Abstract
The ability of Pseudomonas syringae pv. tomato DC3000 to be pathogenic on plants depends on the Hrp (hypersensitive response and pathogenicity) type III protein secretion system and the effector proteins it translocates into plant cells. Through iterative application of experimental and computational techniques, the DC3000 effector inventory has been substantially enlarged. Five homologs of known avirulence (Avr) proteins and five effector candidates, encoded by genes with putative Hrp promoters and signatures of horizontal acquisition, were demonstrated to be secreted in culture and/or translocated into Arabidopsis in a Hrp-dependent manner. These 10 Hrp-dependent outer proteins (Hops) were designated HopPtoC (AvrPpiC2 homolog), HopPtoD1 and HopPtoD2 (AvrPphD homologs), HopPtoK (AvrRps4 homolog), HopPtoJ (AvrXv3 homolog), HopPtoE, HopPtoG, HopPtoH, HopPtoI, and HopPtoS1 (an ADP-ribosyltransferase homolog). Analysis of the enlarged collection of proteins traveling the Hrp pathway in P. syringae revealed an export-associated pattern of equivalent solvent-exposed amino acids in the N-terminal five positions, a lack of Asp or Glu residues in the first 12 positions, and amphipathicity in the first 50 positions. These characteristics were used to search the unfinished DC3000 genome, yielding 32 additional candidate effector genes that predicted proteins with Hrp export signals and that also possessed signatures of horizontal acquisition. Among these were genes encoding additional ADP-ribosyltransferases, a homolog of SrfC (a candidate effector in Salmonella enterica), a catalase, and a glucokinase. One ADP-ribosyltransferase and the SrfC homolog were tested and shown to be secreted in a Hrp-dependent manner. These proteins, designated HopPtoS2 and HopPtoL, respectively, bring the DC3000 Hrp-secreted protein inventory to 22.
The organism Pseudomonas syringae pv. tomato DC3000 is an important model in molecular plant pathology whose pathogenicity depends on effector proteins injected into host plant cells by the Hrp (hypersensitive response and pathogenicity) type III protein secretion system. We have reported the development of a draft sequence of the DC3000 genome (http://www.tigr.org/tdb/mdb/mdbinprogress.html) and the use of that sequence for genomewide identification of virulence-related genes in the Hrp regulon (1). We now have extended our genomewide investigation of the Hrp system by identifying characteristics that enable the prediction of DC3000 proteins that may travel the Hrp pathway and, therefore, are candidate effectors.
Type III protein secretion systems are central to the virulence of many bacteria, including animal pathogens in the genera Salmonella, Yersinia, Shigella, and Escherichia and plant pathogens in the genera Pseudomonas, Erwinia, Xanthomonas, Ralstonia, and Pantoea (2). Loss of the secretion system usually abolishes pathogenicity in mutants, whereas mutation of a single effector gene commonly has little or no effect because of apparent redundancies among the effectors (3). This observation highlights both the collective importance of effectors in pathogenesis and the difficulty in identifying them through loss of function. Given this problem, effector genes have been alternatively sought through the identification of proteins secreted to the medium and of genes coordinately regulated with those encoding the secretion machinery (4). However, some effectors are poorly secreted in culture and/or expressed independently of the secretion system regulon (5, 6). Thus, despite the availability of genomic sequence data for several pathogens that use type III secretion systems, we still have only a fragmentary inventory of the effectors underlying their pathogenicity.
Our genomewide analysis of the Hrp regulon in DC3000 revealed that several of the 48 Hrp promoters with highly significant hidden Markov model E values expressed homologs of known effector proteins, which are designated as Avr (avirulence) or Hop (Hrp-dependent outer proteins) (1). Our analysis also revealed limitations in such a promoter-based search for P. syringae effector genes: (i) Some Hrp promoters express regulatory or toxin biosynthesis proteins rather than effectors. (ii) Some avr genes are preceded by Hrp promoters with relatively poor E values, which substantially enlarges the set of candidate promoters. (iii) Many of the ORFs downstream of Hrp promoter sequences encode unknown proteins. And, as noted above, some effector genes may not be associated with Hrp promoters.
Thus, additional criteria are needed to winnow from the many thousands of potential ORFs in an unfinished bacterial genome a collection of candidate effectors that is usefully comprehensive but small enough for experimental testing. One such criterion is evidence of horizontal acquisition. G+C content, codon preference, or location near mobile DNA elements have been widely used to help the identification of virulence genes (7, 8). This criterion is applicable with P. syringae because most avr genes have atypical G+C content and are associated with mobile genetic elements (9–11). A second potentially useful criterion involves the identification of export-associated signals.
The first 15 or 17 aa of YopE or YopH, respectively, are sufficient to direct a CyaA (Bordetella pertussis adenyl cyclase) reporter to the type III pathway although no consensus secretion sequence has been recognized in these or other Yops (12). Some frameshifts altering the amino acid sequence in this region do not prevent secretion of an Npt reporter, which suggests an mRNA targeting signal (13). However, recent observations with native YopE suggest that targeting information resides in the protein rather than the mRNA and that amphipathicity in amino acids 2–11 is important (14). The mechanism by which the secretion machinery recognizes proteins with amphipathic N termini is unknown, and this property is too general to support efficient genomewide searches for novel effector genes.
P. syringae has a single type III secretion system. The Hrp system is known to secrete harpins (HrpZ and HrpW), the HrpA pilus subunit, and effector proteins. Deletion mutations have demonstrated that the N-terminal 10–15 residues are required for the secretion of AvrPto and AvrB, respectively (15). Fusions with an AvrRpt2 reporter have further demonstrated that additional signals for translocation into plant cells reside within the N-terminal 58 residues of the Xanthomonas campestris pv. vesicatoria AvrBs2 protein (16). In general, the effector proteins of plant and animal pathogens appear to carry targeting information in the N-terminal portion (15–150 residues), but the nature of that information is unclear.
Our genomewide identification of ORFs in the Hrp regulon yielded both a founding inventory of candidate effectors with homology to known Avr proteins or Hops and a collection of effector candidates (1). Here, we enlarged the inventory by demonstrating that five effector homologs and five proteins travel the Hrp pathway. Analysis of the N-terminal sequences of these and other known effector proteins yielded six distinct features or “rules” potentially useful in predicting Hrp secretion substrates. These rules were then used in a genomewide analysis of DC3000 that yielded 32 ORFs that were not preceded by Hrp promoters but had a strong likelihood of encoding novel effectors. Two of these candidate proteins were tested and shown to travel the Hrp pathway.
Materials and Methods
Strains and Media.
Escherichia coli strain DH5α was used for cloning experiments, and P. s. tomato DC3000 or derivatives and P. s. phaseolicola 3121 were used for secretion or translocation assays, respectively. Routine culture conditions for bacteria are similar to those described (6). Antibiotics were used at the following concentrations: 100 μg/ml ampicillin, 20 μg/ml chloramphenicol, 10 μg/ml gentamicin, 100 μg/ml rifampicin, 10 μg/ml kanamycin, and 20 μg/ml tetracycline.
Secretion Assays.
All of the secretion assays used P. s. tomato DC3000 strains carrying a pML123 derivative containing a PCR-cloned ORF (encoding a candidate Hrp-secreted protein) fused to nucleotide sequences that encoded either the hemagglutinin or FLAG epitopes along with their native ribosome binding sites (17). Details about the primers and the constructs are provided in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). Constructs carrying different epitope-tagged ORFs were electroporated into DC3000 and a DC3000 hrcC mutant and grown in Hrp-inducing conditions (18). Additionally, all of the DC3000 strains also carried pCPP2318, a construct that contains blaM lacking signal peptide sequences (19). DC3000 cultures were separated into cell-bound and supernatant fractions as described (6). Proteins were separated with SDS/PAGE by standard procedures (20), transferred to polyvinylidene difluoride membranes, and immunoblotted by using anti-FLAG (Sigma), anti-hemagglutinin (Roche Molecular Biochemicals), or anti-β-lactamase (5 Prime → 3 Prime) as primary antibodies. Primary antibodies were recognized by goat anti-rabbit IgG-alkaline phosphatase conjugate (Sigma), which were visualized by chemiluminescence by using a Western-Light chemiluminescence detection system (Tropix, Bedford, MA) and X-Omat x-ray film.
Plant Materials and Translocation Assays.
Arabidopsis thaliana accession Columbia (Col-0) and rps2–201 (21) mutant plants were grown in a growth chamber with 12 h of light at 24°C (22°C at night) and 70% relative humidity. Details about the primers and constructs described here are listed in Table 3. The partial avrRpt2 gene with the N-terminal 40 codons deleted was amplified by using standard PCR procedures and cloned into pMOD (Epicentre Technologies, Madison, WI). After confirmation by sequence analysis, it was cloned into the KpnI and SalI sites of the broad-host-plasmid pLK, resulting in pΔavrRpt2. DNA fragments spanning 200 bp upstream of the Hrp boxes and the complete ORFs for hopPtoK and hopPtoG were cloned into pΔavrRpt2 to produce phopPtoK-ΔavrRpt2 and phopPtoG-ΔavrRpt2, respectively. Each construct was introduced in P. s. phaseolicola 3121 by electroporation. Bacterial strains in 10 mM MgCl2 at a cell density of 108 colony-forming units/ml were infiltrated into A. thaliana Col-0 and Col-0 rps2–201 plants with a needleless syringe.
Bioinformatic Techniques.
Routine DNA analysis of the draft nucleotide sequence of P. s. tomato DC3000 (http://www.tigr.org/tdb/mdb/mdbinprogress.html) used blast searches (http://www.ncbi.nlm.nih.gov/BLAST/index.html), the Artemis genome viewer and annotation tool (http://www.sanger.ac.uk/Software/Artemis/), and lasergene software (DNAStar, Madison, WI). The two core motifs, described for simplicity in standard Prosite syntax (22), of the algorithm used to identify ORFs that shared general features with Hrp-secreted proteins was written as follows: <M-[CGHKNPQRSTY]-[ILV]-{DEFWY}-{DEMILVFWY}-{DE}-{DE}-{DE}-{DE}-{DE}-{DE}; and <M-[CGHKNPQRSTY]-[CGHKNPQRSTY]-[ILV]-{DEMILVFWY}-{DE}-{DE}-{DE}-{DE}-{DE}-{DE}. Briefly, the < at the left of the Met indicates that the following pattern must appear at the N terminus of the peptide (i.e., an ORF needed to start with Met). Characters in square brackets are alternatives for a single position (i.e., [ILV] denotes a single Ile, Leu, or Val residue). Characters in curly brackets are excluded (i.e., {DE} denotes any single residue except Asp or Glu). Dashes are used to separate residues. Other requirements were as follows: First, for an ORF to be selected it needed a minimum overall length of 150 residues. Second, to select ORFs that encoded polar amino termini, ORFs were required to have a minimum combined number of Ser, Thr, and Gln residues of 7 within the first 50 residues. Finally, the candidate sequences were screened to eliminate those containing the following Prosite patterns: [MIVLFYW]-[MIVLFYW]-[MIVLFYW]; [MLIV]-[MLIV]; [FYW]-[FWY]; [AG]-[AG];[KR]-[KR]; and [NQ]-[NQ]. This step simply eliminates sequences containing runs of residues from containing these particular residue classes. This genomewide search yielded genes that were manually screened for validity as ORFs by using the Artemis genome viewer.
Results
Demonstration that Several P. s. tomato DC3000 Proteins with Homology to Known Avr Proteins Are Secreted in Culture or Translocated into Plant Cells via the Hrp Secretion System.
Our genomewide analysis of ORFs preceded by Hrp promoter sequences yielded eight ORFs with homology to proteins with avirulence activity in other P. syringae pathovars (Fig. 1). We tested four of these DC3000 proteins for their ability to be secreted by the native Hrp system and found all of them to be secreted (Fig. 2A). Because the secretability of these proteins was demonstrated (and the avirulence activity of these DC3000 homologs is unknown), the proteins were renamed as HopPtoC (AvrPpiC2 homolog), HopPtoD1 and HopPtoD2 (AvrPphD homologs), and HopPtoJ (AvrXv3 homolog). We also used the AvrRpt2 translocation assay to test whether the DC3000 ORF that is similar to AvrRps4 (23) was translocated into Arabidopsis plant cells (16, 24). P. s. phaseolicola carrying a broad-host-range plasmid expressing the AvrRps4 homolog fused to the Avr domain of AvrRpt2 (but lacking the secretion signals of AvrRpt2) elicited an RPS2-dependent hypersensitive response (HR) on A. thaliana Col-0 (Fig. 2D), indicating that the amino terminus of the AvrRps4 homolog supplied sufficient information to direct translocation of the fusion protein into plant cells. Consequently, the AvrRps4 homolog was renamed HopPtoK. P. s. phaseolicola expressing HopPtoK did not elicit an HR, indicating that although translocated into host cells, HopPtoK is probably not recognized by the RPS4 protein present in A. thaliana Col-0, in contrast to its P. s. pisi 151 homolog (23) (Fig. 2D).
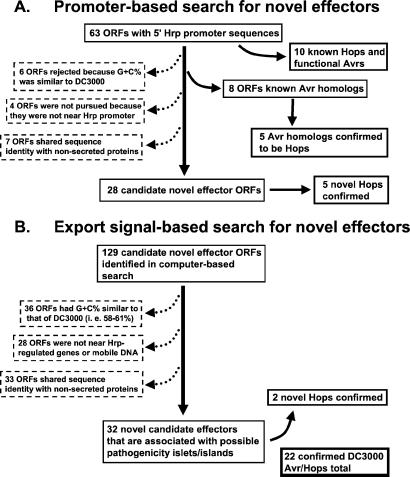
Figure 1.
Flowchart of the two independent searches for novel Hrp effectors. (A) ORFs with 5′ Hrp promoters were analyzed for horizontal transfer indicators to yield a set enriched in candidate effectors for secretion testing. (B) Valid ORFs found by the genomewide search for export signals were analyzed for indicators of horizontal acquisition and association with possible pathogenicity islands or islets to yield a set enriched in candidate effectors for secretion testing.
Figure 2.
Assays for Hrp system-dependent secretion in culture or translocation in planta of candidate effector proteins. P. s. tomato DC3000 and a Hrp secretion mutant derivative were used for tests of Avr homologs (A) or unknown candidate effectors (B and C). (A–C) DC3000 or a DC3000 hrcC mutant (18) carrying test ORFs (i.e., candidate effectors) fused to either the FLAG (F) or hemagglutinin (HA) epitopes were grown in Hrp-inducing media, and cultures were separated into cell (lanes 1–3) and supernatant (lanes 4 and 5) fractions and analyzed by SDS/PAGE and immunobloting. Lanes 1 and 4, wild-type DC3000; lanes 2 and 5, wild-type DC3000 (pTestORF); lanes 3 and 6, DC3000 hrcC mutant (pTestORF). As an additional control against leakage, we included pCPP2318 in all strains, which encodes the mature form of β-lactamase (β-lac). The presence of an epitope-tagged protein in the supernatant fraction of the wild type (lane 5), but absence in the hrcC secretion mutant (lane 6), indicated that the test ORF encoded a secreted product. (D and E) AvrRpt2 translocation assays performed with a DC3000 AvrRps4 homolog (D) or HopPtoG (E). Test strains were infiltrated into A. thaliana Col-0 (RPS2) and Col-0 rps2–201 (rps2) plants (for E only). Plant responses were scored 18 h after inoculation for hypersensitive response (HR) or no visible response (N).
Identification of Additional Hops Encoded by Candidate Effector Genes with 5′ Hrp Promoter Sequences.
Our previous genomewide identification of Hrp promoter sequences with hidden Markov model E values <1e-3 had yielded many that were upstream of unknown ORFs (1). From this group we identified 28 candidate effector ORFs that were not homologs of known Avr proteins/Hops or of any proteins unlikely to be secreted and whose low G+C% content and association with mobile genetic elements suggested horizontal acquisition (Fig. 1). Several of the predicted proteins shared amino acid identity with proteins likely to be effectors. For example, ORF5 yields several ADP-ribosyltransferases in blastp searches (highest blast E value 1e-5), including a type III-secreted ADP-ribosyltransferase from P. aeruginosa (25), and ORF2 is homologous to an ORF adjacent to the avrPpiC2 avr gene of P. s. pisi (26) (Table 1 and Table 4, which is published as supporting information on the PNAS web site). To test whether these proteins travel the Hrp pathway, we cloned the ORFs into a broad-host-range vector fused to either the hemagglutinin or FLAG epitope. DC3000 wild-type and Hrp mutant cultures carrying these constructs were separated into supernatant and cell fractions and analyzed with SDS/PAGE and immunoblots. Five of the eight proteins tested were secreted via the DC3000 Hrp system (Fig. 2B) and consequently were designated as HopPtoE, HopPtoG, HopPtoH, HopPtoI, and HopPtoS1, respectively. Although three ORFs were not detectably secreted in culture, they may still be effectors because AvrB similarly is not secreted in culture although translocated in planta (6, 27).
Table 1.
ORFs with 5′ Hrp promoter sequences and encoding proteins demonstrated to be secreted by the P. syringae Hrp system
| Initial designation* | New designation | Size, bp | % G+C | Homolog (blastpE value) | GenBank accession no. and/or reference |
|---|---|---|---|---|---|
| ORF1 | HopPtoI | 1,899 | 48.9 | None | NA |
| ORF2 | HopPtoH | 657 | 47.2 | ORF3 from P. s. pisi avrPpiC2 locus (1e-114) | CAC16702 (26) |
| ORF3 | HopPtoE | 636 | 50.7 | None | NA |
| ORF4 | HopPtoG | 1,482 | 43.7 | Hypothetical protein from R. solanacearum (1e-137) | NP_521884 |
| ORF5† | HopPtoS1 | 852 | 46.5 | Chicken ADP-ribosyltransferase (1e-5) | P55807 (39) |
NA, not available.
Nucleotide sequences of all ORFs (including ORFs 6–8, which encode proteins that failed the secretion tests) are provided as supporting information on the PNAS web site.
Determined to possess an ART domain (pfam1129), further confirming its similarity to ADP-ribosyltransferases.
AvrRpt2 Translocation Assay Indicates That at Least One of the Additional Hops Is Translocated into Plant Cells.
We chose HopPtoG to test for translocation into plant cells because it shared no similarities with any sequences in the databases and was shown to be secreted (Fig. 2B). P. s. phaseolicola carrying a plasmid expressing hopPtoG–ΔavrRpt2 elicited an RPS2-dependent hypersensitive response in A. thaliana Col-0 (Fig. 2E), indicating that targeting information in HopPtoG directed translocation of the AvrRpt2 fusion protein into plant cells. Thus, HopPtoG appears to be a Hrp-injected effector protein.
Identification of Predictive Patterns in the N-Terminal Regions of Proteins Secreted by the P. syringae Hrp System.
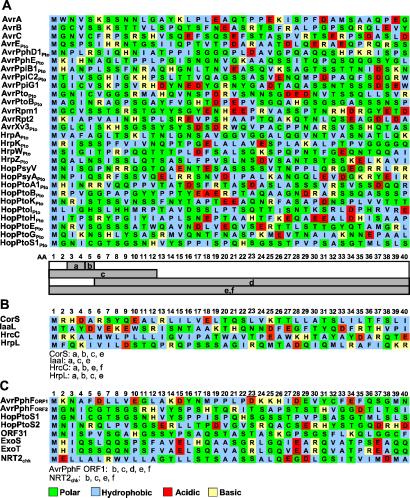
To enable genomewide identification of Hrp effector genes (regardless of the presence of 5′ Hrp promoter sequences), we examined the N-terminal regions of our enlarged set of Hrp-secreted proteins for conserved patterns and properties. We constructed a training set of 28 nonredundant proteins thought to be secreted by the P. syringae Hrp system (as indicated by previous avirulence or secretion tests), and whenever possible we represented a protein family with a homolog from P. s. tomato DC3000 (Fig. 3A). Attempts to align or find motifs in the first 50 aa of these proteins using known programs failed. However, when these amino acids were examined on the basis of their biophysical properties and solvent-exposed substitutability, several patterns emerged, which are expressed as predictive rules in Fig. 3A. In general, the rules define a specific pattern of solvent-exposed, equivalent amino acids that occur in the first five positions, an absence of acidic residues in the first 12 positions, and an overall amphipathicity and richness in polar amino acids in the N-terminal 50 or so residues. Notably, each of four representative proteins that are expressed by the Hrp regulon but are not secreted by the Hrp system (CorS, IaaL, HrcC, and HrpL) failed multiple rules (Fig. 3B). Thus, these export signal rules appeared sufficiently specific to support a genomewide search for additional candidate effector genes, which could then be submitted to Hrp secretion tests.
Figure 3.
Sequence of the first 40 aa of P. syringae Hrp-secreted proteins and other relevant proteins and patterns that are predictive of export signals in secreted proteins. (A) The first 40 aa of a nonredundant set of Hrp-secreted proteins and Avr proteins from P. syringae pathovars are color-coded by functional class. Below the aligned amino acids are bars indicating which positions are pertinent to the various export-associated patterns, expressed as six predictive rules: (a) Ile, Leu, Val, Ala, or Pro are found in positions 3 or 4, but not in both, and often preceded by a Pro, polar, or basic amino acid; (b) position 5 is rarely occupied by a Met, Ile, Leu, Phe, Tyr, or Trp; (c) Asp or Glu do not occur within the first 12 positions; (d) Cys rarely occurs after position 5; (e) the first 50 aa are amphipathic and rich in polar residues, especially Ser and Gln (only 40 residues shown); and (f) no more than three consecutive residues consisting of either Met, Ile, Leu, Val, Phe, Tyr, or Trp occur in the first 50 residues. The sequences of proteins from P. s. tomato DC3000 are indicated with a Pto subscript. (B) Representative P. s. tomato DC3000 proteins associated with the Hrp regulon but not secreted by the Hrp system: CorS (coronatine biosynthesis regulator), IaaL (N-(indole-3-acetyl)-l-lysine synthetase); HrcC, (outer membrane Hrp translocator); and HrpL (alternative sigma factor). (C) Application of the export-signal rules to two ORFs in the AvrPphF locus and six ADP-ribosyl transferases produced by P. s. tomato DC3000 (HopPtoS1, HopPtoS2, and ORF31), P. aeruginosa (ExoS and ExoT), and chicken (NRT2chk). Proteins violating the export signal rules are listed below B and C (with the violations). The GenBank accession numbers are M15194 (AvrA), M21965 (AvrB), M22219 (AvrC), Z21715 (AvrRpt2), NC_002759 (AvrRpm1), L20425 (AvrPto), AJ277495 (AvrPpiG1), AF232005 (HopPsyV), AF232004 (AvrE, HopPtoB (EEL ORF1), HopPtoA1 (CEL ORF5), HrpA, HrpK, HrpW, HrpZ), AAF67151 (AvrPphF ORF1), AAF67152 (AvrPphF ORF2), AAC34756 (HrcC), NP_252530 (ExoS), NP_248734 (ExoT), and P55807 (NRT2Chk). The full sequences for all other P. syringae proteins are published as supporting information on the PNAS web site.
Global Analysis of the P. s. tomato DC3000 Genome for ORFs Predicted to Be Secreted by the Hrp System.
An algorithm based on the export signal rules permitted a computer-based search for candidate Hrp-secreted proteins. The DC3000 genome was searched in all six reading frames for ORFs (at least 150 aa in length) with N termini (starting with Met) that satisfied the export rules. The large number of contigs and ambiguous nucleotides in the DC3000 draft sequence precluded an exhaustive search. The search process was based entirely on the direct translation of the contiguous sequences of unambiguous nucleotide codes, and no attempt was made to restrict the search to ORFs as defined by various gene-finding packages. This genomewide search yielded 400 hits that were manually screened for redundancy with known effectors and for validity as ORFs by using the Artemis genome viewer, based on the presence of blastp hits (28), Glimmer 2.0 ORF calls (29), ribosome binding sites, and transcription termination sequences (30). The resulting 129 apparently valid, additional ORFs were then analyzed for features characteristic of known effectors in pathogenicity islands (or islets), such as atypical G+C% content and presence in the same region of known virulence factors, Hrp promoters, or mobile genetic elements. A flow chart shown in Fig. 1B summarizes how we reduced the pool of 129 ORFs to 32 effector candidates that shared several characteristics of Hrp-secreted proteins. The nucleotide sequence of all 32 ORFs is published as supporting information on the PNAS web site, and an abbreviated list of the six most interesting ORFs based on blastp hits to other virulence genes is provided along with other relevant features in Table 2. Interestingly, our search found a putative effector, SrfC, that is predicted to travel the type III pathway encoded by SPI2 of S. enterica (4). A further indicator of the efficacy of the search was the finding of three additional ADP-ribosyltransferases, ORF 30, 31, and 32, all with significant amino acid sequence identity to HopPtoS1 (Table 2).
Table 2.
Selected ORFs encoding candidate effector proteins that were identified by the genomewide search based on export-signal patterns
| Designation* | New designation† | Size, bp | % G+C | Hrp promoters within 10 kb‡ | Mobile DNA within 10 kb§ | Homolog (blastpE value) | GenBank accession no. and reference |
|---|---|---|---|---|---|---|---|
| ORF29 | HopPtoL | 2700 | 61.0 | n | n | SPI-2 regulated SrfC (1e-21) | AAF74575 (4) |
| ORF30¶ | HopPtoS2 | 795 | 46.5 | y | n | Clostridium exoenzyme C3 ADP-ribosyltransferase (1e-5); 20.5% identical to HopPtoS1‖ | NP_346979 (40) |
| ORF31¶ | NA | 897 | 49.8 | y | y | Chicken ADP-ribosyltransferase (5e-3); also 71.7% identical to HopPtoS1‖ | P55807 (39) |
| ORF32¶ | NA | 507 | 54.2 | y | y | Chicken ADP-ribosyltransferase (5e-3); also 51.3% identical to HopPtoS1‖ | P55807 (39) |
| ORF33 | NA | 2823 | 55.2 | n | y | SepC insecticidal toxin (1e-128) | NP_065279 (41) |
| ORF34 | NA | 534 | 63.5 | y | n | Lytic enzyme (3e-36) | BAA83137 (42) |
NA, not available; n, no; y, yes.
The nucleotide sequences for these ORFs as well as the other ORFs identified in the export-signal-based search are available as supporting information on the PNAS web site.
If protein was determined to be Hrp-secreted by either secretion or translocation assays the protein was given a Hop name.
Indicates that the ORF is within 10 kb of a HrpL-responsive Hrp promoter identified in Fouts et al. (1).
Indicates that a transposon, plasmid, or a phage-related sequence is within 10 kb.
ORF was determined to possess an ART domain (pfam1129), further confirming its similarity to ADP-ribosyltransferases.
Sequence comparisons were carried out with Emboss software (http://bioweb.pasteur.fr/seqanal/interfaces/needle.html).
Confirmation That Two ORFs Identified in the Export Signal-Based Search Encode Hrp-Secreted Proteins.
To determine whether our genomewide search had identified any novel Hrp-secreted proteins we performed secretion assays on two ORFs, 29 and 30, which seemed to be particularly promising candidates. As noted above, the products encoded by ORFs 29 and 30 share similarity with a putative type III effector from S. enterica, SrfC, and ADP-ribosyltransferases, respectively. Both ORFs were PCR-cloned into a broad-host-range vector fused to the FLAG epitope, and each construct was introduced into DC3000 wild-type and Hrp mutant strains. The epitope-tagged ORF29 and ORF30 proteins were secreted by DC3000 in a Hrp-dependent manner without leakage of a cytoplasmic marker protein (Fig. 2C), and consequently they were designated as HopPtoL and HopPtoS2, respectively.
Discussion
We have used two general approaches to mine the DC3000 genome for genes encoding novel Hops. In the first, we identified ORFs that were downstream of Hrp promoters and also appeared to be horizontally acquired. Experimental testing of a subset of these yielded five Hops. In the second approach we used our enlarged set of Hrp-secreted P. syringae proteins to identify common characteristics in the first 50 aa of these proteins. These characteristics have permitted genomewide identification of novel proteins predicted to travel the Hrp pathway in P. s. tomato DC3000. Two of these ORFs were then tested and both were found to be Hrp-secreted. We also demonstrated that several DC3000 proteins with homology to Avr proteins in other P. syringae strains are, in fact, secreted in a Hrp-dependent manner, and these were consequently designated as Hops. The iterative process of sequence pattern-based prediction and experimental testing we pursued has yielded 22 confirmed Hrp-secreted proteins and an orderly process for eventual completion of the inventory of effector proteins.
The export signal rules also permitted us to predict which of the two ORFs in the AvrPphF locus is the effector. This locus was previously described in P. s. phaseolicola (31) and is also present in DC3000. We found that AvrPphF locus ORF1 violates all of the rules, but AvrPphF locus ORF2 none (Fig. 3C). Furthermore, ORF1 shares many of the general characteristics of type III chaperones (3). We were unable to detect secretion of the ORF1 product in secretion assays, and only ORF2 was shown to be translocated into plant cells on the basis of its delivery of an AvrRpt2 reporter (L.S., M. Guo, J.R.A., and X.T., unpublished data). Another demonstration of the selectivity of the export signal rules is that only the chicken ADP-ribosyltransferase NRT2CHK shows major violations of the rules even though this protein is more similar to HopPtoS1 and S2 than either of the type III-secreted ADP-ribosyltransferases from P. aeruginosa, ExoS and ExoT (Fig. 3C).
The observation that there is no overall difference in the N-terminal residue patterns of effectors that are chaperone-associated or chaperone-independent suggests that entry into the pathway is the same for nascent and preformed effectors. Also, we observed no significant difference in the N-terminal residue patterns between effectors and accessory secretion factors such as the HrpA pilus subunit and the harpin-like proteins. Perhaps the simplest method for sorting proteins to be secreted from those to be injected is by timing, with those proteins entering the pathway before the Hrp pilus has connected with host cells being preferentially released to the milieu.
There is presently little knowledge of how effector proteins (particularly those lacking chaperones) are targeted for entry into type III secretion pathways and what component(s) of the secretion machinery serve as gatekeepers. Recently, Lloyd et al. (32) have elegantly demonstrated the importance of amphipathicity in the first 8 aa in type III secretion signals. Our study suggests that positional effects in the first few amino acids of the export signal are also important. The patterns we observed suggest that solvent-exposed amino acids in the N terminus function as a “key” that is engaged by a receptor “lock” in the Hrp machinery. The key-way in the lock is likely to have a net negative charge (as suggested by the lack of acidic amino acids in the first 12 residues of Hops) and appears to recognize a specific pattern in the first five residues. This pattern occurs in almost all Hops. The subsequent 6–50 or more residues of Hops have the general property of amphipathicity (which seems to be the universal characteristic of type III effector proteins) without any positional specificity. Unlike the Salmonella-translocated effectors secreted by SPI2 (33), many of the P. syringae Hops do not appear to be homologs. Thus, the pattern in the first five residues likely represents convergent evolution to fit a Hrp system receptor.
To determine whether the algorithm that we used to search for export-associated patterns would be useful in identifying type III-secreted proteins in other pathogens that use type III secretion systems we searched the genomes of P. aeruginosa PAO1 (http://www.Pseudomonas.com/) and R. solanacearum GMI1000 (http://sequence.toulouse.inra.fr/R.solanacearum.html). Whereas the algorithm yielded 129 ORFs in DC3000, it identified 54 and 73 ORFs in P. aeruginosa and R. solanacearum, respectively. Several, but not all known, type III effector genes were identified in these organisms. For example, the type III effectors ExoS and ExoT both were identified in P. aeruginosa as well as several proteins secreted by the flagellar type III system. In R. solanacearum, the algorithm identified the P. syringae Avr homologs AvrPphD, AvrA, and AvrPpiC2, the Ralstonia PopA harpin-like protein, a hypothetical protein that is similar to PopC, and interestingly, HrpV, a protein encoded within the hrp/hrc cluster, whose function is unknown. We also searched the genomes of two nonpathogens, E. coli K12 MJ1655 (http://www.genome.wisc.edu/k12.htm) and Bacillus subtilis 168 (http://genolist.pasteur.fr/SubtiList/) and identified 54 and 40 ORFs, respectively. These latter bacteria do not have type III secretion systems other than the flagellar system, and it seems unlikely that all of these ORFs represent secreted proteins. Thus, genomewide searches with the current algorithm yield a collection of ORFs that is only enriched in type III-secreted proteins. However, as we have demonstrated, winnowing of this collection using other characteristics associated with effector genes, such as signatures of horizontal acquisition, can efficiently yield a subset (independent of 5′ Hrp promoter sequences) that can be systematically tested for secretion. This process yields DC3000 effector genes unlikely to be found by other means, as we have demonstrated.
HopPtoS1 and HopPtoS2 share sequence similarity with ADP-ribosyltransferases, proteins that have long been implicated in bacterial pathogenesis in animals through the modification of host signal transduction pathways (34), but until now have not been implicated in the bacterial pathogenesis of plants. The DC3000 genomic studies described in an earlier paper clearly show that several of the effectors in DC3000 are redundant (1). By using the pattern-based export prediction described here we have identified three ADP-ribosyltransferase genes (in addition to hopPtoS1) in the genome of DC3000 that have N-termini putative export signals. One of these, ORF32, may not be a functional gene because the ORF is truncated. The other two, HopPtoS2 and ORF31, are full-length genes based on sequence alignments (data not shown). HopPtoS2 is secreted by the Hrp system (Fig. 2C) and ORF31 shares high amino acid sequence identity with the Hrp-secreted HopPtoS1. Interestingly, HopPtoS1 contains putative myristoylation and palmitoylation sites at its N terminus (as does AvrPphF ORF2; Fig. 3C), whereas the other two do not, indicating that HopPtoS1 may be localized to the plasma membrane. Thus, there appear to be at least three Hrp-secreted ADP-ribosyltransferases and these may localize to different regions of the plant cell. The existence of these proteins in P. syringae is particularly noteworthy given that ADP-ribosyltransferase genes have not been identified in the bacterial plant pathogen genomes that have been published thus far (35–38). Significantly, our genomewide search for export signals yielded a homolog of the S. enterica candidate effector SrfC, further adding to the growing list of effectors shared between plant and animal pathogens. It is also noteworthy that one of the 32 ORFs found by the genomewide search (ORF48) is a homolog of a bacterial catalase (blastp 1e-126), and another (ORF49) is a glucokinase homolog (blastp 3e-42). These putative effectors could have a role in oxidative stress and regulation of sugar metabolism, respectively.
We have increased the inventory of DC3000 Hrp-secreted proteins to 22 with our characterization of HopPtoC, HopPtoD1, HopPtoD2, HopPtoE, HopPtoG, HopPtoH, HopPtoI, HopPtoJ, HopPtoK, HopPtoL, HopPtoS1, and HopPtoS2. This enlarged collection of candidate effectors should help us better understand how these proteins promote plant pathogenesis, and further investigation of their export signal patterns should aid our understanding of type III secretion mechanisms and the targeting of proteins to the pathway.
Supplementary Material
Acknowledgments
We thank K. Berry, N. Fedorova, T. Feldblynm, M. Gwinn, D. Haft, H. Khouri, W. Nelson, J. Peterson, D. Russell, B. Tran, L. Umayam, T. Utterback, and S. Van Aken of The Institute for Genomic Research for their efforts to sequence, close, and analyze the P. s. tomatoDC3000 genome. This work was supported by National Science Foundation Plant Genome Research Program Cooperative Agreement DBI-0077622, National Science Foundation Grant MCB-9982646 (to A.C.), and the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture Grant 01-35319-10019 (to J.R.A.). Part of this work was conducted by using the resources of the Cornell Theory Center, which receives funding from Cornell University, New York State, federal agencies, and corporate sponsors.
Abbreviations
- Hrp
hypersensitive response and pathogenicity
- Avr
avirulence
- Hop
Hrp-dependent outer protein
References
- 1.Fouts D E, Abramovitch R B, Alfano J R, Baldo A M, Buell C R, Cartinhour S, Chatterjee A K, D'Ascenzo M, Gwinn M, Lazarowitz S G, et al. Proc Natl Acad Sci USA. 2002;99:2275–2280. doi: 10.1073/pnas.032514099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galán J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis G, van Gijsegem F. Annu Rev Microbiol. 2000;54:734–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 4.Worley M J, Ching K H L, Heffron F. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- 5.Knoop V, Staskawicz B, Bonas U. J Bacteriol. 1991;173:7142–7150. doi: 10.1128/jb.173.22.7142-7150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochman H, Lawrence J G, Groisman E A. Nature (London) 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 8.Ochman H, Moran N A. Science. 2001;292:1096–1098. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 9.Kim J F, Charkowski A O, Alfano J R, Collmer A C, Beer S V. Mol Plant-Microbe Interact. 1998;11:1247–1252. [Google Scholar]
- 10.Alfano J R, Charkowski A O, Deng W, Badel J L, Petnicki-Ocwieja T, van Dijk K, Collmer A. Proc Natl Acad Sci USA. 2000;97:4856–4861. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson R W, Athanassopoulos E, Tsiamis G, Mansfield J W, Sesma A, Arnold D L, Gibbon M J, Murillo J, Taylor J D, Vivian A. Proc Natl Acad Sci USA. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sory M-P, Boland A, Lambermont I, Cornelis G R. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson D M, Schneewind O. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 14.Lloyd S A, Norman M, Rosqvist R, Wolf-Watz H. Mol Microbiol. 2001;39:520–531. doi: 10.1046/j.1365-2958.2001.02271.x. [DOI] [PubMed] [Google Scholar]
- 15.Anderson D M, Fouts D E, Collmer A, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12839–12843. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mudgett M B, Chesnikova O, Dahlbeck D, Clark E T, Rossier O, Bonas U, Staskawicz B J. Proc Natl Acad Sci USA. 2000;97:13324–13329. doi: 10.1073/pnas.230450797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labes M, Puhler A, Simon R. Gene. 1990;89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 18.Yuan J, He S Y. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charkowski A O, Huang H-C, Collmer A. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Kunkel B N, Bent A F, Dahlbeck D, Innes R W, Staskawicz B. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann K, Bucher P, Falquet L, Bairoch A. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinsch M, Staskawicz B. Mol Plant-Microbe Interact. 1996;9:55–61. doi: 10.1094/mpmi-9-0055. [DOI] [PubMed] [Google Scholar]
- 24.Guttman D S, Greenberg J T. Mol Plant-Microbe Interact. 2001;14:145–155. doi: 10.1094/MPMI.2001.14.2.145. [DOI] [PubMed] [Google Scholar]
- 25.Yahr T L, Goranson J, Frank D W. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 26.Arnold D L, Jackson R W, Fillingham A J, Goss S C, Taylor J D, Mansfield J W, Vivian A. Microbiology. 2001;147:1171–1182. doi: 10.1099/00221287-147-5-1171. [DOI] [PubMed] [Google Scholar]
- 27.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delcher A L, Harmon D, Kasif S, White O, Saltzberg S L. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ermolaeva M D, Khalak H G, White O, Smith H O, Saltzberg S L. J Mol Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 31.Tsiamis G, Mansfield J W, Hockenhull R, Jackson R W, Sesma A, Athanassopoulos E, Bennett M A, Stevens C, Vivian A, Taylor J D, Murillo J. EMBO. 2000;19:3204–3214. doi: 10.1093/emboj/19.13.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd S A, Sjostrom M, Andersson S, Wolf-Watz H. Mol Microbiol. 2002;43:51–59. doi: 10.1046/j.1365-2958.2002.02738.x. [DOI] [PubMed] [Google Scholar]
- 33.Miao E A, Miller S I. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finlay B B, Falkow S. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson A J G, Reinach F C, Arruda P, Abreu F A, Acencio M, Alvarenga R, Alves L M C, Araya J E, Baia G S, Baptista C S, et al. Nature (London) 2000;406:151–159. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- 36.Wood D W, Setubal J C, Kaul R, Monks D E, Kitajima J P, Okura V K, Zhou Y, Chen L, Wood G E, Almeida N F, et al. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 37.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman B S, Cao Y, Askenazi M, Halling C, et al. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 38.Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, Billault A, Brottier P, Camus J C, Cattolico L, et al. Nature (London) 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchiya M, Hara N, Yamada K, Osago H, Shimoyama M. J Biol Chem. 1994;269:27451–27457. [PubMed] [Google Scholar]
- 40.Nolling J, Breton G, Omelchenko M V, Markarova K S, Zeng Q, Gibson R, Lee H M, Dubois J, Qiu D, Hitti J, et al. J Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurst M R, Glare T R, Jackson T A, Ronson C W. J Bacteriol. 2000;182:5127–5138. doi: 10.1128/jb.182.18.5127-5138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kaynaya S, Ohnishi M, Murata T, Mori H, Hayshi T. Mol Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.