Abstract
No microorganism capable of anaerobic growth on methane as the sole carbon source has yet been cultivated. Consequently, information about these microbes has been inferred from geochemical and microbiological observations of field samples. Stable isotope analysis of lipid biomarkers and rRNA gene surveys have implicated specific microbes in the anaerobic oxidation of methane (AOM). Here we use combined fluorescent in situ hybridization and secondary ion mass spectrometry analyses, to identify anaerobic methanotrophs in marine methane-seep sediments. The results provide direct evidence for the involvement of at least two distinct archaeal groups (ANME-1 and ANME-2) in AOM at methane seeps. Although both archaeal groups often occurred in direct physical association with bacteria, they also were observed as monospecific aggregations and as single cells. The ANME-1 archaeal group more frequently existed in monospecific aggregations or as single filaments, apparently without a bacterial partner. Bacteria associated with both archaeal groups included, but were not limited to, close relatives of Desulfosarcina species. Isotopic analyses suggest that monospecific archaeal cells and cell aggregates were active in anaerobic methanotrophy, as were multispecies consortia. In total, the data indicate that the microbial species and biotic interactions mediating anaerobic methanotrophy are diverse and complex. The data also clearly show that highly structured ANME-2/Desulfosarcina consortia are not the sole entities responsible for AOM at marine methane seeps. Other microbial groups, including ANME-1 archaea, are capable of anaerobic methane consumption either as single cells, in monospecific aggregates, or in multispecies consortia.
Large reservoirs of methane lie buried beneath the seafloor dissolved in pore fluids, crystallized in solid phase gas hydrates, or as free gas. Typically, little of the methane released from this enormous natural gas reservoir reaches the oxic water column. Instead, it is oxidized to CO2 by microbes living in anoxic marine sediments. The amount of methane consumed by anaerobic oxidation of methane (AOM) each year is approximately equivalent to 5 to 20% of the total annual methane flux to the atmosphere (1, 2). Geochemical evidence, isotopic data, and microbial process measurements all indicate that AOM is directly coupled to the reduction of sulfate in marine sediments. The specific microbes, biochemical pathways, and physiological interactions regulating AOM however, are not well understood.
Recent studies combining stable isotopic analyses of lipid biomarkers with 16S rRNA gene surveys have identified several candidate microbial groups involved in AOM (3, 4). Initially, 13C-depleted archaeal and bacterial biomarkers were discovered in marine methane-seep sediments (3–5). Several of these 13C-depleted lipid biomarkers were characteristic of specific types of methanogenic archaea. In the same samples (3, 4), two archaeal phylogenetic groups (ANME-1 and ANME-2) were found to be abundant among the recovered rRNA gene clones. Subsequently, 13C-depleted archaeal lipid biomarkers have been recovered from other anoxic, methane-rich marine sedimentary habitats, and have been shown to be heterogeneous in both their structure and isotopic composition (6–9). Lipid biomarkers of archaeal origin are comprised of at least 6 distinct compounds, suggesting their production by more than one source organism (5–9). The diversity of archaeal 16S rRNA genes in methane-seep sediments also suggested the presence of multiple groups of putative methane-oxidizing Archaea (3, 4, 10, 11). The occurrence of ANME-1 and ANME-2 in a wide variety seep environments indicates that both archaeal groups may be involved in AOM, and could serve as a source of the 13C-depleted archaeal lipids found in methane-seep sediments.
Microscopic evidence has indicated that the archaeal ANME-2 group is physically associated with a sulfate-reducing bacterial partner (4, 7). In this association, the ANME-2 archaea are located in the central core of each microbial aggregate and are surrounded by a shell of Desulfosarcina spp. This physical association provided direct evidence for microbial consortia that couple AOM with sulfate reduction. In this case, the archaeon (ANME-2) is thought to mediate “reverse methanogenesis” (7, 12, 13). The bacterial partner is presumed to couple the oxidation of incompletely oxidized byproducts of AOM (e.g., H2, acetate or formate; 1, 12, 14) to the reduction of sulfate. Combined phylogenetic identification using fluorescent in situ hybridization (FISH) and secondary ion mass spectrometry (SIMS) provided isotopic evidence directly implicating ANME-2/Desulfosarcina consortia in AOM (13).
Isotopic analyses of biomarkers suggest that a diversity of microbial groups may be associated with AOM at both methane seeps (3–9, 13), as well as in subseafloor diffusion-limited environments (15). In this study the involvement of additional microbial groups in the anaerobic oxidation of methane was investigated, by using coupled isotopic and phylogenetic analyses at the single-cell level. Our results, which combine phylogenetic and isotopic analyses (FISH–SIMS) of single cells, demonstrate a greater diversity of microbes and microbial associations in AOM than has been previously reported.
Methods
Sample Collection.
Methane-seep sediments from Eel River Basin (ERB) were collected in October 1999 and August 2000 by ROV-operated push cores. High levels of methane and sulfide in pore waters in Eel River Basin sediments foster the growth of chemolithotrophic microorganisms. Sediments underlying chemosynthetic clams (Calyptogena pacifica, sample PC-21) and sulfur-oxidizing filamentous bacterial mats (sample PC-45) were used for FISH–SIMS studies described here. Sediments were processed in 3-cm-thick sections as described (13) and subdivided for chemical porewater analysis (measured on site) or molecular studies. Samples for FISH or nucleic acid extractions were frozen in liquid nitrogen or stored in ethanol/PBS (1:1, vol/vol) at −20°C (PBS is 10 mM Na2HPO4/130 mM NaCl, pH 7.2).
Sample Processing for FISH and FISH–SIMS.
Sediment samples processed in three different ways were all successfully used for FISH–SIMS. Small aliquots of methane-seep sediment (0.5 g) were fixed overnight at 4°C in 3.7% formaldehyde in PBS, washed 2 times with PBS, and stored (−20°C) in ethanol: PBS (1:1, vol/vol). Alternatively, sediments were stored at −20°C in ethanol/PBS (1:1, vol/vol) without the formaldehyde fixation step. In a third procedure, sediments frozen at −80°C for up to 2 years were thawed in ethanol: PBS (1:1, vol/vol) and used in the whole-cell FISH assays. Successful assays were obtained by using samples prepared by each of these procedures. Before processing, sediments were diluted 1:20 (vol/vol) in PBS to a final volume of 0.5 ml, sonicated on ice with a Vibra Cell sonicating wand (Sonics and Materials, Danbury, CT) for 20 s at an amplitude setting of 30, and overlaid on top of a pre-established Percoll gradient (see below).
Separation of Cells from Sediment–Percoll Density Gradients and Filtration.
Percoll gradients were established before sample addition. Percoll was initially diluted 1:1 (vol/vol) with filter-sterilized PBS and 10-ml aliquots were distributed into Sorvall 15-ml centrifuge tubes. Gradients were established by centrifuging tubes at 17,800 rpm in a Sorvall RC26+ centrifuge for 30 min at 4°C in a Sorvall SS-34 rotor. Sediments were then overlaid on top of the gradient and spun again for 15 min at 4,780 rpm in a swinging bucket rotor (Sorvall HS-4). The total supernatant from gradients was filtered through a 3 μm polycarbonate filter (Millipore) under low vacuum (<5 psi; 1 psi = 6.89 kPa) and the filters were subsequently washed twice with ≈3 ml of PBS to remove any traces of Percoll. Filters were sectioned in two pieces and transferred onto untreated 1-inch round glass slides used in SIMS analysis. Filter transfer of cell concentrates onto glass slides has been described (16). Briefly, 5 μl of PBS solution was spotted onto an untreated glass slide and 1/2 of the freshly prepared filter was then inverted sample side down onto the slide and air-dried. A graphite or cultured cell standard was later added to the other half of the glass slide not used in the cell transfer for calibration in the SIMS instrument. Slides were then treated with an EtOH dehydration series (50%, 75%, and 100% EtOH), dried, and stored at −20°C for later hybridization.
Whole-Cell FISH Assays.
Hybridization and wash buffers were made as described (4, 13). Twenty microliters of hybridization buffer containing 30% formamide was added to cell concentrates immobilized on 1-inch round glass slides. Cy-3 or FITC-labeled oligonucleotide probes were added to the hybridization buffer to a final concentration of 5 ng/μl. Oligonucleotide probes targeting the bacterial Desulfosarcina–Desulfococcus group (DSS658) and the ANME-2 -targeted (EelMSMX932), as well as general archaeal and bacterial probes (Ar915 and Eub338), have been described (7, 17). Newly designed oligonucleotide probes targeting the ANME-1 group (ANME1–862, 5′-GGCGGGCTTAACGGGCTTC-3′) are detailed in Table 1 and Table 2.
The hybridization mixtures on the slides were covered with a coverslip and incubated for 1.5 h at 46°C in a premoistened chamber. After hybridization, slides were transferred into preheated wash buffer and incubated for an additional 15 min at 48°C. Samples were treated with a dilute 4′,6-diamidino-2-phenylindole (DAPI) solution (5 μg/ml) for 1 min, rinsed in distilled water, and air-dried in the dark. To preserve the sample for SIMS analysis, cells were visualized under epifluorescence using an 80× dry objective (Leitz) without the addition of a mounting medium.
Mapping of Cell Location for SIMS Analysis.
Glass slides containing adequate cell densities (minimum 1–2 targets per field) were scored with an “X” through the center of the slide by using a diamond knife (for greater ease in locating target cells in the SIMS instrument), and target cells were then mapped in reference to the score mark. Digital images of individual cells and cell aggregates hybridizing with group specific oligonucleotide probes were captured by using a Spot SP100 cooled, charge coupled device camera (Diagnostic Instruments, Sterling Heights, MI). Images were recorded by using both epifluroescence and phase microscopy with 80× and 10× objectives. Images were then analyzed in ADOBE PHOTOSHOP 6.0, and potential targets for analysis with the SIMS instrument were located and highlighted on the image.
SIMS Analysis.
Carbon isotopic compositions (12C/13C) of individual cells and cell aggregates previously identified with FISH were determined by using the UCLA CAMECA IMS 1270 ion microprobe as described (13). Secondary C ions were ejected from the surface of the target biomass by using a 10- to 15-μm cesium ion beam and analyzed by using a multicollector arrangement consisting of two electron multipliers (12C13C− detected on-axis and 12C
ions were ejected from the surface of the target biomass by using a 10- to 15-μm cesium ion beam and analyzed by using a multicollector arrangement consisting of two electron multipliers (12C13C− detected on-axis and 12C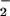 detected off axis). For this study, a relatively low intensity primary Cs+ beam was used (0.03 nanoampere; nA) to allow for data collection from single ANME-1 rods. Instrumental mass fractionation was measured by the analysis of graphite powder (USGS 24–4) dried onto a glass slide. Single cells and cell clusters of Pyrobaculum and Methanosarcina species were additionally analyzed by SIMS to determine an appropriate correction to account for any matrix effects arising from differences between the graphite standard and cell biomass. Average 13C values of individual cultured cells obtained with SIMS were compared with the bulk δ13C composition of the same culture determined by using conventional gas source mass spectrometry (C. H. H., unpublished data). In this case, a +5‰ correction was applied to account for matrix effects.
detected off axis). For this study, a relatively low intensity primary Cs+ beam was used (0.03 nanoampere; nA) to allow for data collection from single ANME-1 rods. Instrumental mass fractionation was measured by the analysis of graphite powder (USGS 24–4) dried onto a glass slide. Single cells and cell clusters of Pyrobaculum and Methanosarcina species were additionally analyzed by SIMS to determine an appropriate correction to account for any matrix effects arising from differences between the graphite standard and cell biomass. Average 13C values of individual cultured cells obtained with SIMS were compared with the bulk δ13C composition of the same culture determined by using conventional gas source mass spectrometry (C. H. H., unpublished data). In this case, a +5‰ correction was applied to account for matrix effects.
Molecular-Isotopic Analyses of Archaeal Lipids.
Molecular-isotopic analyses of archaeal lipids were performed according to protocols reported elsewhere (2–4, 13).
Results
Oligonucleotide Probe Design for the Archaeal ANME-1 Group.
A probe targeting the ANME-1 group was designed by using available archaeal rRNA sequences including those originating from methane-rich marine sediments. The ANME-1 specific probe (ANME1–862) targeted region 862–879 within the 16S rRNA gene (Escherichia coli numbering system). Relative to the ANME-2 group and most other Archaea, it contained two or three centrally located mismatches at positions 870, 875, and 876. The newly designed probe was used to further characterize the archaeal community structure and potential associations between ANME-1 and other bacteria. Dual-label FISH experiments using the ANME-1 targeted probe and the less specific EelMSMX932 probe revealed that in sediment samples containing both EelMSMX932-positive cocci and rods, only the rod-shaped Archaea hybridized strongly with the ANME1–862 probe.
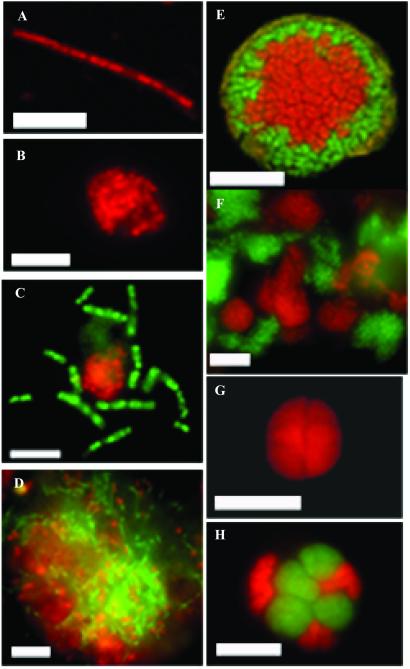
FISH surveys of ANME-1 cells in seven different ERB seep cores containing 13C-depleted lipid biomarkers revealed an abundance of segmented ANME-1 rods in addition to ANME-2 and ANME-2/Desulfosarcina aggregations (Fig. 1). These rods existed singly, as individual rectangular segments 1–2 μm in length; as short segmented rods (5–10 μm); or as long, segmented chains reaching lengths >70 μm (Fig. 1A). ANME-1 rods were also observed in mat-like aggregations, sometimes as tightly associated monospecific clusters (Fig. 1B). In addition to these aggregations, the archaeal ANME-1 cells occasionally formed associations with Bacteria, including the sulfate-reducing Desulfosarcina (Fig. 1 C and D). The ANME-1/bacteria associations were less organized than those described for ANME-2/Desulfosarcina aggregates (7, 13). ANME-1 aggregates were most frequently found in randomized, mat-like structures (Fig. 1D) rather than the organized, layered associations observed for ANME-2 archaea (Fig. 1E).
Figure 1.
Individual cells and cell aggregates of ANME-1 and ANME-2 archaea from ERB sediments, visualized with fluorescent-labeled oligonucleotide probes. Unless otherwise indicated, scale bar represents 5 μm. (A) An ANME-1 archaeal filament stained with Cy-3 labeled ANME1–862 probe, portraying the characteristic features of the ERB ANME-1 group including a segmented ultrastructure and square shaped cell termini. (B) Tightly associated cluster of ANME-1 rods, stained with the same ANME-1 oligonucleotide probe. (C) Color overlay of archaeal ANME-1 rods visualized with the ANME1–862 probe labeled with fluorescein (in green), and Desulfosarcina spp. stained with the DSS_658 probe labeled with Cy-3 (in red). (D) Large archaeal ANME-1/sulfate-reducing Desulfosarcina aggregate, showing an apparently random association of the two groups. Scale bar = 10 μm. (E) Color overlay of a layered ANME-2/DSS aggregate showing a core of ANME-2 Archaea (hybridized with EelMSMX932 probe), surrounded by sulfate-reducing Desulfosarcina (hybridized with DSS658 probe) imaged by laser scanning confocal microscopy. (F) Large aggregation of loosely associated, individual microcolonies of ANME-2 (EelMSMX932_cy3), and Desulfosarcina (DSS658_FITC). (G) Monospecies aggregate of archaeal ANME-2 (EelMSMX932_cy3) not affiliated with a bacterial partner. (H) Color overlay documenting an association of the ANME-2 archaea with a bacterial partner not affiliated with the sulfate-reducing Desulfosarcina. The aggregate was simultaneously hybridized with three oligonucleotide probes, including two FITC labeled probes for the ANME-2 (EelMSMX932) and Desulfosarcina (DSS658), in combination with a Cy-3 labeled probe for bacteria (EUB338). In this aggregate, microcolonies of the ANME-2 were stained with FITC, but the bacterial Desulfosarcina were not detected. Instead, clusters of an unidentified Bacteria (in red), hybridizing with the general bacterial probe labeled with Cy-3, were associated with the archaeal ANME-2 cells (stained in green).
ANME-2/Desulfosarcina aggregates from ERB sediments were usually highly organized structures, with diameters as great as 150 μm (Fig. 1E). A small percentage of ANME-2/Desulfosarcina aggregates also formed more random associations, with individual ANME-2 and Desulfosarcina cells juxtaposed against one another or loosely associated as separate microcolonies within a larger exopolysaccharide matrix (Fig. 1F). ANME-2 aggregates devoid of bacterial partners were also frequently observed (Fig. 1G). In a number of the samples screened from different seep sites in the ERB, monospecific clusters of ANME-2 were observed in samples that also contained ANME-2/Desulfosarcina spp consortia, and ANME-1 filaments. The tightly packed ANME-2 monospecific aggregates tended to have a pseudosarcina-like morphology, and were on average between 4–10 μm in diameter.
We also found evidence for the association of other bacterial types with ANME-2 archaea. A combination of three oligonucleotide probes (including fluorescein-labeled ANME-2-targeted EelMSMX932, fluorescein-labeled Desulfosarcina/Desulfococcus DSS658 probe, and Cy-3-labeled Bacteria probe, Eub338), revealed archaeal/bacterial aggregates containing ANME-2 archaea and an unidentified bacterial partner (Fig. 1H). With this approach, we were able to distinguish bacterial cells related to Desulfosarcina spp. (those cells hybridizing with the green-fluorescing DSS658 and the red-fluorescing Eub338), from other types of Bacteria (cells staining only in red). In contrast to the ANME-2/Desulfosarcina consortia described in previous reports (4, 7, 13), Fig. 1H illustrates a different association between the archaeal ANME-2 and an unidentified bacterial coccus.
δ13C Values for the Archaeal ANME-1 Group: Analysis of Single Cells Using FISH–SIMS.
FISH–SIMS analyses were conducted and the δ13C values for individual and aggregated ANME-1 rods were determined. By using the EelMSMX932 probe in combination with the Desulfosarcina/Desulfococcus probe (DSS658), ANME-1 and ANME-2 cells and cell aggregates were identified (based on probe binding and cell morphology) for SIMS analysis. Microbial cells from core PC-45 originated from methane-seep sediments (3–5 cm) underlying a dense community of C. pacifica clams. These sediments had low oxidation-reduction potentials, with H2S concentrations of ≈2.3 mM, and contained diverse archaeal and bacterial lipid biomarkers depleted in 13C, to δ values of −104‰. Prominent archaeal biomarkers included archaeol, sn-2- and sn-3-hydroxyarchaeol, a tentatively identified sn-2/sn-3-dihydroxyarchaeol, and crocetane. Isotopically depleted mono- and diethers, likely of bacterial origin, were also present. Sediments from push core PC-21 were sampled from a core depth of 7–10 cm at a different ERB seep site displaying active methane venting. Like PC-45, this sample also contained strongly depleted archaeal biomarkers (δ13C to −107‰) and 13C-depleted DIC (−27 versus −21.7 per mil at a control site) suggestive of methane oxidation. The δ13C value for dissolved methane in this sample (−49.5‰) was similar to the δ13C average recorded for methane from a number of seeps within ERB (C. Paull and B. Ussler, personal communication).
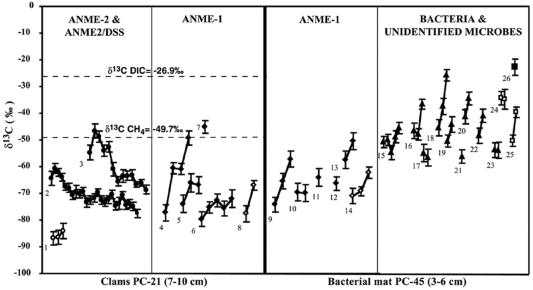
By using FISH–SIMS, profiles of δ13C values for 10 ANME-1 related cells and cell aggregates and three ANME-2 associated aggregates (from PC-45 and PC-21) were determined. Values of δ13C for ANME-1 cells (as low as −83 per mil, Fig. 2) were equal to or less than that of methane in the same sediments (Fig. 3). These ANME-1 δ13C cell carbon values are comparable to those found in the Methanosarcinales-related ANME-2 group (ref. 13; Fig. 2), suggesting that the ANME-1 group is also capable of anaerobic consumption of methane. The cell-to-cell variation of δ13C values recorded for ANME-1 was large, with greater than 30‰ variation between individual ANME-1 cells. There was no significant difference in δ13C values for ANME-1 cells recovered from within different ERB methane-seep locations (samples PC-45 and PC-21). Additionally, the 13C values obtained for individual ANME-1 rods and filaments were similar to those recorded for mono-specific ANME-1 aggregates (Figs. 2 and 3). This finding indicates that observed values were not an artifact of probing single cells, a methodological concern in the newly developed FISH–SIMS technique (13).
Figure 2.
Ion microprobe measurements of δ13C profiles for individual cells and cell aggregates recovered from methane-seep samples underlying clams or bacterial mats (PC-21 and PC-45). The x axis represents the time course Cs+ ion beam exposure for each individual cell profile. Individual cell profiles are indicated by a line connecting the δ13C values measured over time during Cs+ ion-beam exposure. Dashed lines show δ13C values for DIC and methane in sample PC-21 as indicated. (○) Mono-species ANME-2 aggregate (no. 1). (●) ANME-2/DSS aggregates (nos. 2 and 3). (⧫) Individual ANME-1 rods (nos. 4–7 and 9–13). (⋄) ANME-1 rod aggregates (nos. 8 and 14). (σ) Bacterial filaments hybridized with general bacterial oligonucleotide probe Eub338 (nos. 15–23). (□) Unidentified microbial aggregate stained with DAPI (nos. 24 and 25). (■) Diatom frustule (no. 26). The analytical precisions shown (1σ) are appropriate within each depth profile, but do not account for the uncertainty in the calibration of the y axis (±5‰ in this case).
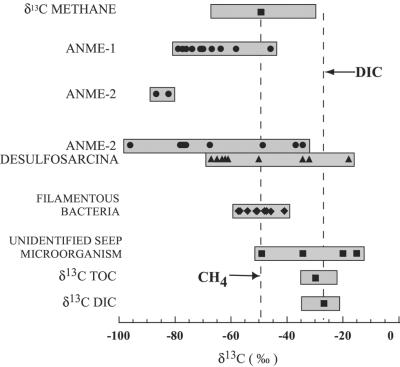
Figure 3.
Bar graph comparing the range of δ13C values measured for individual methanotrophic archaea, sulfate reducing bacteria, or other seep-associated microorganisms as measured by SIMS. Individual points represent either the initial SIMS values recorded for single cells or cell aggregates (ANME-1, ANME-2 alone, bacterial rods, and Desulfosarcina shells), or the eventual value obtained during depth profiling (ANME-2 cores). The plotted SIMS values are pooled from this study with those of a published report (13). Values of δ13C for methane, dissolved inorganic carbon (DIC), and total organic carbon (TOC) all originate from samples collected at the ERB during the 1999 ERB cruise (C. Paull and B. Ussler, personal communication). Bars represent the range of values measured for the different cell types, methane, DIC and TOC from ERB methane seeps. The methane data point corresponds to the pore-water methane concentrations measured in sample PC-21, with the range of ERB methane δ13C values representing all those measured during the 1999 ERB cruise.
ANME-2 and ANME-2/Desulfosarcina cell clusters were also depleted in 13C (with δ13C values as low as −92‰) confirming earlier findings (13). As reported, δ13C profiles progressing from the exterior to the interior of ANME-2/Desulfosarcina aggregates showed an isotopic trend. The central ANME-2 core was significantly more depleted in 13C than the outer, Desulfosarcina shell (Figs. 1–3). This isotopic pattern was unique to the ANME-2/Desulfosarcina consortia, and was not observed in monospecific aggregates of ANME-2 or in any analyses of ANME-1 archaea (Fig. 3). FISH–SIMS analysis of rods and rod clusters (ANME-1 and bacterial rods), always showed the most 13C-depleted values initially. Heavier values were observed as the cell or cells were sputtered away, a result consistent with increasing contributions from background organic material (δ13C of TOC = −30.8‰), possibly exaggerated by partial sample charging.
13C Values of Other Bacteria.
The FISH–SIMS approach was also used to measure the δ13C values of individual bacterial cells and unidentified microbial aggregates. Large filamentous bacteria (approximately 80 μm length and 2–3 μm wide) identified with oligonucleotide probe Eub338, were recovered from the same sediments harboring abundant methane-oxidizing Archaea (sample PC-45). Values of δ13C for these bacterial filaments ranged from −41 to −57‰ (n = 6; Fig. 2). In addition, two unidentified microbial aggregates (stained with DAPI) from the same sample and from sample PC-21 were similarly depleted in 13C (−35 to −49‰). These δ values are significantly more depleted than the average δ13C value for plankton derived organic carbon (−22 to −20‰; ref. 18), including values obtained with SIMS for a partially degraded diatom frustule (δ13C = −22‰) in the PC-45 sample (Fig. 2). Corresponding with the microbial community depletion in 13C, measurements of δ13C of total organic carbon (TOC) from the PC-21 sample yielded also significantly lower values of −30.8‰ compared with a control site (−23.5‰; C. Paull, personal communication).
Discussion
Recent studies have provided convincing evidence for the involvement of the methanogen-related ANME-2 archaeal group in AOM at marine methane seeps (7, 13). In a study of methane-seep microbes from Hydrate Ridge, Oregon, over 90% of the archaeal and sulfate-reducing bacterial biomass was reportedly caused by ANME-2/Desulfosarcina spp. consortia (7). Based on these findings, it was concluded that these consortia are principally responsible for anaerobic methanotrophy at methane seeps. Other reports indicate the possibility of additional methane-oxidizing archaeal types (4, 9, 15, 19), but these suggestions are based mainly on circumstantial evidence.
Our data indicate that the ANME-2 group is probably involved in more diverse activities and associations than originally supposed. ANME-2 archaea appear to exist as monospecific aggregates in some circumstances (Fig. 1G). The monospecific ANME-2 aggregates are as depleted in 13C as the ANME-2/bacterial consortia, suggesting that they are actively involved in AOM. ANME-2 aggregations can also exist in consortia with bacterial partners other than the Desulfosarcina species that are targeted by the DSS658 oligonucleotide probe. These data suggest that ANME-2 archaea are not obligately associated with Desulfosarcina species. Although the non-Desulfosarcina bacteria have not been identified, their physical association suggests a tight metabolic coupling, as has been proposed for the ANME-2/Desulfosarcina consortia. These unidentified bacteria may also be sulfate-reducers, serving the same metabolic role as Desulfosarcina spp. Bacterial 16S rRNA gene surveys from methane-seep areas have identified delta proteobacterial groups that appear to be specifically associated with methane-seep communities (Eel-1 and Eel-2 groups; ref. 4). These bacterial groups comprised a significant percentage of total sequence types recovered from multiple sediment samples within ERB and were also reported to co-occur with the ANME groups in methane-rich sediments from Guaymas Basin (19). These bacterial groups may also form partnerships with ANME-2 archaea.
Although previous microscopic surveys of methane-rich sediments from Hydrate Ridge failed to detect ANME-1 archaea, our observations from ERB sediments using a general archaeal probe (Ar915) suggested the presence of morphologically diverse Archaea (data not shown). FISH assays using the newly designed ANME-1 targeted oligonucleotide probe revealed that this group was abundant in a variety of ERB methane-seep samples, including sediments where ANME-1 16S rRNA genes were first discovered and recovered in high numbers (3). The distribution and abundance of the ANME-1 group within ERB, as well as their appearance at other methane-seep sites (10, 11, 19, 20), strongly suggest that ANME-1 archaea are central players in AOM.
Before this report, ANME-1 archaea had not been microscopically identified. However, one earlier study reported carbonate-associated, mat-forming microorganisms from the Black Sea that were morphologically similar to the ANME-1 filaments we report here. The rod-shaped, carbonate-associated microbes were reported to oxidize 14C-labeled methane under anoxic conditions (21). Further, it was demonstrated that these carbonate-associated microbial mats were moderately depleted in 13C (−58.2‰), and contained isotopically light (−75‰) archaeal biomarkers (9). These earlier studies, combined with the observations reported here, suggest that the carbonate-associated archaea from Black Sea microbial mats may belong to the ANME-1 group, potentially mediating AOM in these and similar environments. In addition to methane seeps, 16S rRNA gene surveys have recovered the ANME-1 group from a number of other methane-rich marine habitats, including two sediment-covered hydrothermal vent areas (19, 20), methane hydrates (11), and shallow marine sediments (10). This finding suggests that the ANME-1 group is an important contributor to anaerobic methane consumption in diverse anoxic marine environments.
FISH–SIMS analysis of ANME-1 archaea indicates their involvement in methane consumption. All ANME-1 cells were substantially depleted in 13C (Figs. 2 and 3). In addition, individual aggregates of archaeal ANME-2/Desulfosarcina consortia and monospecific clusters of ANME-2 were also extremely depleted in 13C, corroborating earlier findings which linked this specific archaeal/bacterial consortium to methane oxidation (13). Combined with previous work, our study now provides strong evidence that at least two distinct groups of Archaea (ANME-1 and ANME-2) are capable of assimilating methane under anoxic conditions (Fig. 3).
Most recent AOM research has focused on the necessity of syntrophic coupling that involves more than one microbial group. Both laboratory and field-based experiments have suggested the interaction of methanogen-relatives with sulfate-reducing bacteria in AOM (12, 22). The data we report here suggest that single archaeal cells and monospecies aggregates can oxidize methane anaerobically, without a tightly associated syntrophic partner. Although we observed ANME-1 cells in association with bacteria, these archaea more frequently occurred as monospecific mats, or single filaments. The possibility that monospecific aggregations and single cells are artificially generated during sample preparation must also be considered. To test for these potential artifacts, we examined ERB sediment microbes by means of FISH in the absence of potentially disruptive treatments (e.g., without sonication or Percoll gradient separation). Monospecific ANME-1 and ANME-2 cell aggregations were also observed in these samples, suggesting that bacteria-free ANME cells are a naturally occurring phenomenon. Despite the absence of a tight bacterial association, ANME-1 aggregates were highly depleted in 13C (on average −75‰), supporting their involvement in anaerobic methane assimilation. In contrast to the ANME-2 group, however, ANME-1 archaea appear less dependent on the activities of a closely associated bacterial partner.
For a single, unassociated microorganism to catalyze AOM, it would need to internally couple methane oxidation to the reduction of a suitable electron acceptor (e.g., sulfate). No cultured, sulfate-reducing bacterium or methanogen has ever been shown to be capable of anaerobic growth on methane as a sole carbon source. However, some isolates can co-oxidize small amounts of methane when growing on other carbon sources, indicating they may possess the enzymatic machinery needed for AOM (22, 23). With so few microbial representatives in pure culture, the possibility that an archaeon (e.g., ANME-1) has developed a more effective strategy for coupling methane oxidation to sulfate reduction should not be discounted. Physiological studies of some methanogenic isolates have shown that lithotrophic growth on CO2 and molecular hydrogen can be accompanied by reduction of elemental sulfur to sulfides (24, 25). The archaeon Archaeoglobus fulgidus, which primarily derives its energy from sulfate reduction, contains some of the metabolic machinery common to methanogenic pathways (e.g., methanofurans and coenzyme F-420). A. fulgidus is also capable of producing minor amounts of methane during growth (26, 27). It is possible that ANME-1 archaea may employ elements of both sulfur and methane-based energetic pathways, oxidizing methane independently of a sulfate-reducing partner. Genomic approaches (28, 29) should help to identify some of the biochemical pathways used by anaerobic methanotrophic archaea. Several genes previously known to exist only in methanogens have already been found within the genomes of ANME-1 and ANME-2 methanotrophic archaea (P. Richardson, S. Hallam, and E.F.D., unpublished data).
Diverse anaerobic methane-oxidizing microorganisms may contribute significantly to the carbon flux in anaerobic microbial communities fueled by methane. Carbon isotopic compositions of individual bacterial cells resembling large, filamentous, sulfide-oxidizing microorganisms, as well as other morphotypes thought not to be directly linked to the anaerobic oxidation of methane, exhibited moderately 13C-depleted cell carbon, approaching −61‰ (Fig. 3). These findings suggest that some members of the seep assemblage use carbon sources originally derived from methane. Possible sources of isotopically depleted carbon for nonmethanotrophic microbes in seep sediments include CO2 (δ 13C of DIC in sample = −27‰), carbon intermediates generated from AOM such as formate or acetate (14), or recycled 13C-depleted archaeal and/or sulfate-reducing bacterial biomass. Low δ13C-values (−61 to −53‰) have also been reported for sulfide-oxidizing bacterial mat total biomass overlying gas hydrate-containing sediments and in seeps at the Florida Escarpment (30, ‖). The methane-derived, 13C-depleted signatures in nonmethanotrophic microbes indicate the influence of anaerobic methanotrophy on carbon flux at multiple trophic levels. The data suggest that anaerobic methanotrophy mediated by ANME-1 and ANME-2 archaea (and possibly other groups) plays a crucial role in the establishment and success of methane-seep communities by converting methane into more readily accessible carbon and energy substrates.
Although similar anaerobic methane cycling appears to occur in sub-seafloor environments (31, 32), the microbes living in these habitats have not yet been well characterized. Available evidence suggests that in these diffusion-limited subseafloor environments, resident microbes may differ substantially in community composition compared with those found at cold seeps (15). It is likely that a greater diversity of microbial species and interactions remain to be discovered and described, especially in habitats that are biogeochemically distinct from methane seeps, such as those found in the deep subsurface.
Acknowledgments
We thank Steve Hallam, Chris Preston and Peter Girguis for advice assistance, and Bill Sullivan for use of the University of California Santa Cruz Confocal Microscope Facility. We also thank Tori Hoehler and an anonymous reviewer for helpful comments and suggestions. This work was supported by grants from the David and Lucile Packard Foundation, the Penn State Astrobiology Research Center, and the University of California, Los Angeles, (UCLA) Center for Astrobiology, National Aeronautics and Space Administration National Astrobiology Institute. The UCLA ion microprobe is partially supported by a grant from the National Science Foundation Instrumentation and Facilities Program.
Abbreviations
- AOM
anaerobic oxidation of methane
- FISH
fluorescent in situ hybridization
- SIMS
secondary ion mass spectrometry
- ERB
Eel River Basin
Footnotes
Gebruk, A., Lein, A. & Krylova, E. (2000) in 9th Annual Deep-Sea Biology Symposium, Galway, Ireland, p. 25.
References
- 1.Valentine D L, Reeburgh W S. Environ Microbiol. 2000;2:477–484. doi: 10.1046/j.1462-2920.2000.00135.x. [DOI] [PubMed] [Google Scholar]
- 2.Hinrichs K-U, Boetius A. In: Ocean Margin Systems. Wefer G, Billett D, Hebbeln D, Jørgensen B B, Schlüter M, van Weering T, editors. Heidelberg: Springer; 2002. , in press. [Google Scholar]
- 3.Hinrichs K-U, Hayes J M, Sylva S P, Brewer P G, DeLong E F. Nature (London) 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 4.Orphan V J, Hinrichs K-U, Ussler W, Paull C K, Taylor L T, Sylva S A, Hayes J M, DeLong E F. Appl Environ Microbiol. 2001;67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinrichs K-U, Summons R E, Orphan V J, Sylva S P, Hayes J M. Org Geochem. 2000;31:1685–1701. [Google Scholar]
- 6.Pancost R D, Sinninghe Damste J S, de Lint S, van der Maarel M J E C, Gottschal J C the Medinaut Shipboard Scientific party. Appl Environ Microbiol. 2000;66:1126–1132. doi: 10.1128/aem.66.3.1126-1132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boetius A, Ravenschlag K, Schubert C J, Rickert D, Widdel F, Gieseke A, Amann R, Jorgensen B B, Witte U, Pfannkuche O. Nature (London) 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 8.Elvert M, Suess E, Greinert J, Whiticar M J. Org Geochem. 2000;31:1175–1187. [Google Scholar]
- 9.Thiel V, Peckmann J, Richnow H, Luth U, Reitner J, Michaelis W. Marine Chem. 2001;73:97–112. [Google Scholar]
- 10.Thomsen T R, K, Finster K, Ramsing N B. Appl Environ Microbiol. 2001;67:1646–1656. doi: 10.1128/AEM.67.4.1646-1656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanoil B D, Sassen R, La Duc M, Sweet S, Nealson K. Appl Environ Microbiol. 2001;67:5143–5153. doi: 10.1128/AEM.67.11.5143-5153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoehler T M, Alperin M J, Albert D B, Martens C S. Global Biogeochem Cycles. 1994;8:451–463. [Google Scholar]
- 13.Orphan V J, House C, Hinrichs K-U, McKeegan K, DeLong E F. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 14.Sørensen K, Finster K, Ramsing N. Microbial Ecol. 2001;42:1–10. doi: 10.1007/s002480000083. [DOI] [PubMed] [Google Scholar]
- 15.Bian L Q, Hinrichs K-U, Xie T M, Brassell S C, Beck J P, Iversen N, Fossing H, Jørgensen B B, Hayes J M. Geochem Geophys Geosys. 2001;2:2000. .GC000112. [Google Scholar]
- 16.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amann R I, Ludwig W, Schleifer K-H. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossman E. In: Manual of Environmental Microbiology. Hurst C, editor. Washington, DC: Am. Soc. Microbiol.; 1997. pp. 565–576. [Google Scholar]
- 19.Teske A, Hinrichs K-U, Edgcomb V, Gomez A d V, Kysela D, Sylva S P, Sogin M L, Jannasch H W. Appl Environ Microbiol. 2002;68:1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takai K, Horikoshi K. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pimenov N, Rusanov I, Poglazova M, Miyushina L, Sorokin D, Khmelenina V, Trotsenko Y. Mikrobiologiya. 1997;66:354–360. [Google Scholar]
- 22.Zehnder A J B, Brock T D. J Bacteriol. 1979;137:430–432. doi: 10.1128/jb.137.1.420-432.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis J, Yarborough H. Chem Geol. 1966;1:137–144. [Google Scholar]
- 24.Stetter K, Gaag G. Nature (London) 1983;305:309–311. [Google Scholar]
- 25.Jeanthon C, L'Haridon S, Reysenbach A L, Vernet M, Messner P, Sleytr U B, Prieur D. Int J Syst Bacteriol. 1998;48:913–919. doi: 10.1099/00207713-48-3-913. [DOI] [PubMed] [Google Scholar]
- 26.Beeder J, Nilsen R, Rones J, Torsvik T, Lein T. Appl Environ Microbiol. 1994;60:1227–1231. doi: 10.1128/aem.60.4.1227-1231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White R. J Bacteriol. 1988;170:4594–4597. doi: 10.1128/jb.170.10.4594-4597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Béjà O, Aravind L, Koonin E, Suzuki M, Hadd A, Nguyen L, Jovanovich S, Gates C, Feldman R, Spudich J, Spudich E, DeLong E F. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 29.Béjà O, Suzuki M, Koonin E, Aravind L, Hadd A, Nguyen L, Villacorta R, Amjadi M, Garrigues C, Jovanovich S, Feldman R, DeLong E F. Environ Microbiol. 2000;2:516–529. doi: 10.1046/j.1462-2920.2000.00133.x. [DOI] [PubMed] [Google Scholar]
- 30.Paull C, Chanton J, Neumann A, Coston J, Martens C. Palaios. 1992;7:361–375. [Google Scholar]
- 31.D'Hondt S, Rutherford S, Spivak A. Science. 2002;295:2067–2070. doi: 10.1126/science.1064878. [DOI] [PubMed] [Google Scholar]
- 32.Borowski W, Paull C, Ussler W. Geology. 1996;24:655–658. [Google Scholar]