Abstract
The immunological mechanism of treatment-free remission is not clearly understood. We aimed to identify immune-related genetic differences that predict molecular relapse after tyrosine kinase inhibitor (TKI) discontinuation in patients with chronic phase chronic myeloid leukemia (CML). In this prospective multicenter study, patients who were treated with TKI for at least 3 years discontinued TKI and were monitored for loss of major molecular response. We used NanoString profiling to find gene expression differences associated with relapse. From August 2019 to April 2020, 42 patients were enrolled from five centers in South Korea. During the median follow-up of 16.9 months, 47.6% (20/42) of patients experienced molecular relapse. The 6- and 12-month molecular relapse-free survival (RFS) rates were 52.5% and 50%, respectively. The e14a2 transcript type and longer duration (≥ 50 months) of deep molecular response before TKI discontinuation were associated with longer molecular RFS. NanoString analysis revealed significant differences in immune-related gene expression between relapsed and non-relapsed patients at the time of TKI discontinuation, including T cell-related genes such as SIGLEC1, ARG2, CD160, and IFNG. In conclusion, differences in expression of immune-related genes may provide a prognostic marker for relapse after TKI discontinuation in patients with CML.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00277-025-06514-8.
Keywords: Chronic myeloid leukemia, Tyrosine kinase inhibitor, Treatment-free remission, NanoString method, Generic difference
Introduction
With the remarkable effectiveness of tyrosine kinase inhibitors (TKIs), treatment-free remission (TFR) has become an emerging therapeutic goal in patients with chronic myeloid leukemia (CML). Attempts at TKI discontinuation are supported by the successful results of the Stop Imatinib (STIM1) trial, which showed no disease progression and molecular relapse-free survival (RFS) of 38% at 60 months after imatinib discontinuation in patients who had undetectable BCR::ABL1 transcripts for at least 2 years [1]. Subsequent clinical studies demonstrated comparable molecular RFS of 40–70% in patients who discontinued TKIs after sustained deep molecular response (DMR) for ≥ 2 years [2]. In a recent clinical update, it was suggested that patients who have been on TKI therapy for more than 5 years with durable DMR for longer than 3 years are suitable candidates for TFR [3]. Nevertheless, more than 50% of patients who meet the criteria for TKI discontinuation experience relapse, and several studies have emphasized that additional biologic factors might be linked to sustained TFR [4]. Age, gender, Sokal risk score, previous interferon exposure, and BCR::ABL1 transcript type have been associated with TFR in various cohorts, but the findings have been inconsistent [4–7]. Furthermore, although longer duration of DMR is an essential marker for successful TFR, it is not considered a predictive factor per se [4, 5].
The immunological mechanism of TFR is not clearly understood; however, experimental data suggest that immune cells play a significant role [8]. Increased numbers of immunosuppressive cells, such as regulatory T cells and CD86 + plasmacytoid dendritic cells, were shown to correlate with higher relapse risk, whereas higher numbers of immune effector cells were associated with durable TFR [2]. In particular, natural killer (NK) cells play a key role in controlling relapse. In the IMMUNOSTIM trial, non-relapsing patients had higher numbers of CD56dim NK cells at the time of imatinib discontinuation compared with patients who relapsed [9]. Several experimental studies provided similar findings of the importance of NK cells in CML biology [10, 11]; however, these studies were performed in small cohorts, and their applicability to clinical settings is limited.
NanoString is a technique that uses multiplex nucleic acid hybridization to measure up to 800 target genes [12]. Previous studies used NanoString to find fusion transcripts and prognostic genes in hematologic malignancies [13, 14]; however, there is little information on genetic factors associated with successful TFR in patients with CML [15]. In the present study, we used the NanoString method to search for immune-related genetic markers that were predictive of molecular relapse after TKI discontinuation. We also investigated other prognostic factors for successful TFR after TKI discontinuation in patients with chronic phase CML.
Methods
Patient characteristics
This prospective multicenter study enrolled patients with chronic phase CML in five Korean centers from August 2019 to April 2020. We included patients using the same protocol described in our previous report [16]. Patients who were ≥ 18 years of age, received a standard dose TKI for at least 3 years, and had sustained DMR for at least 1 year were eligible. The standard dose TKIs were imatinib 400 mg once daily, dasatinib 100 mg once daily, nilotinib 300 mg twice daily, or radotinib 300 mg twice daily. BCR::ABL1 transcripts were measured at least every 4 months, and sustained DMR was confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for at least 1 year before study enrollment. In addition, eligible patients had Eastern Cooperative Oncology Group performance status ≥ 2 and no severe dysfunction in any primary organ. Patients who received allogeneic stem cell transplantation, had previously discontinued a TKI, had reduced their TKI dose for any reason, or were diagnosed with an active concomitant malignancy were excluded.
Measurement of BCR::ABL1 transcripts after TKI discontinuation
Molecular response after TKI discontinuation was measured by RT-qPCR using peripheral blood samples at the time of TKI discontinuation, 1 month later, and then every 2 months for 1 year. RT-qPCR with 4.5-log sensitivity was performed in a central laboratory (LAS, Korea). Molecular relapse was confirmed when two consecutive measurements taken within 1 month showed loss of major molecular response (MMR, MR3.0; BCR::ABL1IS
 0.1%). In cases of MMR loss, patients were re-treated with the same TKI that had been discontinued. Patients who lost MMR were assessed for molecular response every 2 months by RT-qPCR until they re-obtained MMR.
0.1%). In cases of MMR loss, patients were re-treated with the same TKI that had been discontinued. Patients who lost MMR were assessed for molecular response every 2 months by RT-qPCR until they re-obtained MMR.
NanoString analysis
Genetic differences between patients who did or did not relapse after TKI discontinuation were analyzed using the NanoString Immune Profiling Panel. RNA samples were run on the NanoString nCounter Analysis System (NanoString Technologies, Inc., Seattle, WA, USA) according to the manufacturer’s directions. Briefly, 5 µL (100–300 ng) of each RNA sample was mixed with 8 µL of Master Mix (Reporter CodeSet and hybridization buffer). Then, 2 µL of the Capture ProbeSet was added, and the solution was mixed, spun down, and placed in a 65 °C thermocycler (Bio-Rad Laboratories Inc, Hercules, California, USA) for 16 h (maximum hybridization time should not exceed 48 h). The samples were then transferred to the preparation station (NanoString Technologies) with a prepared nCounter Master Kit and a cartridge to allow binding of the samples. The preparation station could process 12 lanes per run in approximately 2.5–3.0 h. The cartridges were subsequently transferred to the Digital Analyzer (NanoString Technologies) and scanned at 555 fields of view.
Outcomes
The primary objective of this study was to find differences in expression of immune-related genes between patients who experienced molecular relapse after TKI discontinuation and those who did not. The secondary objectives were to determine the incidence of molecular relapse and identify additional prognostic factors associated with molecular RFS. If a patient lost MMR, we also evaluated the time to re-achieve MMR and the occurrence or absence of disease progression.
Statistical analysis
Molecular RFS (sustained TFR) was defined from the date of TKI discontinuation to the date of loss of MMR or last follow-up. The duration of DMR before discontinuation of TKI was defined as the period of BCR::ABL1IS 0.01% (MR4.0) before TKI discontinuation. The duration of sustained DMR was defined from the date of TKI discontinuation to the date of loss of DMR (MR4.0). Median values were used as cut-offs to classify patients by independent variables, such as age, TKI treatment duration, and duration of DMR before TKI discontinuation. Categorical variables were compared using Pearson’s chi-squared test or Fisher’s exact test, as appropriate. Molecular RFS was analyzed using the Kaplan–Meier method, and differences between groups were assessed using the log-rank test. Clinical variables in univariate analysis with p values of < 0.05 were included in the multivariate analysis, which was performed using the Cox proportional hazard model. All statistical tests were two-sided, and significance was defined as p < 0.05. All analyses were performed using IBM SPSS version 25.0 (IBM, Armonk, NY, USA).
0.01% (MR4.0) before TKI discontinuation. The duration of sustained DMR was defined from the date of TKI discontinuation to the date of loss of DMR (MR4.0). Median values were used as cut-offs to classify patients by independent variables, such as age, TKI treatment duration, and duration of DMR before TKI discontinuation. Categorical variables were compared using Pearson’s chi-squared test or Fisher’s exact test, as appropriate. Molecular RFS was analyzed using the Kaplan–Meier method, and differences between groups were assessed using the log-rank test. Clinical variables in univariate analysis with p values of < 0.05 were included in the multivariate analysis, which was performed using the Cox proportional hazard model. All statistical tests were two-sided, and significance was defined as p < 0.05. All analyses were performed using IBM SPSS version 25.0 (IBM, Armonk, NY, USA).
For NanoString analysis, we used nSolver Analysis software (version 4.0, NanoString Technologies) for gene expression analysis. Background correction and normalization were performed using default options with control probes in each CodeSet. Gene expression data were log2-transformed prior to statistical analysis. Differential expression analysis between the two conditions was performed using Welch’s t-test. Although multiple testing correction was performed using the Benjamini-Hochberg method to control the false discovery rate, genes were considered differentially expressed based on unadjusted p values < 0.05 and absolute log2 fold-changes > 1. This threshold was selected to prioritize sensitivity in exploratory analysis. To compare expression profiles among the samples, the normalized expression values of a few hundred selected differentially expressed genes were subjected to unsupervised clustering. Scatter plot of the gene expression values and volcano plot of the expression fold-changes and p values were drawn using R scripts (Supplementary Fig. 1a and b).
Ethics approval
This study was approved by the ethics committees of each hospital. Written informed consent was obtained from all participants in accordance with the Declaration of Helsinki.
Results
Patient characteristics
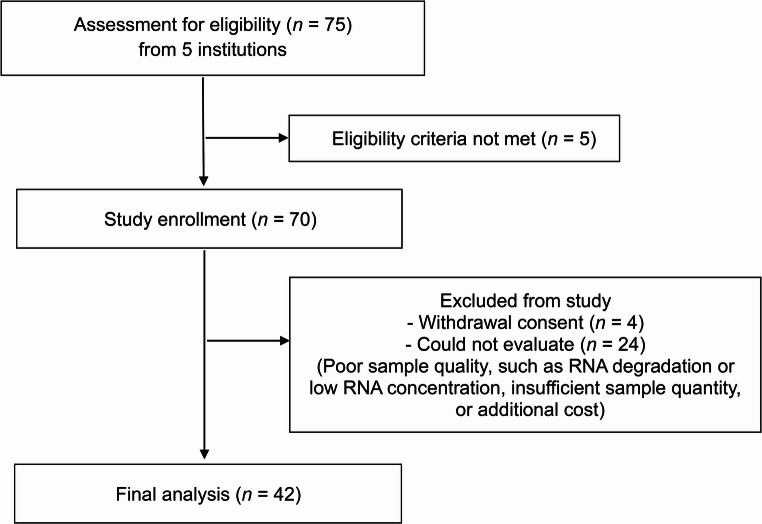
From August 2019 to April 2020, 42 patients from five centers in South Korea were enrolled in this clinical trial (Fig. 1). The patients in this study include some of those from our previous paper [16]. The median follow-up period was 16.9 months (range, 9.8–22.5 months). All patients had received standard doses of TKIs. The median age of the patients was 49 years (range, 20–70 years). At the time of TKI discontinuation, 66.7% (28/42) of the patients were being treated with nilotinib (Table 1). 69% (29/42) of the patients were receiving a first-line TKI treatment, and 31% (13/42) were receiving a second-line (2 patients) or third-line (11 patients) TKI treatment. The baseline characteristics of the patients are shown in Table 1.
Fig. 1.
CONSORT flow diagram
Table 1.
Baseline characteristics of the patients
| Patient characteristics | All patients (N = 42) |
|---|---|
| Median age at diagnosis, years (range) | 49 (20–70) |
| Sex, n (%) | |
| Male | 21 (50.0) |
| Female | 21 (50.0) |
| ECOG, n (%) | |
| 0 | 24 (57.1) |
| 1 | 3 (7.1) |
| Unknown | 15 (35.7) |
| Median white blood cell counts at diagnosis, per µL (range) |
83,235 (9,040–293,000) |
| BCR::ABL1 transcript type, n (%) | |
| E14a2 | 29 (69.0) |
| E13a2 | 10 (23.8) |
| Unknown | 3 (7.1) |
| Line of treatment at the time of stopping TKI, n (%) | |
| First-line | 29 (69.0) |
| Second- or third-line | 13 (31.0) |
| First-line TKI treatment, n (%) | |
| Imatinib | 16 (38.1) |
| Dasatinib | 5 (11.9) |
| Nilotinib | 18 (42.9) |
| Radotinib | 1 (2.4) |
| Unknown | 2 (4.8) |
| Treatment at the time of stopping TKI, n (%) | |
| Imatinib | 6 (14.3) |
| Dasatinib | 7 (16.7) |
| Nilotinib | 28 (66.7) |
| Radotinib | 1 (2.4) |
| Median BCR::ABL1 IS by RT-qPCR at TKI discontinuation, % (range) | 0 (0–0.01) |
| Duration from TKI treatment initiation to TKI discontinuation, months (range) | 70 (29–147) |
| Duration of DMR before stopping TKI, months | 50 (15–141) |
Abbreviations: ECOG, Eastern Cooperative Oncology Group performance status; TKI, tyrosine kinase inhibitor; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; IS, International Scale; DMR, deep molecular response
Molecular RFS and prognostic factors
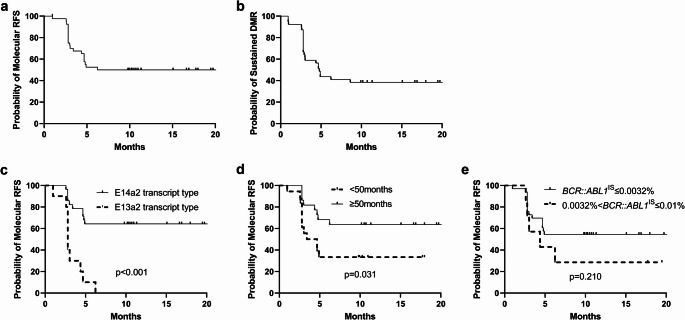
After TKI discontinuation, 52.4% (22/42) of the patients sustained MMR, and 47.6% (20/42) experienced molecular relapse. Molecular RFS at 6 and 12 months was 52.5% and 50%, respectively (Fig. 2a). The probability of sustained DMR after TKI discontinuation was 43.6% at 6 months and 38.5% at 12 months (Fig. 2b).
Fig. 2.
(a, b) The probability of molecular relapse-free survival (a) and sustained DMR after TKI discontinuation (b). (c, d, e) The probability of molecular relapse-free survival according to BCR::ABL1 transcript type (c), duration of DMR (d), and RT-qPCR values at the time of TKI discontinuation (e). Survival curves were estimated using the Kaplan–Meier method and compared using the log-rank test. DMR, deep molecular response; TKI, tyrosine kinase inhibitor; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; IS, International Scale
In a univariate analysis, the e14a2 transcript type (p < 0.001) and longer duration (≥ 50 months) of DMR before TKI discontinuation (p = 0.031) were significant factors for longer molecular RFS (Fig. 2c and d; Table 2). The e14a2 transcript type and longer duration (≥ 50 months) of DMR before TKI discontinuation were also associated with longer molecular RFS in a multivariate analysis (hazard ratio [HR] = 7.249, 95% confidence interval [CI]: 2.682–19.594, p < 0.001 for transcript type; HR = 3.829, 95% CI: 1.487–9.858, p = 0.005 for duration of DMR before TKI discontinuation; Table 2). However, achieving a deep molecular response with BCR::ABL1IS ≤ 0.0032% at the time of TKI discontinuation was not significantly associated with prolonged molecular RFS (p = 0.210; Fig. 2e).
Table 2.
Prognostic factors for molecular RFS after TKI discontinuation
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Variables | 1-year RFS, % (± SE) | p-value | HR | 95% CI | p-value | |||
| Age | ≥ 49 years | 61.9 (± 10.6) | 0.115 | |||||
| < 49 years | 36.8 (± 11.1) | |||||||
| Gender | Male | 45.0 (± 11.1) | 0.406 | |||||
| Female | 55.0 (± 11.1) | |||||||
| WBC at TKI stop | ≥ 100,000/µL | 28.6 (± 12.1) | 0.065 | |||||
| < 100,000/µL | 64.0 (± 9.6) | |||||||
| BCR::ABL1 | E14a2 | 64.3 (± 9.1) | < 0.001 | 1 | < 0.001 | |||
| transcript type | E13a2 | 0 | 7.249 | 2.682–19.594 | ||||
| BCR::ABL1 IS at 3 | ≥ 10% | 42.9 (± 18.7) | 0.719 | |||||
| months, % | < 10% | 50.0 (± 9.4) | ||||||
| TKI treatment line | 1 st line | 46.4 (± 9.4) | 0.598 | |||||
| 2nd or 3rd line | 58.3 (± 14.2) | |||||||
| TKI type at stop | 1 st generation | 40.0 (± 21.9) | 0.898 | |||||
| 2nd generation | 51.4 (± 8.4) | |||||||
| Duration of TKI | ≥ 70 months | 61.9 (± 10.6) | 0.068 | |||||
| treatment | < 70 months | 36.8 (± 11.1) | ||||||
| Duration of DMR | ≥ 50 months | 63.6 (± 10.3) | 0.031 | 1 | 0.005 | |||
| before TKI stop | < 50 months | 33.3 (± 11.1) | 3.829 | 1.487–9.858 | ||||
| RT-qPCR values | 0.0032%<BCR::ABL1 IS≤0.01% | 28.6 (± 17.1) | 0.210 | |||||
| at TKI stop | BCR::ABL1 IS≤0.0032% | 54.5 (± 8.7) | ||||||
Abbreviations: TKI, tyrosine kinase inhibitor; RFS, relapse-free survival; SE, standard error; HR, hazard ratio; CI, confidence interval; WBC, white blood cell; IS, International Scale; DMR, deep molecular response; RT-qPCR, reverse transcription-quantitative polymerase chain reaction
All patients who lost MMR were re-treated with the same TKI that had been discontinued, and all of these patients re-obtained MMR. The median time to re-achievement of MMR was 3.0 months (range, 1.0–5.5 months). No patients experienced disease progression to accelerated or blast phase.
Gene expression differences associated with molecular relapse
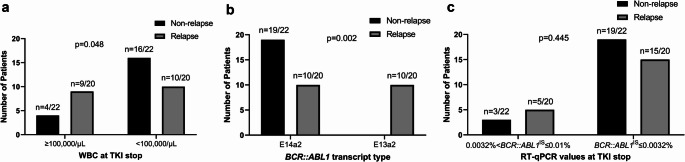
The characteristics of patients who did or did not experience molecular relapse are shown in Table 3. Patients with lower white blood cell counts (< 100,000/µL) at the time of TKI discontinuation (p = 0.048) and those with the e14a2 transcript type (p = 0.002) had significantly lower molecular relapse rates after TKI discontinuation (Fig. 3a and b; Table 3). In contrast, achieving a deep molecular response with BCR::ABL1 ≤ 0.0032% at the time of TKI discontinuation did not significantly reduce the risk of molecular relapse (p = 0.445; Fig. 3c; Table 3).
Table 3.
Characteristics of patients stratified according to molecular relapse
| Variable | Non-relapse N = 22 |
Relapse N = 20 |
p-value | |
|---|---|---|---|---|
| Age, years | ≥ 49 years | 13 (65.0%) | 8 (40.0%) | 0.113 |
| < 49 years | 7 (35.0%) | 12 (60.0%) | ||
| Gender | Male | 10 (45.5%) | 11 (55.0%) | 0.537 |
| Female | 12 (54.5%) | 9 (45.0%) | ||
| WBC at TKI stop | ≥ 100,000/µL | 4 (20.0%) | 9 (47.4%) | 0.048 |
| < 100,000/µL | 16 (80.0%) | 10 (52.6%) | ||
|
BCR::ABL1 transcript type |
E14a2 | 19 (100%) | 10 (50.0%) | 0.002 |
| E13a2 | 0 | 10 (50.0%) | ||
| BCR::ABL1 IS at 3 | ≥ 10% | 14 (82.4%) | 14 (77.8%) | 1.000 |
| months | < 10% | 3 (17.6%) | 4 (22.2%) | |
| TKI treatment line | 1 st line | 14 (63.6%) | 15 (75.0%) | 0.426 |
| 2nd or 3rd line | 8 (36.4%) | 5 (25.0%) | ||
| TKI type at stop | 1 st generation | 3 (13.6%) | 3 (15.0%) | 1.000 |
| 2nd generation | 19 (86.4%) | 17 (85.0%) | ||
| Duration of TKI | ≥ 70 months | 13 (59.1%) | 8 (40.0%) | 0.217 |
| treatment | < 70 months | 9 (40.9%) | 12 (60.0%) | |
| Duration of | ≥ 50 months | 15 (68.2%) | 8 (40.0%) | 0.067 |
| DMR before TKI stop | < 50 months | 7 (31.8%) | 12 (60.0%) | |
| RT-qPCR values | 0.0032%<BCR::ABL1 IS≤0.01% | 3 (13.6%) | 5 (25.0%) | 0.445 |
| at TKI stop | BCR::ABL1 IS≤0.0032% | 19 (86.4%) | 15 (75.0%) | |
Abbreviations: TKI, tyrosine kinase inhibitor; WBC, white blood cell; IS, International Scale; DMR, deep molecular response; RT-qPCR, reverse transcription-quantitative polymerase chain reaction
Fig. 3.
Comparison between molecular relapse and non-relapse groups according to (a) WBC count at TKI stop, (b) BCR::ABL1 transcript type, and (c) RT-qPCR values at TKI stop. Statistical comparisons between groups were performed using Pearson’s chi-squared test or Fisher’s exact test, as appropriate. WBC, white blood cell; TKI, tyrosine kinase inhibitor; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; IS, International Scale
Analysis of immune-related gene expression at the time of TKI discontinuation revealed that BAGE, LTF, CEACAM8, SIGLEC1, and ARG2 were overexpressed in patients who did not experience molecular relapse, whereas CD160 and IFNG were overexpressed in patients who experienced molecular relapse (Table 4 and Supplementary Fig. 2). Furthermore, among the patients who experienced molecular relapse, CEACAM6 and CXCL5 were overexpressed at the time of molecular relapse compared with the time of TKI discontinuation (Table 4).
Table 4.
Gene expression analysis using NanoString
| Differences in gene expression in relapse patients compared with non-relapse patients at the time of TKI discontinuation | ||||
|---|---|---|---|---|
| Gene | Log2 FC | Linear FC | p-value | BH p-value |
| BAGE | −1.430 | −2.688 | 0.007 | 0.088 |
| LTF | −1.050 | −2.075 | 0.021 | 0.157 |
| CEACAM8 | −0.937 | −1.916 | 0.033 | 0.193 |
| SIGLEC1 | −0.826 | −1.773 | 0.043 | 0.227 |
| ARG2 | −0.813 | −1.757 | 0.003 | 0.063 |
| IRF5 | −0.751 | −1.684 | 0.000 | 0.002 |
| CAMP | −0.724 | −1.650 | 0.049 | 0.241 |
| PLAUR | −0.719 | −1.647 | 0.006 | 0.085 |
| POU2AF1 | −0.705 | −1.631 | 0.002 | 0.053 |
| IL21R | −0.685 | −1.608 | 0.000 | 0.011 |
| LCN2 | −0.664 | −1.585 | 0.039 | 0.214 |
| IL11RA | −0.658 | −1.577 | 0.000 | 0.018 |
| CD33 | −0.610 | −1.527 | 0.000 | 0.011 |
| CCR7 | −0.603 | −1.520 | 0.003 | 0.055 |
| PTGS2 | 0.600 | 1.520 | 0.046 | 0.237 |
| FCGR1A | 0.647 | 1.570 | 0.025 | 0.170 |
| CXCL10 | 0.758 | 1.690 | 0.020 | 0.154 |
| CD160 | 0.779 | 1.720 | 0.002 | 0.053 |
| IFNG | 1.000 | 2.010 | 0.000 | 0.011 |
| Differences in gene expression at the time of relapse compared with when TKI was discontinued in relapse patients | ||||
| ENTPD1 | −0.735 | −1.664 | 0.000 | 0.019 |
| FCGR1A | −0.673 | −1.595 | 0.015 | 0.151 |
| CD79A | 0.606 | 1.520 | 0.002 | 0.077 |
| CXCL5 | 0.799 | 1.740 | 0.001 | 0.044 |
| CEACAM6 | 0.802 | 1.740 | 0.034 | 0.221 |
Abbreviations: TKI, tyrosine kinase inhibitor; FC, fold-change
Discussion
We aimed to find prognostic factors and immune-related gene expression differences related to molecular relapse in patients with chronic phase CML who discontinued TKI. We found that (1) the relapse rate after TKI discontinuation was 50% at 12 months; (2) the e14a2 transcript type and longer DMR duration (≥ 50 months) were predictive of successful TFR; and (3) BAGE, LTF, CEACAM8, SIGLEC1, and ARG2 were overexpressed at the time of TKI discontinuation in patients who sustained TFR, whereas CD160 and IFNG were overexpressed in patients who experienced relapse.
Although our criteria for TKI cessation were less stringent than those in the STIM1 trial, the 12-month relapse rate in our study was similar to those in previous clinical trials [6, 17, 18]. We also found that longer DMR duration before TKI discontinuation was related to successful TFR, whereas TKI treatment duration did not affect TFR. Other studies have produced conflicting results. The STIM1, DASFREE, and EURO-SKI trials each emphasized that longer duration of TKI therapy increased the likelihood of successful TFR; however, the EURO-SKI study found that the duration of DMR before TKI cessation was the most relevant factor for successful TFR, whereas the other trials showed inconsistent associations between DMR duration and recurrence [6, 17, 18]. It is noteworthy that current guidelines suggest that TKI treatment > 5 years and sustained DMR duration > 2 years are essential factors for selection of TFR candidates, but approximately 50% of patients experience molecular relapse under these guidelines [19]. Previous reports suggested that biological factors such as low Sokal score, advanced age, low CD4 T cell number, and high NK cell number are relevant factors affecting sustained TFR [18, 20, 21].
In the present study, not only longer DMR duration but also e14a2 transcript type were associated with successful TFR. This finding is similar outcomes in previous research [16]. However, the underlying mechanism for the role of transcript type remains controversial [4, 5]. Some hypotheses are that tyrosine kinase activity, immunological differences, or RT-qPCR artifacts are associated with different outcomes [22–24]. Further solid evidence is needed to confirm the finding that the e14a2 transcript type is associated with molecular relapse.
The primary objective of this study was to find immune-related genes that affect relapse after TKI discontinuation in patients with CML. We found that increased expression of BAGE, LTF, CEACAM8, SIGLEC1, and ARG2 and decreased expression of CD160 and IFNG were related to successful TFR. Among these, SIGLEC1, ARG2, CD160, and IFNG have been associated with T cell functions in several reports [25–28]. CD169 (SIGLEC1), expressed mainly in macrophages, has tumor-suppressing roles in various malignant tumors [29]. The expression of SIGLEC family genes was also correlated with clinical features, immune cell infiltration, and survival outcomes in patients with acute myeloid leukemia [30]. An association was reported between SIGLEC3 and CML cells, but little is known about the association between CML cells and SIGLEC1 [26]. Previous data suggested that SIGLEC1 + macrophages enhanced the proliferation, cytotoxicity, and cytokine production of CD8 + T cells and could affect successful TFR [20, 31]. Similarly, ARG2 (arginase-2) is known to play a role in T cell immunity through a mechanism in which ARG2-specific CD8+ T cells have cytotoxic function against cancer cells [25]. Therefore, it is possible that increased expression of SIGLEC1 and ARG2 reduced the likelihood of relapse after TKI discontinuation by promoting CD8 + T cell activity.
Increased expression of CD160 and IFNG had a negative effect on sustained TFR. CD160 is a negative regulator of NK T cells and reduces cytokine production during early innate immune response [27]. Experimental data showed that CD160-expressing CD8 + T cells had reduced proliferation capacity and IL-2 production, whereas CD160 blockade significantly increased CD8 + T cell proliferation [32]. IFNG, encoding type II interferon, has an impact on tumor-immune interaction, and its expression is positively associated with neutrophil and macrophage infiltration in cancer [33]. However, recent research suggested that IFN-γ release by acute myeloid leukemia cells could induce regulatory T cells in the bone marrow immune microenvironment and was associated with poor overall survival [28]. Additionally, IFN-γ-related gene profiles were associated with chemotherapy resistance in acute myeloid leukemia [34]. Therefore, reduced expression of IFNG might promote successful TFR in CML by reducing regulatory T cell numbers. Previous clinical trials showed that increased numbers of CD8 + T cells and decreased numbers of regulatory CD4 + T cells are key immunologic factors for sustained TFR after TKI discontinuation [20, 21]. Therefore, SIGLEC1, ARG2, CD160, and IFNG may indirectly influence TFR through these immunologic mechanisms.
BAGE, CEACAM8, and LTF were previously reported to be associated with prognosis in solid cancers [35]; however, there is little evidence that these genes are associated with hematologic malignancies, and their immunologic function in sustained TFR is ambiguous [36]. CEACAM8 (CD66b) is known as an activation marker for human granulocytes [37]. Soluble CEACAM8 promotes remission in acute myeloid leukemia but has negative effects in acute lymphoblastic leukemia, and no role of CEACAM8 in CML has been established [36]. Likewise, LTF was related to progression-free survival in patients with prostate cancer, but its role in leukemic cells is ambiguous [38].
CEACAM6 and CXCL5 were overexpressed at the time of relapse compared with the time of TKI discontinuation in patients who experienced molecular relapse. These genes were previously associated with tumor metastasis and poor survival in patients with solid cancers [39, 40]; however, no previous studies have shown associations between these genes and CML. Therefore, further evidence is needed to confirm the function of these genes in CML.
This study has several limitations. First, although this was a multicenter study, the sample size was small for an analysis of genetic differences. Therefore, confirmation of our findings warrants further investigation in larger cohorts. Second, risk scores such as Sokal or EUTOS scores were not analyzed because it was difficult to obtain consistent spleen size measurements in a multicenter study. Third, some clinical data were lost or omitted when patients were transferred between hospitals during treatment. Fourth, this study only measured gene expression levels and did not include a functional analysis, which is needed to establish the exact associations between genetic factors and CML relapse. Nevertheless, the results contribute to our understanding of how immune-related genetic factors are related to successful TFR, especially in Asian populations. In addition, this study demonstrates how the NanoString method can be used to analyze genetic factors related to successful TFR in patients with CML.
In conclusion, 50% of CML patients successfully discontinued TKI therapy. The e14a2 transcript type and longer DMR duration were prognostic markers for successful TFR. There were differences in immune-related gene expression between patients who experienced molecular relapse and those who did not relapse after TKI discontinuation. Especially, T cell-related genes such as SIGLEC1, ARG2, CD160, and IFNG showed different expression levels according to relapse. Immune-related genetic analysis shows promise as a prognostic tool in patients with CML who want to discontinue TKI therapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
IK designed the research study. HP, HJK. SKS, YB, DK, SYL, JHK, HK, JP, DYS, IK and SP conducted the study and collected the data. HP, YB, DK and SYL analyzed the data. HP and SYL wrote the paper. All authors read and approved the final manuscript.
Funding
None.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committees of each hospital. Written informed consent was obtained from all participants in accordance with the Declaration of Helsinki.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Etienne G, Guilhot J, Rea D et al (2017) Long-Term Follow-Up of the French stop Imatinib (STIM1) study in patients with chronic myeloid leukemia. J Clin Oncol 35:298–305. 10.1200/jco.2016.68.2914 [DOI] [PubMed] [Google Scholar]
- 2.Inzoli E, Aroldi A, Piazza R, Gambacorti-Passerini C (2022) Tyrosine kinase inhibitor discontinuation in chronic myeloid leukemia: eligibility criteria and predictors of success. Am J Hematol 97:1075–1085. 10.1002/ajh.26556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jabbour E, Kantarjian H (2022) Chronic myeloid leukemia: 2022 update on diagnosis, therapy, and monitoring. Am J Hematol 97:1236–1256. 10.1002/ajh.26642 [DOI] [PubMed] [Google Scholar]
- 4.D’Adda M, Farina M, Schieppati F et al (2019) The e13a2 BCR-ABL transcript negatively affects sustained deep molecular response and the achievement of treatment-free remission in patients with chronic myeloid leukemia who receive tyrosine kinase inhibitors. Cancer 125:1674–1682. 10.1002/cncr.31977 [DOI] [PubMed] [Google Scholar]
- 5.Claudiani S, Apperley JF, Gale RP et al (2017) E14a2 BCR-ABL1 transcript is associated with a higher rate of treatment-free remission in individuals with chronic myeloid leukemia after stopping tyrosine kinase inhibitor therapy. Haematologica 102:e297–e299. 10.3324/haematol.2017.168740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saussele S, Richter J, Guilhot J et al (2018) Discontinuation of tyrosine kinase inhibitor therapy in chronic myeloid leukaemia (EURO-SKI): a prespecified interim analysis of a prospective, multicentre, non-randomised, trial. Lancet Oncol 19:747–757. 10.1016/s1470-2045(18)30192-x [DOI] [PubMed] [Google Scholar]
- 7.Castagnetti F, Binotto G, Capodanno I et al (2021) Making Treatment-Free remission (TFR) easier in chronic myeloid leukemia: Fact-Checking and practical management tools. Target Oncol 16:823–838. 10.1007/s11523-021-00831-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schütz C, Inselmann S, Saussele S et al (2017) Expression of the CTLA-4 ligand CD86 on plasmacytoid dendritic cells (pDC) predicts risk of disease recurrence after treatment discontinuation in CML. Leukemia 31:829–836. 10.1038/leu.2017.9 [DOI] [PubMed] [Google Scholar]
- 9.Rea D, Henry G, Khaznadar Z et al (2017) Natural killer-cell counts are associated with molecular relapse-free survival after Imatinib discontinuation in chronic myeloid leukemia: the IMMUNOSTIM study. Haematologica 102:1368–1377. 10.3324/haematol.2017.165001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilander M, Olsson-Strömberg U, Schlums H et al (2017) Increased proportion of mature NK cells is associated with successful Imatinib discontinuation in chronic myeloid leukemia. Leukemia 31:1108–1116. 10.1038/leu.2016.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong J, Qin YZ, Zhao XS, Hou Y, Liu KY, Huang XJ, Jiang H (2021) Profiles of NK cell subsets are associated with successful tyrosine kinase inhibitor discontinuation in chronic myeloid leukemia and changes following interferon treatment. Ann Hematol 100:2557–2566. 10.1007/s00277-021-04606-9 [DOI] [PubMed] [Google Scholar]
- 12.Goytain A, Ng T (2020) NanoString nCounter technology: High-Throughput RNA validation. Methods Mol Biol 2079:125–139. 10.1007/978-1-4939-9904-0_10 [DOI] [PubMed] [Google Scholar]
- 13.Hu D, Zhou W, Wang F et al (2016) Development of a NanoString assay to detect leukemogenic fusion transcripts in acute myeloid leukemia. Int J Lab Hematol 38:663–673. 10.1111/ijlh.12555 [DOI] [PubMed] [Google Scholar]
- 14.D’Angelo A, Kilili H, Chapman R et al (2023) Immune-related pan-cancer gene expression signatures of patient survival revealed by NanoString-based analyses. PLoS ONE 18:e0280364. 10.1371/journal.pone.0280364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adnan Awad S, Brück O, Shanmuganathan N et al (2022) Epigenetic modifier gene mutations in chronic myeloid leukemia (CML) at diagnosis are associated with risk of relapse upon treatment discontinuation. Blood Cancer J 12:69. 10.1038/s41408-022-00667-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park H, Kim HJ, Sohn SK et al (2023) Effect of BCR::ABL1 transcript type and droplet digital polymerase chain reaction on successful treatment-free remission in chronic myeloid leukemia patients who discontinued tyrosine kinase inhibitor. Ther Adv Hematol 14:20406207231205637. 10.1177/20406207231205637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahon FX, Réa D, Guilhot J et al (2010) Discontinuation of Imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre stop Imatinib (STIM) trial. Lancet Oncol 11:1029–1035. 10.1016/s1470-2045(10)70233-3 [DOI] [PubMed] [Google Scholar]
- 18.Shah NP, García-Gutiérrez V, Jiménez-Velasco A et al (2020) Dasatinib discontinuation in patients with chronic-phase chronic myeloid leukemia and stable deep molecular response: the DASFREE study. Leuk Lymphoma 61:650–659. 10.1080/10428194.2019.1675879 [DOI] [PubMed] [Google Scholar]
- 19.Hochhaus A, Baccarani M, Silver RT et al (2020) European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 34:966–984. 10.1038/s41375-020-0776-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura S, Imagawa J, Murai K et al (2020) Treatment-free remission after first-line dasatinib discontinuation in patients with chronic myeloid leukaemia (first-line DADI trial): a single-arm, multicentre, phase 2 trial. Lancet Haematol 7:e218–e225. 10.1016/s2352-3026(19)30235-2 [DOI] [PubMed] [Google Scholar]
- 21.Imagawa J, Tanaka H, Okada M et al (2015) Discontinuation of dasatinib in patients with chronic myeloid leukaemia who have maintained deep molecular response for longer than 1 year (DADI trial): a multicentre phase 2 trial. Lancet Haematol 2:e528–535. 10.1016/s2352-3026(15)00196-9 [DOI] [PubMed] [Google Scholar]
- 22.Lucas CM, Harris RJ, Giannoudis A et al (2009) Chronic myeloid leukemia patients with the e13a2 BCR-ABL fusion transcript have inferior responses to Imatinib compared to patients with the e14a2 transcript. Haematologica 94:1362–1367. 10.3324/haematol.2009.009134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baccarani M, Rosti G, Soverini S (2019) Chronic myeloid leukemia: the concepts of resistance and persistence and the relationship with the BCR-ABL1 transcript type. Leukemia 33:2358–2364. 10.1038/s41375-019-0562-1 [DOI] [PubMed] [Google Scholar]
- 24.Kjaer L, Skov V, Andersen MT et al (2019) Variant-specific discrepancy when quantitating BCR-ABL1 e13a2 and e14a2 transcripts using the Europe against Cancer qPCR assay. Eur J Haematol 103:26–34. 10.1111/ejh.13238 [DOI] [PubMed] [Google Scholar]
- 25.Weis-Banke SE, Lisle TL, Perez-Penco M et al (2022) Arginase-2-specific cytotoxic T cells specifically recognize functional regulatory T cells. J Immunother Cancer 10. 10.1136/jitc-2022-005326 [DOI] [PMC free article] [PubMed]
- 26.Herrmann H, Cerny-Reiterer S, Gleixner KV et al (2012) CD34(+)/CD38(-) stem cells in chronic myeloid leukemia express Siglec-3 (CD33) and are responsive to the CD33-targeting drug gemtuzumab/ozogamicin. Haematologica 97:219–226. 10.3324/haematol.2010.035006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim T-J, Park G, Kim J et al (2019) CD160 serves as a negative regulator of NKT cells in acute hepatic injury. Nat Commun 10:3258. 10.1038/s41467-019-10320-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corradi G, Bassani B, Simonetti G et al (2022) Release of IFNγ by acute myeloid leukemia cells remodels bone marrow immune microenvironment by inducing regulatory T cells. Clin Cancer Res 28:3141–3155. 10.1158/1078-0432.Ccr-21-3594 [DOI] [PubMed] [Google Scholar]
- 29.Jiang K-Y, Qi L-L, Kang F-B, Wang L (2022) The intriguing roles of Siglec family members in the tumor microenvironment. Biomark Res 10:22. 10.1186/s40364-022-00369-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi H, Gao L, Zhang W, Jiang M (2022) Identification and validation of a siglec-based and aging-related 9-gene signature for predicting prognosis in acute myeloid leukemia patients. BMC Bioinformatics 23:284. 10.1186/s12859-022-04841-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou X, Chen G, Zhao Y (2021) Research progress on CD169-positive macrophages in tumors. Am J Transl Res 13:8589–8597 [PMC free article] [PubMed] [Google Scholar]
- 32.Viganò S, Banga R, Bellanger F et al (2014) CD160-Associated CD8 T-Cell functional impairment is independent of PD-1 expression. PLoS Pathog 10:e1004380. 10.1371/journal.ppat.1004380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji H, Ba Y, Ma S et al (2021) Construction of Interferon-Gamma-Related gene signature to characterize the Immune-Inflamed phenotype of glioblastoma and predict prognosis, efficacy of immunotherapy and radiotherapy. 12. 10.3389/fimmu.2021.729359 [DOI] [PMC free article] [PubMed]
- 34.Vadakekolathu J, Minden MD, Hood T et al (2020) Immune landscapes predict chemotherapy resistance and immunotherapy response in acute myeloid leukemia. Sci Transl Med 12. 10.1126/scitranslmed.aaz0463 [DOI] [PMC free article] [PubMed]
- 35.Kouno J, Nagai H, Nagahata T et al (2004) Up-regulation of CC chemokine, CCL3L1, and receptors, CCR3, CCR5 in human glioblastoma that promotes cell growth. J Neurooncol 70:301–307. 10.1007/s11060-004-9165-3 [DOI] [PubMed] [Google Scholar]
- 36.Zaghloul A, Morsi H, Ismail M, Teama S, Abdulateef N (2018) Soluble carcinoembryonic antigen cell adhesion molecule 1,6 and 8 in acute myeloid leukemia: their relation to survival and prognosis. Life Sci J 15:11. 10.7537/marslsj150418.01 [Google Scholar]
- 37.Yoon J, Terada A, Kita H (2007) CD66b regulates adhesion and activation of human eosinophils. J Immunol 179:8454–8462. 10.4049/jimmunol.179.12.8454 [DOI] [PubMed] [Google Scholar]
- 38.Shaheduzzaman S, Vishwanath A, Furusato B et al (2007) Silencing of lactotransferrin expression by methylation in prostate cancer progression. Cancer Biol Ther 6:1088–1095. 10.4161/cbt.6.7.4327 [DOI] [PubMed] [Google Scholar]
- 39.Deng J, Jiang R, Meng E, Wu H (2022) CXCL5: A coachman to drive cancer progression. Front Oncol 12:944494. 10.3389/fonc.2022.944494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey R, Zhou M, Islam S et al (2019) Carcinoembryonic antigen cell adhesion molecule 6 (CEACAM6) in pancreatic ductal adenocarcinoma (PDA): an integrative analysis of a novel therapeutic target. Sci Rep 9:18347. 10.1038/s41598-019-54545-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.