Abstract
Naturally competent bacteria have the ability to take up free DNA from the surrounding medium and incorporate this DNA into their genomes by homologous recombination. In naturally competent Streptococcus pneumoniae, and related streptococcal species from the mitis phylogenetic group, the competent state is not a constitutive property but is induced by a peptide pheromone through a quorum-sensing mechanism. Recent studies have shown that natural genetic transformation is an important mechanism for gene exchange between streptococci in nature. A prerequisite for effective gene exchange is the presence of streptococcal donor DNA in the environment. Despite decades of study of the transformation process we still do not know how this donor DNA is released from streptococcal cells to the external milieu. Traditionally, it has been assumed that donor DNA originates from cells that die and fall apart from natural causes. In this study we show that induction of the competent state initiates release of DNA from a subfraction of the bacterial population, probably by cell lysis. The majority of the cells induced to competence take up DNA and act as recipients, whereas the rest release DNA and act as donors. These findings show that natural transformation in streptococci provides a natural mechanism for genetic recombination that resembles sex in higher organisms.
Induction of the competent state in streptococci belonging to the mitis phylogenetic group is controlled by the extracellular concentration of a secreted peptide pheromone called the competence-stimulating peptide (CSP) (1–3). This type of autoinduction, often called quorum-sensing, constitutes a mechanism by which bacteria can communicate at an intercellular level to monitor their own population density (4). Different strains and species of naturally competent streptococci produce CSPs with different primary structures. Two different types of competence pheromones, CSP-1 and CSP-2, predominate in strains of Streptococcus pneumoniae, whereas strains belonging to, for instance, Streptococcus mitis, produce a large variety of pheromone types (pherotypes) (refs. 2 and 3 and unpublished results). In the present work we have used different mutants of S. pneumoniae strain Rx, which responds to the CSP-1 pheromone (1).
In S. pneumoniae competence is triggered when CSP reaches an extracellular concentration of 1–10 ng/ml, which corresponds to a population density of about 107 cells per ml (1). The signaling cascade, which leads to induction of the competent state, begins with the interaction of CSP with its histidine kinase receptor (ComD), which responds by transferring a phosphate group to its cognate response regulator (ComE) (5, 6). After phosphorylation the response regulator activates transcription of several genes containing a ComE-binding site in their promoter regions (7–9). As the promoter region of comX, a gene encoding an alternative sigma factor, contains a sequence motif similar to the ComE-binding site, it is assumed that comX is among the genes controlled by ComE–P (10). Expression of ComX activates transcription of the late genes that together constitute a competence regulon containing the proteins involved in binding, processing, uptake, and recombination of DNA (8–11).
Our knowledge of the different steps in the transformation process, such as regulation of competence, DNA uptake, and recombination, has increased enormously during the last decade (1, 4, 8–10). There has been, however, little progress in our understanding of how gene exchange takes place between streptococci under natural conditions. Presumably, streptococci participate in gene exchange when situated in biofilms in locations such as the oral cavity and the nasopharynx (12). Analogous to autoinduction of competence in liquid culture, streptococcal cells growing in biofilms will undergo competence development when a certain cell density is reached. Most likely, the purpose of this quorum-sensing mechanism is to monitor the concentration of potential gene donors in the immediate neighborhood. The aim of the present study was to determine whether release of donor DNA and its subsequent uptake by recipients are separate or coordinated events. In other words, do competent streptococci depend on DNA that is released to the environment at random when other streptococci die and fall apart, or is release of DNA from donor cells coordinated in time and space with uptake by the recipients? In 1960, Ottolenghi and Hotchkiss (13) addressed similar questions with in vitro gene exchange between mixed growing populations of two mutants of S. pneumoniae strain R6, resistant to sulfonamid/amethopterin and streptomycin/micrococcin, respectively. They did not succeed, however, in determining whether the DNA released in their cocultivation experiments came from dead cells as a result of normal population turnover or from active release triggered by induction of the competent state. We decided to reinvestigate this important question and reached a clear conclusion. Our data show that DNA is actively released from pneumococci during competence development by competence-induced lysis of a subfraction of the cells.
Materials and Methods
Bacterial Strains and Growth Conditions.
Strains CP1415 and CP1500, which have been described previously (14, 15), are both derived from S. pneumoniae strain Rx (Table 1). Bacterial strains were grown in casein tryptone medium (CAT) (16), which was also used in transformation and β-galactosidase assays. CAT contains (per liter) 5 mg of choline chloride, 5 g of tryptone, 10 g of enzymatic casein hydrolysate, 1 g of yeast extract, and 5 g of NaCl. After sterilization, 0.2% glucose and 167 mM K2HPO4 were added before use. Unless specified otherwise, all incubations were carried out at 37°C, and all optical density measurements of bacterial cultures were done spectrophotometrically at 550 nm.
Table 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Relevant characteristics | Source/ref. |
|---|---|---|
| Strain | ||
| S. pneumoniae Rx | R36A derivative | 37 |
| CP1200 | Rx, but malM511 str-1 | 15 |
| CP1415 | CP1200, but comA− by insertion of the ermAM cassette into the ClaI site of comA | 14 |
| CP1500 | hex nov-r1 bry-r str-1 ery-r1 ery-r2 | 38 |
| R262 | R800, but negative for endogenous β-galactosidase activity (ebg−) | 19 |
| R319 | R800, but comE∷pXF518 by transformation with plasmid DNA | 19 |
| EK100 | CP1415, but egb− by transformation with genomic DNA from R262 | This work |
| EK4166 | EK100, but hirL∷pEVP3 by transformation with plasmid DNA | This work |
| EK4167 | EK4166, but NovR by transformation with CP1500 genomic DNA | This work |
| H1 | CP1415, but NovR by transformation with CP1500 genomic DNA | This work |
| H2 | H1, but comE∷pXF518 by transformation with R319 genomic DNA | This work |
| H3 | H1, but lytA∷pEVP3 by transformation with plasmid DNA | This work |
| Plasmid | ||
| pEVP3 | Nonreplicative vector (CmR, lacZ) | 18 |
Construction of S. pneumoniae Rx Mutants Used in This Study.
The EK4166 mutant (Table 1) was originally made to study the effect of disrupting a pneumococcal gene of unknown function termed hirL (GenBank accession no. AAL00764). The gene product of hirL shows strong homology to a family of plant defense proteins named hypersensitive-induced reaction (HIR) (17). It turned out that the hirL gene is relatively strongly expressed throughout the logarithmic growth phase, but disruption of this gene did not affect natural competence or the growth rate of the cells. The EK4166 mutant, containing a transcriptional fusion between hirL and lacZ, could therefore be used to look for competence-induced release of β-galactosidase.
PCR was performed with the degenerated primers eiv.3 (5′-TAATATGCATGARACIAARACIAARGAYAAYG-3′) and eiv.4 (5′-TAATGGATCCGCIGCRTTIATYTCRTTTAC-3′) containing an NsiI and a BamHI site at their 5′ ends, respectively. DNA from strain CP1415 was used as template, and the annealing temperature in the PCR reaction was 60°C. After 29 cycles the product was analyzed by agarose gel electrophoresis, and a fragment of the expected size (∼325 bp) was detected. The rest of the PCR reaction mix was used in a TOPO-cloning (Invitrogen), as described by the manufacturer. The cloned PCR-fragment was excised from the TOPO vector with the restriction enzymes BamHI and NsiI, purified by standard methods, and ligated into the plasmid pEVP3 (18) precleaved with BamHI and NsiI. This ligation reaction was used to transform Escherichia coli Top 10 cells (Invitrogen), and purified pEVP3 plasmid from a transformant containing the cloned PCR fragment was then used to transform strain EK100 (NovS, ComA−, ebg−) by natural transformation. A transformant with the pEVP3 plasmid, which does not replicate in pneumococci, integrated into the hirL gene, was isolated by selection on agar plates containing 2.5 μg/ml of chloramphenicol. This insertion-duplication mutant, termed EK4166, contains a transcriptional fusion between hirL and lacZ located on the chromosome.
The EK100 strain (Table 1), used in the transformation experiment described above, was made ebg− (lacking endogenous β-galactosidase activity) by transforming strain CP1415 with chromosomal DNA from S. pneumoniae strain R262 (ebg−) (19) and selecting for white mutants on 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) agar.
The EK4167 (NovR) mutant was made by transforming EK4166 (NovS) with chromosomal DNA from CP1500 (NovR), and selecting for transformants by plating on CAT-agar containing 10 μg/ml of novobiocin (see Table 1). Similarly, the H2 (NovR, ComA−, ComE−) mutant was made by transforming strain H1 (NovR, ComA−) with chromosomal DNA from strain R319 (ComE−) (19) and selecting transformants by plating on CAT-agar containing 2.5 μg/ml of chloramphenicol (see Table 1). Finally, a mutant (H3) lacking the pneumococcal autolysin (LytA) was constructed by transforming the H1 strain with the pEVP3 plasmid containing a ∼350-bp internal fragment of the lytA gene and selecting for transformants able to grow in the presence of 2.5 μg/ml of chloramphenicol. PCR, carried out with the LytA1 (5′-ATATTGGTACCGCGGTTGGAATGCTGAGACCTATGCAGC-3′) and LytA2 (5′-ATATTGGATCCGTACCAGTAGCCAGTGTCATTCTTCTGCC-3′) primers, and subsequent cloning of the resulting DNA fragment into the pEVP3 plasmid were carried out as described above. To verify that the correct insertion-duplication mutant had been made, two primers, LytA3 (5′-GAACAGATTTGCCTCAAGTCGGCG-3′) and LytA4 (5′-CCAGTTGCCGTCTGTGTGCTTCCTCC-3′), complementary to sequences in the lytA gene upstream and downstream of the site of integration, were synthesized. PCR with these primers in combinations with primers complementary to sequences in the pEVP3 vector, upstream and downstream of the pEVP3 polylinker, gave rise to PCR fragments of the expected size, thus confirming that the lytA gene had been disrupted. To confirm further that the LytA autolysin is no longer functional in the H3 mutant, this strain and the H1 strain were treated with 0.1% and 1% deoxycholate. This treatment caused the H1 strain to lyse within 10 min, whereas the H3 strain remained unaffected. The same result was obtained when 0.1% and 1% Triton X-100 was used instead of deoxycholate. In sum, these results show that the H3 strain is not producing LytA (20).
β-Galactosidase Assays.
To determine whether competence induction in S. pneumoniae strain Rx causes cell lysis, supernatants and cell lysates from cultures induced to competence with the peptide pheromone CSP-1 (NH2-EMRLSKFFRDFILQRKK-COOH) (1) were assayed for β-galactosidase activity. Uninduced samples, receiving no peptide pheromone, were always used as negative controls. In some assays the peptide pheromones CSP-2 (NH2-EMRISRIILDFLFLRKK-COOH) (21) and SpiP (NH2GWWEELLHETILSKFKITKALELPIQL-COOH) (22, 23), which do not induce competence in strain Rx, were also included.
An overnight culture of strain EK4166 or EK4167 grown in CAT was diluted to OD550 = 0.05 in preheated CAT (37°C) and incubated until it reached OD550 = 0.2–0.3. The culture was then diluted once more to OD550 = 0.05 in preheated CAT and incubated in a water bath at 37°C. During logarithmic growth (OD550 = 0.1–0.7), 10-ml samples were withdrawn at regular intervals (OD550 ≅ 0.1, 0.2, etc.), and CSP-1 was immediately added to a final concentration of 250 ng/ml. Uninduced samples, receiving no peptide pheromone, were run in parallel as negative controls. After 30 min at 37°C samples were removed from the water bath and placed on ice. To measure total β-galactosidase activity directly in samples from pneumococcal cultures, the bacterial cells were first lysed by addition of 0.1% (vol/vol) Triton X-100, followed by a 10-min incubation at 37°C. After lysis, all samples were chilled on ice until assayed. To measure β-galactosidase activity in supernatants, samples were first centrifuged at 2,500 × g for 20 min and then filtered (0.2-μm filter) to obtain supernatants that were completely cell-free.
β-Galactosidase assays of supernatants were carried out in Eppendorf tubes at 30°C and contained 240 μl of 5× Z buffer [5 mM MgCl2/250 mM β-mercaptoethanol/50 mM KCl/0.3 M Na2HPO4⋅7H2O; 0.2 M NaH2PO4⋅H2O; 4 mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG), pH 7.0], 800 μl of supernatant, and 160 μl of CAT medium. β-Galactosidase activity in cell lysates was assayed in the same way except that only 100 μl of cell lysate was used in the reactions. After stopping the reactions by adding 0.5 ml of a 1 M Na2CO3 solution, hydrolysis of ONPG was recorded in a spectrophotometer at 420 nm. Enzyme activity was calculated according to Miller (24).
When studying the effect of choline on competence-induced release of β-galactosidase the assay was carried out exactly as described above, except that the overnight culture of strain EK4166 was diluted into CAT containing 2% choline chloride. This choline-containing medium was then used throughout the assay. The negative control, which was run in parallel, was grown in CAT medium.
Transformation Assays.
To detect possible competence-induced release of NovR DNA into the growth medium from strains EK4167, H2, or H3, transformation assays were carried out by adding samples of supernatant from these NovR strains to competent cultures of the novobiocin-sensitive strain CP1415, as specified below. To obtain vigorously growing bacteria, overnight cultures of NovR strains were treated as described for the β-galactosidase assays. Competence was induced by addition of 250 ng/ml of CSP-1 at OD550 = 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, and 0.7. Uninduced negative controls were always run in parallel. Thirty minutes after initiation of competence induction, 10-ml culture samples were withdrawn and centrifuged at 2,500 × g in a prechilled centrifuge (4°C) for 10 min. The supernatants were harvested and put on ice, and were thereafter sterile-filtered (0.2 μm) as quickly as possible (within 5–10 min) and snap-frozen in a bath of liquid nitrogen. When analyzed for the presence of released NovR DNA (see below), these cell-free supernatants were thawed on ice and kept cold until refrozen (−80°C) to suppress nuclease activity.
An overnight culture of strain CP1415 was diluted in preheated CAT to OD550 = 0.05, incubated for 45 min, diluted again 10-fold, and incubated further for 30 min at 37°C. At this stage, the CP1415 culture was ready for transformation, which was always carried out in Eppendorf tubes in a total volume of 1 ml (bacterial culture plus added cell-free supernatant). In most experiments, 0.1 or 0.5 ml of cell-free supernatant, to be tested for the presence of NovR DNA, were added to 0.9 or 0.5 ml of bacterial culture, respectively. Then 250 ng of CSP-1 was immediately added to induce competence in the CP1415 cells. After incubation for 120 min at 37°C, growth was stopped by placing the samples in an ice bath. For each sample the number of transformants (colony-forming units per ml of supernatant) was estimated by plating on CAT-agar plates containing 10 μg/ml of novobiocin.
Time Course Study of β-Galactosidase Release.
To study the kinetics of β-galactosidase release a time course experiment was carried out. An overnight culture of strain EK4167 was diluted to OD550 = 0.1 in preheated CAT (37°C) and incubated until it reached OD550 = 0.3. The culture was then diluted again to OD550 = 0.1 and incubated further at 37°C until it reached OD550 = 0.4. At this OD, the cells were induced to competence by addition of 250 ng/ml of CSP-1. Samples of 1.5 ml of bacterial culture were collected every 5 min for a period of 60 min after competence induction. Immediately after the samples were withdrawn, they were centrifuged at ∼12,000 × g for 2 min and filtered (0.2 μm) to obtain a cell-free extract. Then the supernatants were placed on ice until assayed. Samples from an uninduced culture (without CSP-1 added) were run in parallel as a negative control.
Results
Competence-Induced Release of β-Galactosidase from S. pneumoniae Cells.
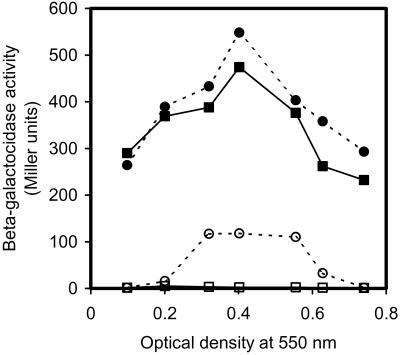
To determine whether some of the cells lyse when competence is induced in a growing population of S. pneumoniae, we used a mutant constructed from the EK100 strain (NovS, ComA−, ebg−) that expresses E. coli β-galactosidase from a strong constitutive promoter. This mutant, designated EK4166, is negative for endogenous β-galactosidase activity (ebg−). To make EK4166, an internal 325-bp PCR fragment from a pneumococcal gene of unknown function, termed hirL (GenBank accession no. AAL00764), was cloned into the pEVP3 plasmid. The pEVP3 plasmid carries an E. coli lacZ reporter gene suitable for expression in transcriptional fusions in pneumococcus (18). Transforming strain EK100 with this construct resulted in integration of the recombinant plasmid into the hirL gene by an insertion-duplication mechanism, giving rise to a chromosomally located transcriptional fusion between hirL and lacZ (hirL∷lacZ). Disruption of the hirL gene did not have any influence on competence development or transformability, as EK4166 cells transformed with the same efficiency as wild type in all transformation assays carried out. The EK4166 mutant lacks a functional CSP transporter (ComA−) and can for this reason develop the competent state only if synthetic CSP-1 is added to the culture (1, 25). We could therefore use this mutant to measure the amount of β-galactosidase produced in total cell extracts of competence-induced and uninduced EK4166 cells at different population densities, and compare these values to the β-galactosidase activity in corresponding sterile-filtered (0.2 μm) supernatants. The result of this experiment clearly shows that cytoplasmic β-galactosidase is released from pneumococcal cells to the supernatant in competent EK4166 cultures when competence is induced at OD550 = 0.3–0.55 (Fig. 1). The β-galactosidase activity in corresponding supernatants from noncompetent cultures was about 50-fold lower. Based on the total β-galactosidase activity present in the cultures (cells plus supernatant) and the activity present in the corresponding supernatants alone, a percentage of the cells undergoing lysis can be calculated. In the experiment shown in Fig. 1, ≈1/5 of the total β-galactosidase activity is found in the supernatant when the EK4166 culture is induced to competence at OD550 = 0.4, indicating that about 20% of the cells have lysed. Interestingly, competence-induced lysis seems to be influenced by growth phase and cell density. In the particular experiment shown in Fig. 1, competence induction does not result in a significant release of β-galactosidase from the EK4166 cells in the beginning of logarithmic phase (OD550 = 0.1–0.2) and in the transition between logarithmic and stationary phase (OD550 ≈ 0.7). The loss of cell lysis toward the end of logarithmic phase is reproducible from one experiment to another, and is probably the result of loss of competence induction at this stage of the growth phase (1). The level of cell lysis obtained at lower cell densities (OD550 = 0.1–0.3), however, is less reproducible. Most often no competence-induced cell lysis is seen at OD550 ≤ 0.1; however, between OD550 = 0.2–0.3, the results will vary between experiments.
Figure 1.
Competence-induced release of cytoplasmic β-galactosidase from S. pneumoniae cells. Samples of the bacterial culture were collected at different cell densities (OD550 = 0.1–0.8) and induced to competence by adding 250 ng/ml of synthetic CSP-1. Corresponding uninduced samples were collected as negative controls. A cell lysate (lysed cells plus supernatant) and cell-free supernatant were made from each sample, and the β-galactosidase activity was measured in both. Closed circles, cell lysates from competent cells; closed squares, cell lysates from noncompetent cells; open circles, supernatants from competent cells; open squares, supernatants from noncompetent cells. β-Galactosidase activities are given in Miller units.
Competence-Induced Release of Chromosomal DNA from S. pneumoniae Cells.
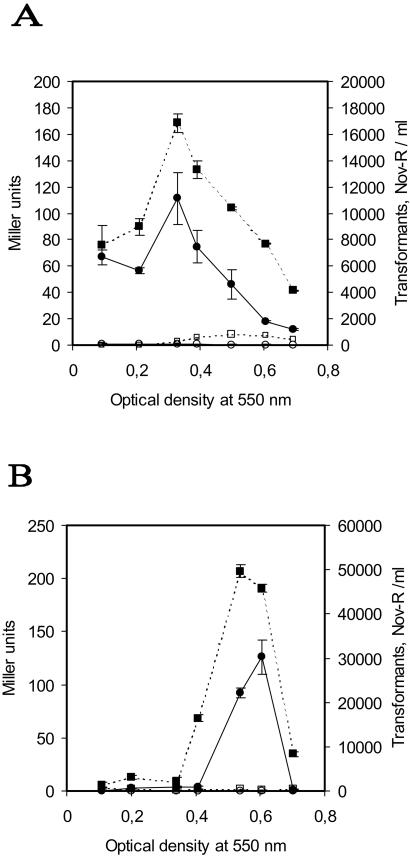
Having demonstrated release of β-galactosidase from competence-induced pneumococcal cultures, we wanted to determine whether chromosomal DNA is liberated from the cells as well. For this purpose we made a novobiocin-resistant mutant of EK4166 by transforming this strain with chromosomal DNA from S. pneumoniae CP1500 (NovR) and selecting for novobiocin resistance. Cultures of the resulting NovR mutant (EK4167) were induced to competence at different cell densities, and supernatants were harvested by centrifugation (2,500 × g) and filtration (0.2 μm). These supernatants and corresponding supernatants from noninduced cultures of EK4167 were assayed for both β-galactosidase activity and the presence of DNA containing a NovR marker. A standard transformation assay, using CP1415 cells, was carried out to detect NovR DNA. The results show that a low number (0–100 per ml) of novobiocin-resistant transformants were obtained when transforming CP1415 cells with supernatants from noncompetent EK4167 cells. In contrast, a maximum of about 3 × 104 transformants per ml of supernatant were obtained with supernatants from competent EK4167 cells (Fig. 2B). Compared with noncompetent cells, the number of transformants obtained with supernatants from competence-induced cells was on average 100-fold higher. In both of the separate but otherwise identical experiments depicted in Fig. 2 A and B, the maximum number of transformants obtained coincides with the maximum level of β-galactosidase activity present in the same supernatant. These results show that chromosomal DNA is released from pneumococci during competence development and suggest that the release of chromosomal DNA and β-galactosidase is brought about by the same cell-lysing mechanism.
Figure 2.
Analysis of β-galactosidase activity and NovR DNA contents in the same supernatant. Samples were taken at regular intervals during growth in CAT and induced to competence by adding 250 ng/ml of CSP-1. Uninduced samples, receiving no peptide pheromone, were run in parallel as negative controls. A and B show the results of two separate experiments that otherwise are identical. Closed squares and open squares represent β-galactosidase activity in cell-free supernatants from competent and noncompetent cultures, respectively. Closed circles represent the number of transformants obtained per ml when novobiocin-sensitive pneumococci were transformed with cell-free supernatant from competence-induced EK4167 (NovR) cultures. Open circles represent the negative control, showing the number of transformants obtained with cell-free supernatants from noncompetent EK4167 (NovR) cultures. β-Galactosidase activities are given in Miller units and are the means ± SEs of triplicate samples. Where error bars are not visible, the SE was within the area occupied by the symbol.
Kinetics of β-Galactosidase Release.
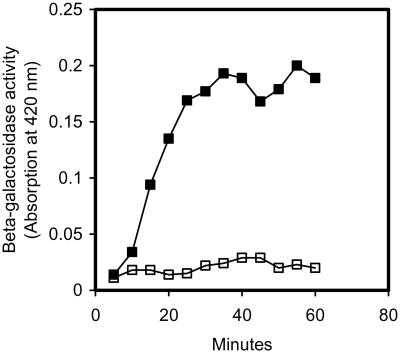
In S. pneumoniae strain Rx competence reaches a maximum about 20 min after pheromone induction and then declines during the next 20–30 min until most of the cells in the population have returned to the noncompetent state (9). In a recent article by Peterson et al. (9) the kinetics of competence induction was studied by monitoring β-galactosidase expression in S. pneumoniae cells harboring a comX∷lacZ reporter fusion. Their results showed that increasing amounts of β-galactosidase are produced during the first 25 min after competence induction. After 25 min the β-galactosidase activity leveled off, demonstrating that expression of the alternative sigma factor ComX was shut down. In a similar experiment, we studied the kinetics of competence-induced release of β-galactosidase from EK4166 cells. The results of this experiment showed that the kinetics of ComX expression and β-galactosidase release are strikingly similar (Fig. 3). After about 25–30 min there was no further release of β-galactosidase, showing that cell lysis had stopped. Together these data show that DNA uptake and DNA release reach their maximums at about the same time, ensuring high efficiency of gene transfer.
Figure 3.
Kinetics of competence-induced release of cytoplasmic βgalactosidase to the external medium. Samples assayed for β-galactosidase activity were collected from an S. pneumoniae EK4166 culture every 5 min for a period of 60 min after competence induction. Closed squares, supernatants from a culture that received 250 ng/ml of CSP-1 at time 0; open squares, supernatants from a corresponding noncompetent culture.
Competence Development and Cell Lysis Are Both Controlled by the Same Quorum-Sensing Mechanism.
Competence-induction experiments have shown that S. pneumoniae strain Rx, which belongs to the CSP-1 pherotype, responds very poorly to CSP-2. Similarly, no competence-inducing activity is detected when the peptide pheromone SpiP, which regulates bacteriocin production in the Rx strain (22, 23), is added to noncompetent cultures (unpublished results). If competence development and cell lysis are regulated by the same quorum-sensing mechanism (ComCDE), it would be expected that both processes are equally CSP-1-specific. To determine whether this is the case we added 250 ng/ml of CSP-1, CSP-2, or SpiP to EK4167 cultures at OD550 = 0.4 and measured the β-galactosidase activity in the supernatants after a 30-min induction. The results clearly showed that only CSP-1 triggers cell lysis (Table 2). However, even though cell lysis depends on CSP-1, it is possible that it does not act through the two-component regulatory system (ComDE) that controls competence development. To address this question we disrupted ComE in the H1 strain by transforming with genomic DNA from S. pneumoniae stain R319 (ComE−) (19) and selecting for chloramphenicol resistance. The resulting mutant H2 (NovR, ComA−, ComE−), which is completely deficient in competence, was then tested in the same type of DNA-release assay as described above. CSP-1 was added to cultures of this mutant at ODs ranging from OD550 = 0.1–0.7 and incubated for 30 min. Negative controls, receiving no competence pheromone, were run in parallel. Sterile-filtered supernatants were subsequently used to transform strain CP1415. The same low number of transformants (0–100 per ml of supernatant) was obtained with supernatants from all cultures, regardless of whether they had been treated with CSP-1. These results demonstrate that CSP-1-induced DNA release does not take place when ComE is disrupted and show that the ComCDE signal transduction pathway controls both induction of the competent state and the cell-lysis mechanism.
Table 2.
Comparison of β-galactosidase activity in cell lysates and corresponding cell-free supernatants from pneumococcal cultures treated with different peptide pheromones
| Pheromone added* | Competence-induced | Cell lysates† | Supernatants† |
|---|---|---|---|
| None | No | 634.1 ± 19.6 | 0.63 ± 0.41 |
| CSP-1 | Yes | 817.8 ± 26.4 | 38.4 ± 3.1 |
| CSP-2 | No | 700.4 ± 22.7 | 0.26 ± 0.2 |
| SpiP | No | 699.2 ± 18.2 | 0.41 ± 0.2 |
Peptide pheromones were added at a concentration of 250 ng/ml.
β-Galactosidase activities are given in Miller units and are the means ± SEs of triplicate samples.
Cell Lysis Depends on a Choline-Binding Protein (CBP).
CBPs bind noncovalently to the phosphorylcholine moiety of the S. pneumoniae cell wall through a conserved choline-binding domain (26, 27). A search of the S. pneumoniae genome with the amino acid sequence of the choline-binding domain of CbpA revealed that there are 12 members of the CBP family in the S. pneumoniae type 4 strain (28). Several of these proteins are involved in host cell recognition and colonization of the nasopharynx, one bears sequence similarity to a serine protease, and three (LytA, LytB, and LytC) are murein hydrolases (28). In has been known for many years that a concentration of 2% choline chloride in the broth inhibits the action of the pneumococcal autolysin LytA (29, 30). The same seems to be the case for LytB and LytC, as CBPs in general do not attach to the cell wall in the presence of high concentrations of choline (28, 31). To determine whether any of the three murein hydrolases could be involved in competence-induced cell lysis, we set up an experiment to examine the effect of choline on the release of β-galactosidase from EK4166 cells. The results presented in Table 3 show that addition of 2% choline to the growth medium completely abolishes enzyme release. Control experiments carried out to check whether the addition of choline interferes with competence development demonstrated that its presence in the growth medium has no negative effect on the transformability of the pneumococcus. However, the presence of 2% choline in the enzyme reaction mix resulted in a 20% reduction of β-galactosidase activity compared with standard reaction conditions. This modest reduction in enzyme activity cannot explain the results in Table 3, which show that the presence of choline totally blocks release of β-galactosidase. Consequently, choline most likely inhibits release of β-galactosidase by directly interfering with the binding of CBPs to the cell wall, demonstrating that at least one of these proteins is involved in the cell-lysis mechanism.
Table 3.
CSP-1-induced release of β-galactosidase in the presence of 2% choline
| OD at 600 nm | β-Galactosidase activity (Miller units) in
supernatants from EK4166 cells*
|
|||
|---|---|---|---|---|
| Growth in
CAT plus 2% choline
|
Growth in CAT
|
|||
| CSP-1 added | No pheromone added | CSP-1 added | No pheromone added | |
| 0.1 | 0.36 | 15.4 | 26.9 | 11 |
| 0.2 | 1.8 | 2.4 | 28.2 | 5.2 |
| 0.3 | 1.0 | 0.9 | 78.7 | 4.5 |
| 0.4 | 0.82 | 0.9 | 325.0 | 4.0 |
| 0.5 | 0.55 | 0.9 | 91.3 | 4.6 |
| 0.6 | 0.60 | 0.4 | 32.3 | 2.9 |
| 0.7 | 1.1 | 1.7 | 5.60 | 2.6 |
Miller units are the means of two parallels.
Interestingly, transcription of the lytA gene, an amidase that breaks down the N-acetylmuramoyl-l-alanine bonds in the peptidoglycan backbone of S. pneumoniae (32), is up-regulated during competence through the ComCDE signal transduction pathway (33). Thus far, LytA is the only pneumococcal murein hydrolase known with certainty to be competence-induced, i.e., to be a member of the ComE or ComX regulons. That CSP-1-induced up-regulation of LytA expression would not take place in a ComE− background could explain why we failed to detect release of NovR DNA from H2 cells treated with CSP-1. We therefore decided to construct a mutant (H3) with a disrupted lytA gene, to determine whether absence of a functional autolysin would interfere with CSP-1-induced release of DNA. Our results show that competence-induced release of NovR DNA in the H3 mutant is reduced approximately 10-fold compared with wild type, demonstrating that LytA is an important component of the DNA-release mechanism. However, the number of NovR transformants obtained with supernatants from competence-induced H3 cells is still approximately 10-fold higher than the corresponding number obtained with supernatants from uninduced H3 cells.
Discussion
We have shown that DNA uptake and DNA release are both induced by the same signal transduction pathway (ComCDE) in S. pneumoniae. Consequently, proteins involved in the release of DNA must be members of the ComE or ComX regulons. We have also shown that the DNA-release mechanism depends on at least 1 of the 12 different CBPs synthesized by pneumococci. As the pores in the Gram-positive cell wall are too small to allow β-galactosidase and DNA to pass through (34), we assume that the observed leakage of these molecules from the cytoplasm into the growth medium involves degradation of the peptidoglycan. We therefore speculated that LytA, LytB, or LytC is involved in the DNA-release mechanism. All three are murein hydrolases that are anchored to the pneumococcal cell wall through their choline-binding domains (32). The most obvious candidate is LytA, which is a member of the ComX regulon. LytA is expressed in noncompetent cells, but transcription of the lytA gene is further up-regulated (≈6-fold) during competence development (8). As it is constitutively expressed, LytA must be under strong negative regulation to avoid uncontrolled digestion of the cell wall exoskeleton. It is not known how it is activated in stationary phase to cause autolysis of the culture. Disruption of the lytA gene significantly lowered competence-induced release of DNA from the H3 mutant cells, demonstrating that LytA plays an important role in the DNA-release mechanism. However, increased expression of LytA cannot by itself account for all known properties of this mechanism. Even in the absence of LytA some competence-induced release of DNA can still be observed, indicating that LytB, LytC, or even a murein hydrolase from a pneumococcal bacteriophage could be involved in addition to LytA. Furthermore, competence-induced release of β-galactosidase and DNA is cell density-dependent, a property that is difficult to reconcile with autolysis caused by increased LytA activity alone. In addition, only a subfraction of the cells is lysed, indicating that some finely balanced control system is operating to prevent lysis of the whole population. Clearly, much remains to be learned about the DNA-release mechanism and how it is regulated during competence development.
It seems to be important for naturally competent bacteria to avoid taking up too much DNA from distantly related or unrelated bacteria. This DNA uptake could overload the mismatch-repair system and lead to damages in the recipient's chromosome (35). Naturally competent Gram-negative bacteria such as Neisseria gonorrhoeae and Haemophilus influenzae have solved this problem by having short DNA uptake sequences (10–11 bp) distributed throughout their genomes (36). When exposed to a mixture of homologous and foreign DNA these Gram-negative bacteria will preferentially take up homologous DNA that contains their own uptake sequence. In streptococci intercellular communication by pheromones, quorum-sensing regulation of competence induction, and the DNA-release mechanism reported in the present work, are used to ensure that homologous DNA will predominate in the environment at the time when competent cells are ready to take up DNA. The quorum-sensing mechanism, which depends on specific competence pheromones, will inhibit competence development until a sufficient number of related streptococci, belonging to the same pherogroup, are present in a certain volume of space. A subfraction of the bacteria belonging to this pherogroup will then act as donors and release DNA that can be taken up by the others. Our experiments, which have been carried out with planctonic cells in liquid culture, indicate that 5–20% of the cells in a competent population will lyse and act as donors of DNA (Fig. 1 and Table 2). However, the exact percentage will probably vary with growth conditions and might be different in biofilms that represent a radically different environment for the bacteria.
Presumably, the pheromone-dependent gene-exchange mechanism described in this study will be one of the factors that drives speciation in naturally competent streptococci. If a competent Streptococcus, through random mutations or recombination, acquires a new pherotype that is unique and as a result loses the ability to communicate with other related streptococci, it becomes genetically isolated and will probably gradually evolve into a separate species. Supporting this theory is the fact that some of the more well defined species within the mitis phylogenetic group, Streptococcus gordonii, Streptococcus sanguis, and Streptococcus crista, produce few pheromone types that are distinct for each species and that are unrelated to pheromones produced by the other species belonging to the mitis group (2).
Acknowledgments
We thank professor Jean-Pierre Claverys (Univ. Paul Sabatier, Toulouse, France) for kindly providing S. pneumoniae strains R262 and R319. This work was supported by a grant from the Research Council of Norway.
Abbreviations
- CSP
competence-stimulating peptide
- CAT
casein tryptone medium
- CBP
choline-binding protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Håvarstein L S, Coomaraswamy G, Morrison D A. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Håvarstein L S, Hakenbeck R, Gaustad P. J Bacteriol. 1997;179:6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whatmore A M, Barcus V A, Dowson C G. J Bacteriol. 1999;181:3144–3154. doi: 10.1128/jb.181.10.3144-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Håvarstein L S, Morrison D A. In: Cell–Cell Signaling in bacteria. Dunny G M, Winans S C, editors. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 9–26. [Google Scholar]
- 5.Håvarstein L S, Gaustad P, Nes I F, Morrison D A. Mol Microbiol. 1996;21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 6.Pestova E V, Håvarstein L S, Morrison D A. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 7.Ween O, Gaustad P, Håvarstein L S. Mol Microbiol. 1999;33:817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- 8.Rimini R, Jansson B, Feger G, Roberts T C, de Francesco M, Gozzi A, Faggioni F, Domenici E, Wallace D M, Frandsen N, Polissi A. Mol Microbiol. 2000;36:1279–1292. doi: 10.1046/j.1365-2958.2000.01931.x. [DOI] [PubMed] [Google Scholar]
- 9.Peterson S, Cline R T, Tettelin H, Sharov V, Morrison D A. J Bacteriol. 2000;182:6192–6202. doi: 10.1128/jb.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M S, Morrison D A. J Bacteriol. 1999;181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell E A, Choi S Y, Masure H R. Mol Microbiol. 1998;27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 12.Li Y-H, Lau P C Y, Lee J H, Ellen R P, Cvitkovitch D G. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ottolenghi E, Hotchkiss R D. Science. 1960;132:1257–1258. [PubMed] [Google Scholar]
- 14.Morrison D A, Trombe M, Hayden M, Waszak G, Chen J D. J Bacteriol. 1984;159:870–876. doi: 10.1128/jb.159.3.870-876.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoemaker N B, Guild W R. Mol Gen Genet. 1974;128:283–290. doi: 10.1007/BF00268516. [DOI] [PubMed] [Google Scholar]
- 16.Morrison D A, Lacks S A, Guild W R, Hageman J M. J Bacteriol. 1983;156:281–290. doi: 10.1128/jb.156.1.281-290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadimpalli R, Yalpani N, Johal G S, Simmons C R. J Biol Chem. 2000;275:29579–29586. doi: 10.1074/jbc.M002339200. [DOI] [PubMed] [Google Scholar]
- 18.Claverys J P, Dintilhac A, Pestova E V, Martin B, Morrison D A. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 19.Alloing G, Martin B, Granadel C, Claverys J-P. Mol Microbiol. 1998;29:75–83. doi: 10.1046/j.1365-2958.1998.00904.x. [DOI] [PubMed] [Google Scholar]
- 20.Mosser J L, Tomasz A. J Biol Chem. 1970;245:287–298. [PubMed] [Google Scholar]
- 21.Pozzi G, Masala L, Iannelli F, Manganelli R, Håvarstein L S, Piccoli L, Simon D, Morrison D A. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reichmann P, Hakenbeck R. FEMS Microbiol Lett. 2000;190:231–236. doi: 10.1111/j.1574-6968.2000.tb09291.x. [DOI] [PubMed] [Google Scholar]
- 23.de Saizieu A, Gardès C, Flint N, Wagner C, Kamber M, Mitchell T J, Keck W, Amrein K E, Lange R. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 25.Hui F M, Morrison D A. J Bacteriol. 1991;173:372–381. doi: 10.1128/jb.173.1.372-381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Puelles J M, Sanz J M, Garcia J L, Garcia E. Gene. 1990;89:69–75. doi: 10.1016/0378-1119(90)90207-8. [DOI] [PubMed] [Google Scholar]
- 27.Wren B W. Mol Microbiol. 1991;5:797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 28.Gosink K K, Mann E R, Guglielmo C, Tuomanen E I, Masure H R. Infect Immun. 2000;68:5690–5695. doi: 10.1128/iai.68.10.5690-5695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giudicelli S, Tomasz A. J Bacteriol. 1984;160:1188–1190. doi: 10.1128/jb.158.3.1188-1190.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balachandran P, Hollingshead S K, Paton J C, Briles D E. J Bacteriol. 2001;183:3108–3116. doi: 10.1128/JB.183.10.3108-3116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia P, Paz González M, Garcia E, Garcia J L, López R. Mol Microbiol. 1999;33:128–138. doi: 10.1046/j.1365-2958.1999.01455.x. [DOI] [PubMed] [Google Scholar]
- 32.Lopez R, González M P, Garcia E, Garcia J L, Garcia P. Res Microbiol. 2000;151:437–443. doi: 10.1016/s0923-2508(00)00172-8. [DOI] [PubMed] [Google Scholar]
- 33.Mortier-Barrière I, de Saizieu A, Claverys J P, Martin B. Mol Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 34.Dijkstra A J, Keck W. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humbert O, Prudhomme M, Hakenbeck R, Dowson C G, Claverys J P. Proc Natl Acad Sci USA. 1995;92:9052–9056. doi: 10.1073/pnas.92.20.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith H O, Gwinn M L, Salzberg S L. Res Microbiol. 1999;150:603–616. doi: 10.1016/s0923-2508(99)00130-8. [DOI] [PubMed] [Google Scholar]
- 37.Ravin A W. J Bacteriol. 1959;77:296–309. doi: 10.1128/jb.77.3.296-309.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cato A, Guild W R. J Mol Biol. 1968;37:157–178. doi: 10.1016/0022-2836(68)90080-6. [DOI] [PubMed] [Google Scholar]