG, catalytic domain; Zn, zinc finger; α, α-helix; H-T-H, helix–turn–helix. (A) Domains of I-TevI. Original and current models are shown. Boxes indicate the number of residues in each segment of the protein. Residue numbers indicate domain boundaries. Numbers in ovals demarcate the three segments of the linker. (B) Distance-constraining activity of the zinc finger. At large deletions and insertions, wild-type I-TevI defaults to cleave at wild-type distance (Left), whereas ΔZn cleaves predominantly at native cleavage sequence (Right). Labels are as in Fig. 5. (C) Models for directing catalytic domain to a fixed distance. Thin lines depict hypothetical interactions. The catalytic-clamp model shows interactions between the zinc finger and the catalytic domain, whereas the linker-organizer model shows the zinc finger interacting with the linker, to position the catalytic center of I-TevI at the CS. In both models the zinc finger is proposed to be anchored on the DNA, and in both cases, protein–protein interactions limit flexibility and therefore promote distance-specific cleavage by the sequence-tolerant catalytic domain.
These studies not only reassign the role of the zinc finger, but they also change the relative proportions of the different domains of I-TevI. The flexible linker was originally estimated at 55 aa, connecting the 92-aa catalytic domain to a 98-aa DNA-binding domain (14) (Fig. 6A). According to the current model, in which the zinc finger is a component of the linker, the linker length is extended to 75 aa, whereas the domain responsible for DNA-binding affinity is limited to 78 residues.
The Lengthy Interdomain Linker with the Zinc Finger as a Distance Determinant for Cleavage.
Linkers between domains of many proteins have been shown to have specific structures and/or functions. For example, the length, and to some extent the sequence, of the 26-residue flexible interdomain linker of the α subunit of E. coli RNA polymerase confers promoter class-dependent activity (18–20). In some cases the flexible linkers show little defined structure or sequence requirements, as in the case of the 21-residue linker of the yeast heat shock transcription factor (21). Here the linker is needed to maintain spacing between the DNA-binding and trimerization domains and to orient the linked domains.
The unusual linker of I-TevI, residues 93–167, makes up one-third of the protein and, at 75 residues, can itself be divided into three sections. The N-terminal portion of the linker, residues 93–116, is thought to be unstructured in the absence of DNA, yet required for catalytic activity (14). The specific role of residues 117–148, a region susceptible to proteolysis by various proteases (11) and tolerant to 1- and 2-aa deletions (14), is not yet known, but this region may function in the ability of I-TevI to extend and contract to find the CS. Finally, the zinc finger, residues 149–167, appears to act to place a distance constraint on the catalytic domain. Thus, besides its extreme length, the functional properties of the interdomain linker set it apart from junction sequences in other proteins.
To probe the hypothesis that the zinc finger may function as the distance-sensing component of the linker, two series of zinc-finger mutants were used, with deletions and point mutations (Fig. 3A). These were tested in a matrix with DNA homing-site variants containing insertions and deletions (Figs. 3 C and D and 4). Like wild-type I-TevI, the zinc-finger mutants are still sensitive to helical face, indicating that the linker has substantially maintained its fundamental disposition to the helix, rather than becoming grossly disordered. Furthermore, the combination of protein and substrate mutants allowed the conclusion that a dynamic feature of the linker is restricted to a specific distance range by the zinc finger, and when the distance restriction is compromised, the sequence selectivity of the catalytic domain takes over.
Interestingly, ΔZn and CZnA mutants can maintain activity over a greater range of homing-site variants than can partial deletions or double point mutations of the zinc finger. These partial mutants may be less ordered on the DNA, with increased entropy possibly accounting for at least some of the decrease in activity. Regardless, all of the zinc-finger mutants are considerably more effective at overcoming deletions than insertions in effecting cleavage, suggesting that the mutant linker is better able to retract than extend. In either event, all zinc-finger mutants are less able to cleave the wild-type substrate, as well as insertions and deletions of the homing site, than is wild-type I-TevI, suggesting that the zinc finger has a role in enhancing cleavage activity.
Most importantly, CS mapping indicated that, like wild-type I-TevI, the ΔZn and CZnA mutants cleaved the wild-type homing site accurately, and they cleaved the 4-bp deletion mutant at the correct sequence (Fig. 5). However, unlike the wild-type enzyme, which reverts to distance with a lengthy insertion or deletion (10), the zinc-finger mutants exhibit sequence over distance preference (Fig. 5, mutants −10, +5, and +10). It seems counterintuitive that ΔZn (19-aa deletion) extends farther than does wild-type I-TevI to cleave +5 and +10 homing sites at the correct sequence, and that the potentially unfolded CZnA mutant retracts to cleave at the correct sequence on the −10 homing site. The major conclusion to be drawn from these observations is that the zinc finger acts as a dominant measuring device that directs cleavage at a fixed distance (Fig. 6B). In the absence of this device, the zinc-finger mutants lose the ability to cleave at the preset distance, and they cleave at low efficiency at the displaced CS sequence, which becomes favored.
The Zinc Finger as a Molecular Constraint in I-TevI.
Zinc fingers are multifunctional structure elements. The classical zinc fingers are major-groove DNA-binding proteins typified by the TFIIIA-type Cys-2–His-2 zinc fingers (22, 23). However, the zinc finger of I-TevI makes no apparent contribution to the binding of its DNA substrate (Figs. 1 and 2), consistent with the fact that T-even phage DNA is highly modified in the major groove (9). The DNA of phage T4 contains glucosylated 5-hydroxymethylcytosine, effectively blocking the major groove. Although there are rare examples of zinc-finger domains that contact bases and bind in the minor groove (24), such interactions are not seen in I-TevI. Furthermore, zinc fingers that reach across the minor groove, making only backbone contacts and acting as spacers, are seen in TFIIIA fingers 4 and 6 (25–27), but the distance-determining function of the zinc finger of I-TevI argues against its being a purely bridging entity.
Zinc-binding moieties are also involved in protein–protein interactions, RNA binding, and lipid binding (28). The GATA-1-type Cys-4 zinc fingers mediate protein–protein interactions (29, 30). Although I-TevI also has four cysteine ligands to zinc, it is a much smaller zinc finger. Further, I-TevI acts as a monomer and does not apparently require other proteins for endonuclease activity (9). The possibility exists, however, that the zinc finger is involved in novel intramolecular protein–protein interactions, and as such has evolved the function of directing cleavage at a fixed distance from the DNA-binding site.
Although the zinc finger does not contribute to the binding energy of the I-TevI/homing-site interaction, the potential for it to make contacts with the DNA exists. The crystal structure of the DNA-binding domain complexed with the homing site shows hydrogen bonds between Tyr-162 and Ser-165 of the zinc finger and the phosphate backbone, and a potential hydrogen bond between Gln-158 and A-9 (12). In addition, methylation and ethylation interference experiments (8) suggest that the full-length protein in the vicinity of the zinc finger blocks G-7 (Fig. 3D, bottom strand), both in the major groove and at the phosphate backbone. These data are consistent with the existence of zinc finger–DNA interactions that could provide an anchor for the zinc finger while it performs its function.
How might the zinc finger position the catalytic domain at a specific distance from the DNA-binding domain, which is fixed by being bound to the DNA substrate around the IS? This flexible locking mechanism could be mediated by intramolecular protein–protein interactions between the zinc finger and the catalytic domain, in a “catalytic-clamp” model (Fig. 6C Upper). Alternatively, interactions between the zinc finger and other components of the linker could control folding of the linker, in a “linker organizer” model (Fig. 6C Lower). We thus propose that, by anchoring itself on the DNA and constraining the catalytic domain directly or indirectly through intramolecular interactions, the zinc finger acts as a distance determinant for cleavage. According to either model, protein–protein interactions, as suggested by the Cys-4 composition of the zinc finger, would serve to position the active center of the catalytic domain of I-TevI at the CS. Clearly, the catalytic-clamp and linker-organizer models are not mutually exclusive. Although there is no direct physical evidence for intramolecular protein–protein interactions, it is apparent from the data that the zinc finger is able to impart global properties on I-TevI function and, therefore, cross-talk between it and other regions of the enzyme is likely.
Modular Evolution of I-TevI.
The segmented arrangement of structural elements suggests I-TevI to be a result of modular evolution. First, the catalytic GIY-YIG module was envisaged as a cartridge that was fused to many different DNA-binding domains (11, 14, 31). Second, the DNA-binding domain of I-TevI was hypothesized to comprise multiple binding modules strung together like beads on a string (12). In support of modular evolution, a closely related GIY-YIG homing endonuclease from Bacillus mojavensis, I-BmoI, has two α-helical subdomains and an H-T-H subdomain, but no zinc finger (12, 31).
In contrast to I-TevI, I-BmoI cleaves close to its IS, in strictly sequence-specific fashion (31). Like I-BmoI, other GIY-YIG endonucleases, I-TevII (32), SegF (33), and SegG (Q. Q. Liu, A. Belle, D. Edgell, D. Shub, and M.B., unpublished work), also appear to be sequence specific. Furthermore, none of these enzymes, nor any other GIY-YIG endonuclease, have been found to have a zinc finger. These observations, together with the facts that I-TevI is a much more active, yet sequence-tolerant enzyme, that its free catalytic domain does not bind substrate, and that there is consistently a 4- to 7-fold reduction in cleavage activity when the zinc finger is mutated suggest the following scenario. The distance determinant for cleavage arose as a means of ensuring efficient binding of preferred nucleotides at the CS, by increasing the effective concentration of the weak binding catalytic domain. This adaptation of the zinc finger, to position the catalytic domain for cleavage and thereby to promote activity at the preferred sequence, introduced the distance constraint. In the wild-type protein, the zinc finger would dominate and prevent the weak-binding catalytic domain from finding the correct sequence in the substrate mutants. However, unfolding or removal of the finger would abolish the constraint and allow the binding of the catalytic domain to occur at the correct CS. With the wild-type protein, if the preferred CS is out of range, a default to distance would still allow cleavage, because of the sequence tolerance of I-TevI. In contrast to I-TevI, the sequence-specific endonucleases, which have no zinc finger, would not tend to evolve a distance preference, because no catalysis would occur without the correct DNA sequence.
Perhaps the zinc finger was inserted by a domain-fusion event that was futile for I-TevI because the zinc finger could not engage in customary interactions in the major groove. The zinc finger might then have evolved to function rather as a distance determinant, to position the catalytic domain for efficient cutting at the CS. Thus, the zinc finger would facilitate the dissemination of the host intron, not only by enhancing activity at its natural CS, but also by promoting cleavage at a favored distance when the native site is absent.
Acknowledgments
We thank David Edgell and Kenji Ichiyanagi for helpful discussions and technical advice and Maryellen Carl for expert manuscript preparation. We also thank Fred Gimble, Alan Lambowitz, David Edgell, Kenji Ichiyanagi, Joe Kowalski, Markus Landthaler, George Silva, and Wei Wu for critical readings of the manuscript. We acknowledge the contribution of the Molecular Genetics Core Facility at the Wadsworth Center. National Institutes of Health Grants GM44844 and GM39422 (to M.B.) and GM56966 (to P.V.R.) funded this work.
Abbreviations
- CS
cleavage site
- IS
intron insertion site
- EMSA
electrophoretic mobility-shift assay
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 27, 1999.
References
- 1.Lambowitz A M, Belfort M. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 2.Belfort M, Roberts R J. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambowitz A M, Caprara M G, Zimmerly S, Perlman P S. In: The RNA World. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 451–485. [Google Scholar]
- 4.Gimble F S. FEMS Microbiol Lett. 2000;185:99–107. doi: 10.1111/j.1574-6968.2000.tb09046.x. [DOI] [PubMed] [Google Scholar]
- 5.Chevalier B S, Stoddard B L. Nucleic Acid Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belfort M, Derbyshire V, Cousineau B, Lambowitz A. In: Mobile DNA II. Craig N, Craigie R, Gellert M, Lambowitz A, editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 761–783. [Google Scholar]
- 7.Jurica M S, Stoddard B L. Cell Mol Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryk M, Quirk S M, Mueller J E, Loizos N, Lawrence C, Belfort M. EMBO J. 1993;12:2141–2149. doi: 10.1002/j.1460-2075.1993.tb05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller J E, Smith D, Bryk M, Belfort M. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryk M, Belisle M, Mueller J E, Belfort M. J Mol Biol. 1995;247:197–210. doi: 10.1006/jmbi.1994.0133. [DOI] [PubMed] [Google Scholar]
- 11.Derbyshire V, Kowalski J C, Dansereau J T, Hauer C R, Belfort M. J Mol Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- 12.Van Roey P, Waddling C A, Fox K M, Belfort M, Derbyshire V. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell-Pedersen D, Quirk S M, Aubrey M, Belfort M. Gene. 1989;82:119–126. doi: 10.1016/0378-1119(89)90036-x. [DOI] [PubMed] [Google Scholar]
- 14.Kowalski J C, Belfort M, Stapleton M A, Holpert M, Dansereau J T, Pietrokovski S, Baxter S M, Derbyshire V. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell-Pedersen D, Quirk S, Clyman J, Belfort M. Nucleic Acids Res. 1990;18:3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey J. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 17.Bell-Pedersen D, Quirk S M, Bryk M, Belfort M. Proc Natl Acad Sci USA. 1991;88:7719–7723. doi: 10.1073/pnas.88.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita N, Endo S, Ishihama A. Biochemistry. 2000;39:6243–6249. doi: 10.1021/bi000020d. [DOI] [PubMed] [Google Scholar]
- 19.Meng W, Savert N J, Busby S J W, Thomas M S. EMBO J. 2000;19:1555–1566. doi: 10.1093/emboj/19.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jean D, Gendron D, Delbecchi L, Bourgaux P. Nucleic Acids Res. 1997;25:4004–4012. doi: 10.1093/nar/25.20.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flick K E, Gonzalez L, Jr, Harrison C J, Nelson C M. J Biol Chem. 1994;269:12475–12481. [PubMed] [Google Scholar]
- 22.Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 23.Wolfe S A, Nekludova L, Pabo C O. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 24.Zhu L, Wilken J, Phillips N B, Narendra U, Chan G, Stratton S M, Kent S B, Weiss M A. Genes Dev. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]
- 25.Clemens K R, Xiubei L, Wolf V, Wright P E, Gottesfeld J M. Proc Natl Acad Sci USA. 1992;89:10822–10826. doi: 10.1073/pnas.89.22.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes J J, Tullius T D. J Mol Biol. 1992;227:407–417. doi: 10.1016/0022-2836(92)90897-s. [DOI] [PubMed] [Google Scholar]
- 27.Neeley L, Trauger J W, Baird E E, Dervan P B, Gottesfeld J M. J Mol Biol. 1997;274:439–445. doi: 10.1006/jmbi.1997.1411. [DOI] [PubMed] [Google Scholar]
- 28.Laity J H, Lee B M, Wright P E. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 29.Mackay J P, Crossley M. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 30.Leon O, Roth M. Biol Res. 2000;33:21–30. doi: 10.4067/s0716-97602000000100009. [DOI] [PubMed] [Google Scholar]
- 31.Edgell D R, Shub D A. Proc Natl Acad Sci USA. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loizos N, Tillier E R M, Belfort M. Proc Natl Acad Sci USA. 1994;91:11983–11987. doi: 10.1073/pnas.91.25.11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belle A, Landthaler M, Shub D A. Genes Dev. 2002;16:351–362. doi: 10.1101/gad.960302. [DOI] [PMC free article] [PubMed] [Google Scholar]