Abstract
Binding of CBF3, a protein complex consisting of Ndc10p, Cep3p, Ctf13p, and Skp1p, to the centromere DNA nucleates kinetochore formation in budding yeast. Here, we investigate how the Ctf13p/Skp1p complex becomes competent to form the CBF3-centromere DNA complex. As revealed by mass spectrometry, Ctf13p and Skp1p carry two and four phosphate groups, respectively. Complete dephosphorylation of Ctf13p and Skp1p does not interfere with the formation of CBF3-centromere DNA complexes in vitro. Furthermore, deletion of corresponding phosphorylation sites results in viable cells. Thus, in contrast to the current view, phosphorylation of Ctf13p and Skp1p is not essential for the formation of CBF3-centromere DNA complexes. Instead, the formation of active Ctf13p/Skp1p requires Hsp90. Several lines of evidence support this conclusion: activation of heterologous Ctf13p/Skp1p by reticulocyte lysate is inhibited by geldanamycin and Hsp90 depletion. skp1 mutants exhibit growth defects on media containing geldanamycin. A skp1 mutation together with Hsp90 mutations exhibits synthetic lethality. An Hsp90 mutant contains decreased levels of active Ctf13p/Skp1p.
The Saccharomyces cerevisiae kinetochore (for review see ref. 1) is composed of more than 30 different proteins, many of which have been discovered recently (2–11). Several of these proteins have homologues in mammalian and/or Schizosaccharomyces pombe kinetochores, suggesting that the principles of kinetochore function are conserved from yeast to mammals (1). With the exception of Cbf1p, none of the described kinetochore proteins that has its own DNA-binding site can be localized at the centromere when CBF3, a protein complex that consists of Ndc10p, Cep3p, Ctf13p, and Skp1p, and specifically binds to centromere DNA, is inactivated (see refs. 1–11). Thus, assembly of the CBF3-centromere DNA complex is an absolute prerequisite for the formation of the S. cerevisiae kinetochore, and it is of particular interest whether and how the assembly of this complex is regulated.
The formation of protein complexes and protein DNA complexes can be mediated by chaperones. Whereas many chaperones act in nascent protein folding, Hsp90 is known to modulate the structure of fully synthesized proteins and facilitate protein–protein interactions. Hsp82p and Hsc82p are the S. cerevisiae members of the Hsp90 protein family that is highly conserved in all eukaryotes. Again, Hsp82p and Hsc82p seem not to be essential for de novo folding of proteins. Instead, they act on a subset of proteins that require specialized help to reach their active conformation (12). Currently, only a few in vivo substrates of Hsp82p/Hsc82p have been described for S. cerevisiae. These include the kinases Ste11p and Gcn2p (13) as well as the phosphatase calcineurin (14).
Here, we investigate the formation of functional Ctf13p/Skp1p, an essential component of the CBF3-centromere DNA complex that is fundamental for kinetochore existence in S. cerevisiae. We provide substantial evidence that formation of functional Ctf13p/Skp1p requires Hsp90 but does not require phosphorylation.
Materials and Methods
Growth Media and Strain Construction.
Yeast strains and plasmids are described in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Details of plasmid construction are available on request. The skp1–51 allele was generated by PCR-mediated mutagenesis and selected as described (2). A cassette that contained HIS3 cloned into the 3′ noncoding sequences of the skp1–51 allele was used to direct the integration of skp1–51 into the SKP1 locus of YJL20, yielding YJL106.
Protein Expression and Purification.
HisCtf13p/HisSkp1p was copurified from S. cerevisiae as described (15). pOS152 (2μ LEU2 HisCTF13) and pOS253 (2μ URA3 HisSKP1) were used to direct the expression of fusion proteins with MGH10SSGHIEGRH or MAH10 linked to the N-terminal methionine of Ctf13p and Skp1p, respectively.
For HisCtf13p/HisSkp1p copurification from E. coli, pOS234 was constructed that allowed polycistronic translation of MGH10SSGHIEGRH-Ctf13p and MAH10-Skp1p. Expression and purification was performed as described (16). Ndc10p and Cep3p were purified from P. pastoris and E. coli, respectively, as described (15).
Plasmid pOS86 or pOS237 was used to transcribe and then translate Ctf13p or FlagSkp1p, respectively, in vitro in nuclease-treated rabbit reticulocyte lysate (RL; Promega) according to the manufacturer's instructions.
Mutation of Phosphorylation Sites in Ctf13p and Skp1p.
For Ctf13p, site-directed mutagenesis of pJL218 (ProtA-CTF13) was performed with the GeneEditor System from Promega. This procedure resulted in plasmid pOS643 that codes for Ctf13p with the amino acid sequence 216–229 deleted, as confirmed by DNA sequencing. To test for cell viability, YJL33 (ctf13∷TRP1) harboring plasmid pJL114 (CEN6 URA3 CTF13) was transformed with pOS643, and transformants were restreaked onto synthetic complete medium (SC) plates containing 5-fluoroorotic acid (5-FOA) selecting for cells that had lost pJL114.
For Skp1p, the NsiI-HindII fragment of pJL248 (SKP1) was replaced by an NsiI-HindII, PCR-derived fragment that codes for Skp1p sequences downstream of amino acid 64. The resulting plasmid, pJL428, expresses Skp1p with the amino acids 38–64 deleted. To test whether mutant Skp1p supports cell viability, YOS71 (skp1∷HIS3) harboring pOS221 (SKP1 URA3) was transformed with pJL428 and transformants were restreaked onto SC plates containing 5-FOA selecting for cells that had lost pOS221.
Synthetic Lethality.
Strain YJL106 (Matα skp1–51:HIS3) harboring plasmid pOS221 (SKP1 URA3) and strain ΔPCLDa (Mata hsp82∷LEU2 hsc82∷LEU2) harboring the hsp82(T101I) allele or the hsp82(G170D) allele on a pRS314-based plasmid were mated. After sporulation, haploids with a Leu+, Ura+, and His+ phenotype were selected, and cells that had both hsp82 and hsc82 disrupted were identified by their temperature-sensitive phenotype. To test for synthetic lethality, the cells were grown on 5-FOA for 4 days at 23°C to select for cells that had lost pOS221. The skp1–51/hsp82(G170D) double mutant that showed slow growth on 5-FOA was restreaked onto yeast extract/peptone/dextrose (YPD) plates and grown at 29°C for 3 days.
Immunodepletion, Immunoprecipitation, Heterodimer Formation, λ-Phosphatase Treatment, and Electrophoretic Mobility-Shift Assay (EMSA).
For depletion of Hsp90 from RLs, 40 μg of anti-Hsp90 antibody (SPA-830, StressGen Biotechnologies, Victoria, Canada) was bound to 10 μl of Protein G Sepharose. Anti-Hsp90 beads (5 μl) were incubated with 15 μl of RL for 3 h at 4°C. After removal of the beads, this step was repeated with a fresh 5 μl aliquot of anti-Hsp90 beads. For mock-depletion, 40 μg of unspecific IgG bound to 10 μl of Protein G Sepharose was used. For immunoprecipitation, 10 μl of anti-Hsp90 beads (see above) were incubated with 35 μl of in vitro translated, 35S-labeled Skp1p or Ctf13p for 3 h at 4°C, then washed and eluted with SDS sample buffer. For copurification of [35S]Ctf13p with HisSkp1p, 500 ng of HisSkp1p were added to [35S]Ctf13p (35 μl) and incubated for 2 h at 30°C. Subsequently, 30 μl of nitrilotriacetic acid (NTA)-agarose (Qiagen, Chatsworth, CA) were added and incubated for 3 h at 4°C. After washing, the NTA-agarose was eluted with 4% (vol/vol) SDS and 550 mM imidazole.
For dephosphorylation, 300 μl of ResourceQ-purified HisCtf13p/HisSkp1p (about 1 μg of each; ref. 15) was incubated with 2.4 units of λ-phosphatase (New England Biolabs) for 2 h at 30°C. To remove the phosphatase, the mix was incubated with 100 μl of NTA-agarose for 2 h at 4°C. Beads were washed and then eluted with 250 mM imidazole. EMSAs were performed as described (17) with the exception that EGTA was added to a final concentration of 5 mM if nuclease-treated RL was included in the assay.
Mass Spectroscopy.
For LC-MS, dephosphorylated or mock-treated HisCtf13p/HisSkp1p (see above) was incubated with 6 M guanidine-HCl and chromatographed on an RP-C4 HPLC column (2.1/50, Vydac, Hesperia, CA) in an acetonitrile gradient. After flow splitting, 10% of the samples were analyzed online by a Finnigan SSQ 7000 electrospray ionization (ESI) mass spectrometer in the positive mode. The remaining 90% of the sample was used for automated Edman degradation.
For phospho-peptide analysis, 500 ng of HisCtf13p or HisSkp1p were subjected to in-gel digestion by trypsin (18). Peptides were subjected to negative-ion electrospray mass spectroscopy in the precursor ion mode on an API III (Sciex, Thornhill, ON, Canada) or to positive-ion electrospray mass spectroscopy in the fragment ion mode on a Q-Tof (Micromass, Manchester, U.K.) as described (19).
Results
S. cerevisiae-Ctf13p/Skp1p Does Not Require Phosphorylation for CBF3-Centromere DNA Complex Formation in Vitro.
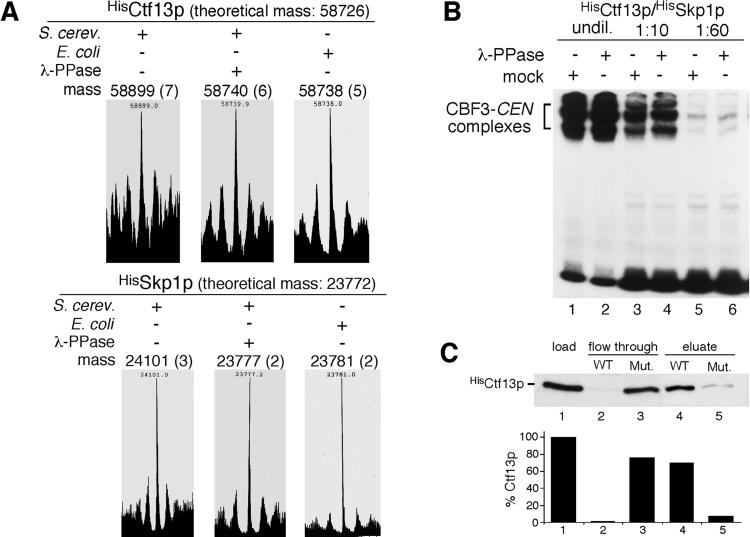
It was reported that Ctf13p expressed in insect cells requires phosphorylation to form CBF3-centromere DNA complexes in vitro (20). Therefore, we investigated the phosphorylation state of a Ctf13p/Skp1p heterodimer, purified from S. cerevisiae, by ESI mass spectrometry. This procedure revealed masses of 58,899 ± 7 Da and 24,101 ± 3 Da for Ctf13p and Skp1, respectively (Fig. 1A). After phosphatase treatment, HisCtf13p/HisSkp1p exhibited masses of 58,740 ± 6 Da and 23,777 ± 2 Da. Thus, dephosphorylation of Ctf13p and Skp1p resulted in a mass shift of 159 Da and 324 Da, respectively, which corresponds well with the loss of two and four phosphate residues. The theoretical average masses of unmodified HisCtf13p and HisSkp1p are 58,726 Da and 23,772 Da, respectively, considering that the proteins were lacking the N-terminal methionine, as had been revealed by automatic Edman degradation (data not shown). Thus, the masses observed for the dephosphorylated proteins were just 14 Da and 5 Da, respectively, above the theoretical average masses of unmodified HisCtf13p and HisSkp1p. These differences approach the accuracy of the method. In the case of HisCtf13p, it also may indicate acetylation or oxidation but not phosphorylation. Furthermore, the masses of E. coli-expressed HisCtf13p (58,738 ± 5 Da) and HisSkp1p (23,781 ± 2 Da) corresponded very well with the masses of dephosphorylated HisCtf13p and HisSkp1p obtained from S. cerevisiae (Fig. 1A). Therefore, we conclude that the phosphatase treatment resulted in complete dephosphorylation, and that the majority of Ctf13p contains two phosphate residues whereas the majority of Skp1p contains four. Some HisCtf13p preparations revealed a more heterogeneous mass distribution before dephosphorylation, which indicated that HisCtf13p in these preparations carried one or two phosphate residues (data not shown).
Figure 1.
Ctf13p/Skp1p phosphorylation is not essential for the formation of CBF3-centromere DNA complexes in vitro. (A) LC-MS of mock-treated or phosphatase-treated HisCtf13p and HisSkp1p purified from S. cerevisiae or untreated proteins purified from E. coli. The molecular masses of the proteins in Daltons after deconvolution of the spectra are shown. SDs are given in brackets. (B) λ-phosphatase or mock-treated HisCtf13p/HisSkp1p (1 ng each at the highest concentration and dilutions thereof as indicated) were incubated with HisNdc10p (60 ng), HisCep3p (15 ng), and 32P-labeled centromere DNA in a total volume of 30 μl and analyzed by EMSA. (C) The majority of the S. cerevisiae-derived preparation is composed of functional HisCtf13p/HisSkp1p. (Upper) HisCtf13p/HisSkp1p (100 ng each) was incubated with HisNdc10p (200 ng), HisCep3p (200 ng), and centromere DNA-affinity matrix containing wild-type (WT) or nonfunctional (Mut.) centromere DNA. After washing, the matrix was eluted with high salt as described (17). Fractions were analyzed by Western blotting using anti-Ctf13p antibody. (Lower) Quantification of Western analysis.
To test whether Ctf13p phosphorylation is required for CBF3-centromere DNA-complex formation, we incubated the dephosphorylated HisCtf13p/HisSkp1p heterodimer with purified Ndc10p, Cep3p, and CEN3 DNA and performed an EMSA. ATP was absent from this experiment, thus ruling out the possibility of rephosphorylation of Ctf13p/Skp1p. This result showed (Fig. 1B) that phosphatase treatment of Ctf13p/Skp1p does not interfere with the formation of CBF3-centromere DNA complexes in vitro. In principle, active Ctf13p/Skp1p might contain additional phosphate residues that were not removed by the phosphatase. If so, this active form should only comprise a small fraction of the total protein because it was not detected by mass spectrometry. To clarify this possibility, we incubated HisCtf13p/HisSkp1p in the presence of Ndc10p and Cep3p with a centromere DNA affinity matrix (Fig. 1C). Of the applied Ctf13p/Skp1p, 97% bound to the matrix, and 70% could be recovered. Only 24% bound to a control matrix that carried nonfunctional centromere DNA and 8% could be recovered. Therefore, the majority of the HisCtf13p/HisSkp1p preparation comprises active material. Taken together, our data strongly suggest that the phosphate groups detected on S. cerevisiae-Ctf13p and Skp1p are not required for the formation of CBF3-centromere DNA complexes in vitro.
Deletion of Ctf13p and Skp1p Phosphorylation Sites Does Not Interfere with Cell Viability.
To determine the position of phosphorylation sites, HisCtf13p/HisSkp1p were subjected to in-gel digestion with trypsin, and phospho-peptides were analyzed by mass spectroscopy. This procedure revealed three Ctf13p peptides with masses of 2,088.5, 2,168.7, and 2,248.7 Da (see Fig. 5A, which is published as supporting information on the PNAS web site), which correlates well with the masses of the tryptic Ctf13p peptide DVDVSGANSDENSSPSSTIK (amino acids 212–231) carrying one, two, and three phosphate groups, respectively. For HisSkp1p, a tryptic peptide with the mass of 3,532.0 Da was identified (Fig. 5B). This mass correlates well with the molecular mass of the tryptic Skp1p peptide NYLNDMHDSNLQNNSDSESDSDSETNHK (amino acids 31–58) carrying four phosphate groups. The identity of these peptides was confirmed by their fragmentation patterns (Fig. 5 C–E). Although the method applied does not guarantee detection of all phospho-peptides, the results are in agreement with Ctf13p carrying one or two and Skp1p carrying four phosphate residues.
To investigate the function of the phosphate groups, we deleted all possible phosphorylation sites within the identified regions by site-directed mutagenesis, in particular, the region S216–T229 of Ctf13p and the region D38–N64 of Skp1p. When proteins carrying either of these mutations were expressed on a CEN-based plasmid in a null background of the corresponding protein, no growth defect was detected. Furthermore, extracts prepared from these strains revealed no defect in CBF3-centromere DNA complex formation (data not shown). We conclude that the phosphate groups identified on Ctf13p and Skp1p (that most likely represent all phosphate residues on the purified proteins) are not required for the formation of the CBF3-centromere DNA complex.
Heterologously Expressed Ctf13p/Skp1p Requires NTP-Dependent Activation That Correlates with Ctf13p/Skp1p Heterodimer Formation.
In the following paragraphs, we will use the term “functional” as a state of Ctf13p/Skp1p that, when incubated with Ndc10p, Cep3p, and centromere DNA, results in formation of CBF3-centromere DNA complexes, as revealed by a gel mobility shift assay. We will use the term “activation” as a process that changes nonfunctional Ctf13p/Skp1p to functional.
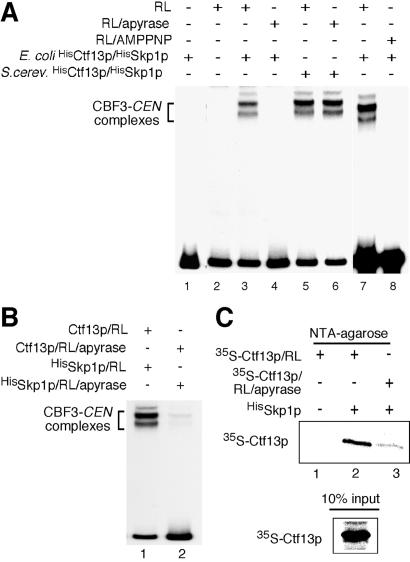
A purified mixture of HisCtf13p and HisSkp1p coexpressed in E. coli was nonfunctional (Fig. 2A, lane 1). Surprisingly, the mixture became functional after preincubation with rabbit RL (Fig. 2A, lanes 3 and 7). The activation in RL required NTP because preincubation of RL with apyrase or AMPPNP prevented activation of Ctf13p/Skp1p (Fig. 2A, lanes 4 and 8) In contrast, preincubation of active Ctf13p/Skp1p (from S. cerevisiae) with apyrase had no effect (Fig. 2A, lane 6).
Figure 2.
Rabbit RL activates HisCtf13p/HisSkp1p and promotes heterodimer formation in an ATP-dependent manner. (A) HisCtf13p/HisSkp1p (10 ng each) purified from E. coli were incubated with 7 μl of RL for 1 h at 30°C. After the addition of HisNdc10p (60 ng), HisCep3p (15 ng), and 32P-labeled centromere DNA, the samples were analyzed by EMSA. Where indicated, RL was preincubated with 1.5 units of apyrase or 10 mM AMPPNP for 15 min at 30°C before adding HisCtf13p/HisSkp1p. (B) Ctf13p and Skp1p have to be coincubated in ATP-containing RL to get activated. (Lane 1) HisSkp1 (100 ng) purified from E. coli was incubated with 3.5 μl of RL for 1 h at 30°C and subsequently combined with Ctf13p that had been synthesized in 3.5 μl of RL by in vitro translation. After further incubation (1 h at 30°C), the sample was mixed with Ndc10p (60 ng), Cep3p (15 ng), and 32P-labeled centromere DNA and analyzed by EMSA. (Lane 2) As described for lane 1, with the exception that the samples were treated with 0.75 units of apyrase for 15 min at 30°C before they were combined. (C) HisSkp1p purified from E. coli (500 ng) was added to [35S]Ctf13p that had been synthesized in RL by in vitro translation (35 μl). Thereafter, HisSkp1p was purified over NTA-agarose, and copurification of [35S]Ctf13p was analyzed by SDS/PAGE and autoradiography. Where indicated, the RL containing [35S]Ctf13p was treated with 6 units of apyrase for 1 h at 30°C before the addition of HisSkp1p.
We then produced Ctf13p by in vitro translation in RL and incubated HisSkp1p from E. coli with RL. When both samples were combined and incubated in the RL, Ctf13p/HisSkp1p became functional (Fig. 2B, lane 1). However, if the Ctf13p-RL and HisSkp1p-RL samples were treated with apyrase after the first incubation step but before combining the two samples, activation of Ctf13p/HisSkp1p was prevented (Fig. 2B, lane 2). Therefore, Ctf13p and Skp1p have to be present together in the RL to get activated. The above data did not reveal whether one protein mediates the activation of the other (and, if so, which is the activated one) or whether both get activated, possibly by forming a functional heterodimer. We favor the latter idea, because apyrase treatment of the RL not only inhibited activation of Ctf13p/Skp1p but also, at the same time, diminished heterodimer formation (Fig. 2C). [35S]methionine-labeled Ctf13p and HisSkp1p copurified only if they were coincubated in NTP containing RL (Fig. 2C, lane 2) but not if they were coincubated in apyrase-treated RL (Fig. 2C, lane 3). Thus, activation of Ctf13p/Skp1p correlates with heterodimer formation.
Hsp90 Is Involved in the Activation of Ctf13p/Skp1p in RL.
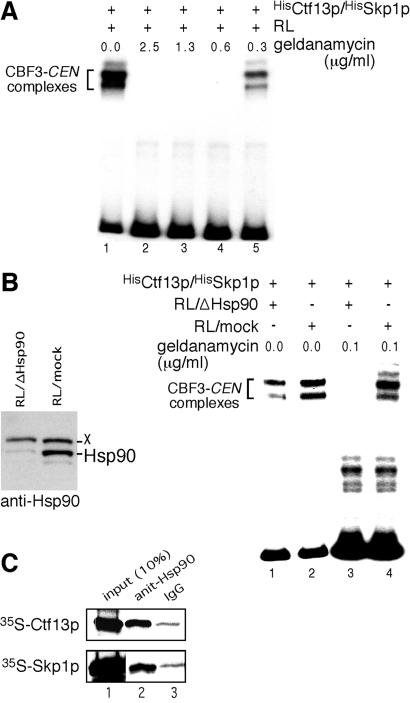
Phosphorylation plays no role for the activation of S. cerevisiae-derived Ctf13p/Skp1p (see above) and activated HisCtf13p/HisSkp1p reisolated from RL was not inactivated by phosphatase treatment (data not shown). Thus, chaperone-mediated protein folding became the prime candidate for the molecular mechanism that underlies the NTP-dependent Ctf13p/Skp1p activation. When RLs were pretreated with geldanamycin, a specific inhibitor of Hsp90 (21), HisCtf13p/HisSkp1p activation was inhibited (Fig. 3A). To support this data, we depleted a major fraction of Hsp90 from RL (Fig. 3B) and checked for HisCtf13p/HisSkp1p activation. As seen in Fig. 3B, depletion of Hsp90 greatly diminished (2.3-fold) the activation of HisCtf13p/HisSkp1p (compare lanes 1 and 2). If geldanamycin was added to the depleted RL at a concentration of 0.1 μg/ml, activation of HisCtf13p/HisSkp1p was completely prevented (Fig. 3B, lane 3). In contrast, 0.1 μg/ml geldanamycin in mock-depleted RL had no detectable effect (Fig. 3B, lane 4). To further support the role of Hsp90, we checked for coimmunoprecipitation of Ctf13p and Skp1p with Hsp90 from RL. [35S]Ctf13p or [35S]FlagSkp1p was produced by in vitro translation, and Hsp90 was precipitated from the corresponding RL. This result revealed that [35S]Ctf13p and [35S]FlagSkp1p copurified specifically with Hsp90 (Fig. 3C, compare lanes 2 and 3). In conclusion, there is strong evidence that Hsp90 is involved in HisCtf13p/HisSkp1p activation in RL.
Figure 3.
Hsp90 is required to promote HisCtf13p/HisSkp1p activation in RL. (A) HisCtf13p/HisSkp1p (10 ng each) purified from E. coli was incubated with 7 μl of RL that had been pretreated with geldanamycin at the indicated concentrations for 30 min at 30°C. Subsequently, the samples were combined with Ndc10p (60 ng), Cep3p (15 ng), and 32P-labeled centromere DNA and analyzed by EMSA. (B Left) Western analysis of depleted and mock-treated RL using anti-Hsp90 antibodies. X, an unknown protein that crossreacts with the anti-Hsp90 antibody. (Right) HisCtf13p/HisSkp1p (10 ng each) was incubated with 7 μl of RL that had been depleted of Hsp90 (RL/ΔHsp90) or mock-depleted (RL/mock). Where indicated, the RL was pretreated with 0.1 μg/ml geldanamycin. Subsequently, the samples were analyzed by EMSA as in A. (C) [35S]Ctf13p and [35S]FlagSkp1p were synthesized by in vitro translation in RL (35 μl) and subjected to immunoprecipitation by using anti-Hsp90 antibody or unspecific IgG. Copurification of [35S]Ctf13p and [35S]FlagSkp1p with Hsp90 was analyzed by SDS/PAGE and autoradiography.
Hsp90 Mediates Ctf13p/Skp1p Activation in S. cerevisiae.
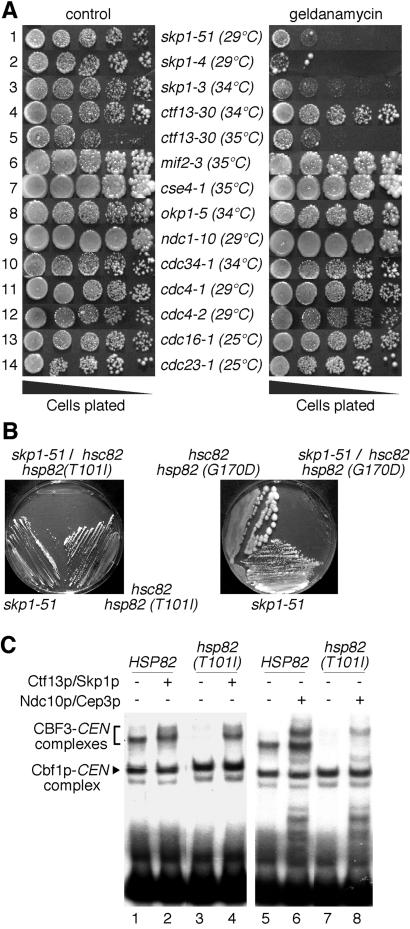
We asked whether skp1 and ctf13 temperature-sensitive mutants exhibited growth defects on media that contained geldanamycin (Fig. 4A). The only ctf13 mutant tested, ctf13–30, showed a weak growth defect on geldanamycin plates in comparison to control plates (Fig. 4A, row 5). Drastic growth defects were observed for skp1 mutants. Most severely affected were skp1–4 and skp1–51 (Fig. 4A, rows 1 and 2), two mutants that exhibit chromosome segregation defects (ref. 22, and J.L., unpublished work). skp1–3, a mutant with a defect in Skp1p/Cullin/F-box complex (SCF) function (22), showed intermediate sensitivity to geldanamycin (Fig. 4A, row 3). Several other mutants with defects in kinetochore proteins or cell-cycle proteins did not reveal growth defects on geldanamycin plates (Fig. 4A, rows 6–14). Therefore, the observed effect is specific for skp1 mutants and possibly ctf13 mutants as well.
Figure 4.
Hsp90 is required to obtain functional Ctf13p/Skp1p in S. cerevisiae. (A) S. cerevisiae strains harboring temperature-sensitive alleles of different kinetochore proteins or cell-cycle regulators were grown on YPD plates containing 20 μg/ml geldanamycin or on YPD control plates. The plates were incubated at temperatures that were ≈ 2–3° below the restrictive temperature of the strain under investigation. (B) skp1 and hsp82 mutations exhibit synthetic lethality. Growth of single and double mutants on synthetic medium containing 5-FOA at 23°C (Left) or on a YPD plate at 29°C (Right) is shown. (C) The hsp82(T1001I) mutant exhibits a decreased amount of functional Ctf13p/Skp1p. Whole-cell extracts from hsp82(T101I) or wild-type cells grown at 23°C were prepared as described (17), incubated with 32P-labeled centromere DNA, and analyzed by EMSA. Where indicated, the extracts were supplemented with purified HisCtf13p/HisSkp1p (1 ng each) from S. cerevisiae or HisNdc10p (60 ng) and HisCep3p (15 ng). Note that the formation of CBF3-centromere DNA complexes is markedly decreased in the hsp82(T101I) mutant, whereas the formation of Cbf1-centromere DNA complexes is not (compare lanes 1 and 3 or lanes 5 and 7). Furthermore, supplementation with HisCtf13p/HisSkp1p leads to an increase in CBF3-CEN signal to wild-type level (compare lanes 2 and 4), whereas supplementation with HisNdc10p and HisCep3p does not (compare lanes 6 and 8).
Next, we investigated whether Hsp82p and Skp1p show genetic interaction. We crossed two temperature-sensitive hsp82 mutants, T101I and G170D (23), with skp1–51 cells harboring plasmid pJL221 (SKP1, URA3). Haploid double mutants harboring pJL221a were streaked onto 5-FOA plates to select for cells that had lost the wild-type copy of SKP1 on pJL221a. When incubated at 23°C, no growth was observed for skp1–51/hsp82(T101I) (Fig. 4B Left). skp1–51/hsp82(G170D) grew slowly at 23°C (data not shown) but not at 29°C (Fig. 4B Right). The fact that hsp82 and skp1 mutations exhibit synthetic lethality implies a role of Hsp82p in the activation of Ctf13p/Skp1p in S. cerevisiae.
If so, one would expect that hsp82 mutants form CBF3-centromere DNA complexes inefficiently. Indeed, extracts prepared from hsp82(T101I) grown at 25°C formed CBF3-centromere DNA complexes considerably less efficiently than wild-type cells (Fig. 4C, lanes 1 and 3). Notably, the formation of another protein–centromere DNA complex, Cbf1p-centromere DNA, was completely unaffected. This result demonstrates that the hsp82 defect specifically compromised CBF3-centromere DNA formation. If the hsp82 defect decreases Ctf13p/Skp1p activity, supplementation of the hsp82(T101I) extract with functional Ctf13p/Skp1p (from S. cerevisiae) should reconstitute the formation of CBF3-centromere DNA complexes to wild-type levels. Indeed, this is the case (Fig. 4C, lanes 2 and 4). In contrast, supplementing the extract with functional Ndc10p and Cep3p did not reconstitute CBF3-centromere DNA formation to wild-type levels (Fig. 4C, lanes 6 and 8). In conclusion, there is strong evidence that Hsp82p is important for the formation of functional Ctf13p/Skp1p in S. cerevisiae.
Discussion
Ctf13p/Skp1p Phosphorylation.
The Ctf13p/Skp1p heterodimer from S. cerevisiae does not require phosphate groups to form CBF3-centromere DNA complexes. This requirement does not conform with the data described for Ctf13p/Skp1p expressed in insect cells (20). In that paper, it was reported that Ctf13p required Skp1p-mediated phosphorylation to form CBF3-centromere DNA complexes in vitro. The reason for this discrepancy is unclear. Phosphorylated Ctf13p from insect cells was reported to form a centromere DNA-binding complex with Ndc10p and Cep3p in the absence of Skp1p (20). In contrast, functional Ctf13p purified from S. cerevisiae always forms a stable dimer with Skp1p, and treatments that dissociate the complex lead to loss of activity (O.S. and J.L., unpublished work). Thus, phosphate groups on Ctf13p from insect cells might substitute for, and even displace, Skp1p in CBF3 complex formation. Alternatively, Ctf13p phosphorylation in insect cells might stabilize an imperfectly folded Ctf13p and thus substitute for Hsp90-dependent activation of Ctf13p in insect cells.
Direct or Indirect Involvement of Hsp90.
Does Hsp90 directly act on Ctf13p/Skp1p or are unknown activators of Ctf13p/Skp1p the direct substrates of Hsp90? The in vitro data argues strongly for a scenario where Hsp90 directly mediates the formation of a functional Ctf13p/Skp1p conformation. Geldanamycin treatment or Hsp90 depletion should not affect the activity of putative Ctf13p/Skp1p activators in the RL because they have been in the presence of functional Hsp90 before Hsp90 depletion. Nevertheless, Ctf13p/Skp1p activation is inhibited when Hsp90 activity is compromised. Furthermore, Ctf13p and Skp1p copurify with Hsp90 from RL.
Hsp90 is known to interact with other chaperones and cochaperones (see ref. 13 and references therein). None of our data exclude the involvement of these proteins. On the contrary, the efficiency of Ctf13p/Skp1p activation in RLs (roughly 5%) seems small in comparison to the large amounts of Hsp90 present in the RL. This finding suggests that additional components, present in the RL in limiting quantity, are required for Ctf13p/Skp1p activation. Sgt1p, a protein that is required to obtain functional Ctf13p/Skp1p in S. cerevisiae (24), could be one of these components. No direct interaction of Sgt1p with Hsp90 or associated proteins has been described. Nevertheless, it is intriguing that the N terminus of Sgt1p exhibits considerable homology (27% identity, 43% similarity over 127 amino acids with 27% gaps) with the human HOP protein, an assembly factor for Hsp90 and Hsp70 (Fig. 6, which is published as supporting information on the PNAS web site).
Hsp90-Dependent Ctf13p/Skp1p Heterodimer Formation.
Heterodimer formation between Ctf13p and Skp1p in RL correlates with Ctf13p/Skp1p activation. This correlation implies that Hsp90 activates Ctf13p/Skp1p by forming a stable heterodimer, which subsequently forms, together with Ndc10p and Cep3p, a DNA-binding unit. This fact is reminiscent of the formation of an active Ecdysone receptor (EcR/USP) in Drosophila where six proteins including Hsp90 mediate the folding of EcR to provide a conformation that interacts with USP, thus forming a DNA-binding unit (25). In contrast to the Ecdysone receptor where Hsp90 and cofactors act on only one of the two subunits, EcR, the Ctf13p/Skp1p activation requires that both proteins are incubated with Hsp90 simultaneously. Furthermore, both proteins interact with Hsp90. Thus, the formation of a Ctf13p/Skp1p heterodimer may involve Hsp90-dependent conformational changes that are stabilized only when the partner protein is present. Alternatively, Hsp90 might promote heterodimer formation first and then further modulate the conformation of the complex.
Hsp90 and the Formation of Heterodimers Between Skp1p and F-Box Proteins.
When not bound to Ctf13p, Skp1p can interact with alternative partners, most of which bind to Skp1p via a so-called F-box motif (for review see ref. 26). Because Ctf13p also contains a weak F-box, the question arises whether the formation of Skp1p/F-box protein heterodimers require Hsp90 in general. It has been reported (27) that E. coli-expressed Skp1p/Cdc4p is competent to form active SCF from purified components. This fact would argue against an Hsp90-facilitated process. On the other hand, skp1–3 cells with a bona fide SCF defect (22) show growth inhibition on geldanamycin plates. This fact argues for the participation of Hsp90. Thus, whether or not Hsp90 is involved in the regulation of SCF complexes needs further investigation.
Supplementary Material
Acknowledgments
We thank R. Deutzmann and E. Hochmuth for LC-MS analysis and automated Edman degradation, and S. Rank for technical assistance. We thank S. Lindquist and J. Buchner for providing yeast strains, U. Gehring for supplying us with geldanamycin, and J. Ortiz for discussing the manuscript. The work was supported by a grant from the Deutsche Forschungsgemeinschaft and by a long-term fellowship of the Human Frontier Science Program (to O.S.).
Abbreviations
- EMSA
electrophoretic mobility-shift assay
- RL
reticulocyte lysate
- SC
synthetic complete medium
References
- 1.Kitagawa K, Hieter P. Nat Rev Mol Cell Biol. 2001;2:678–687. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- 2.Ortiz J, Stemmann O, Rank S, Lechner J. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon H J, Carbon J. Proc Natl Acad Sci USA. 1999;96:13208–13213. doi: 10.1073/pnas.96.23.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng X, Kahana J A, Silver P A, Morphew M K, McIntosh J R, Fitch I T, Carbon J, Saunders W S. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wigge P A, Kilmartin J V. J Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera M M, Schramm C, Schiebel E. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janke C, Ortiz J, Tanaka T U, Lechner J, Schiebel E. EMBO J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He X, Rines D R, Espelin C W, Sorger P K. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 9.Cheeseman I M, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates J R, Huffaker T C, Drubin D G, Barnes G. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Bachant J, Alcasabas A A, Wang Y, Qin J, Elledge S J. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Measday V, Hailey D W, Pot I, Givan S A, Hyland K M, Cagney G, Fields S, Davis T N, Hieter P. Genes Dev. 2002;16:101–113. doi: 10.1101/gad.949302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nathan D F, Vos M H, Lindquist S. Proc Natl Acad Sci USA. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchner J. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 14.Imai J, Yahara I. Mol Cell Biol. 2000;20:9262–9270. doi: 10.1128/mcb.20.24.9262-9270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stemmann O, Lechner J. EMBO J. 1996;15:3611–3620. [PMC free article] [PubMed] [Google Scholar]
- 16.Lechner J. EMBO J. 1994;13:5203–5211. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lechner J, Carbon J. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 18.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Nature (London) 1996a;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 19.Wilm M, Neubauer G, Mann M. Anal Chem. 1996b;68:527–533. doi: 10.1021/ac950875+. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan K B, Hyman A A, Sorger P K. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 21.Whitesell L, Mimnaugh E G, De Costa B, Myers C E, Neckers L M. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connelly C, Hieter P. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nathan D F, Lindquist S. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitagawa K, Skowyra D, Elledge S J, Harper J W, Hieter P. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 25.Arbeitman M N, Hogness D S. Cell. 2000;101:67–77. doi: 10.1016/S0092-8674(00)80624-8. [DOI] [PubMed] [Google Scholar]
- 26.Gieffers C, Schleiffer A, Peters J-M. Protoplasma. 2000;211:20–28. [Google Scholar]
- 27.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.