Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of hospital-acquired infections that are becoming increasingly difficult to combat because of emerging resistance to all current antibiotic classes. The evolutionary origins of MRSA are poorly understood, no rational nomenclature exists, and there is no consensus on the number of major MRSA clones or the relatedness of clones described from different countries. We resolve all of these issues and provide a more thorough and precise analysis of the evolution of MRSA clones than has previously been possible. Using multilocus sequence typing and an algorithm, burst, we analyzed an international collection of 912 MRSA and methicillin-susceptible S. aureus (MSSA) isolates. We identified 11 major MRSA clones within five groups of related genotypes. The putative ancestral genotype of each group and the most parsimonious patterns of descent of isolates from each ancestor were inferred by using burst, which, together with analysis of the methicillin resistance genes, established the likely evolutionary origins of each major MRSA clone, the genotype of the original MRSA clone and its MSSA progenitor, and the extent of acquisition and horizontal movement of the methicillin resistance genes. Major MRSA clones have arisen repeatedly from successful epidemic MSSA strains, and isolates with decreased susceptibility to vancomycin, the antibiotic of last resort, are arising from some of these major MRSA clones, highlighting a depressing progression of increasing drug resistance within a small number of ecologically successful S. aureus genotypes.
Methicillin was introduced in 1959 to treat infections caused by penicillin-resistant Staphylococcus aureus. In 1961 there were reports from the United Kingdom of S. aureus isolates that had acquired resistance to methicillin (methicillin-resistant S. aureus, MRSA) (1), and MRSA isolates were soon recovered from other European countries, and later from Japan, Australia, and the United States. MRSA is now a problem in hospitals worldwide and is increasingly recovered from nursing homes and the community (2, 3). The methicillin resistance gene (mecA) encodes a methicillin-resistant penicillin-binding protein that is not present in susceptible strains and is believed to have been acquired from a distantly related species (4). mecA is carried on a mobile genetic element, the staphylococcal cassette chromosome mec (SCCmec), of which four forms have been described that differ in size and genetic composition (5). Many MRSA isolates are multiply resistant and are susceptible only to glycopeptide antibiotics such as vancomycin and investigational drugs. MRSA isolates that have decreased susceptibility to glycopeptides (glycopeptide intermediately susceptible S. aureus, GISA) (6, 7), reported in recent years, are a cause of great public health concern.
Many studies have characterized MRSA isolates from individual hospitals or countries and have identified strains that appear to be well adapted to the hospital environment, are established in several hospitals within a country, or have spread internationally (epidemic MRSA, EMRSA). MRSA isolates are generally characterized by pulsed-field gel electrophoresis, a powerful technique for identifying the relatedness of isolates from recent outbreaks within a hospital, but are not well suited to long-term global epidemiology, which requires a procedure that is highly discriminatory but that indexes variation that accumulates slowly. Multilocus sequence typing (MLST) provides such a procedure and characterizes isolates of bacteria unambiguously by using the sequences of internal fragments of seven housekeeping genes (8, 9). MLST has been developed and validated for S. aureus (10) and provides a discriminatory method that allows related strains recovered in different countries to be readily identified.
The origins of the major MRSA clones are still poorly understood. Kreiswirth et al. (11) proposed that all MRSAs were descended from a single ancestral S. aureus strain that acquired mecA, but more recent studies (12, 13) show that some MRSAs are very divergent, implying that mecA has been transferred between S. aureus lineages. The data from MLST can be used to probe the evolutionary and population biology of bacterial pathogens and to predict ancestral genotypes and patterns of evolutionary descent within groups of related genotypes. We have applied MLST to an international collection of 359 MRSA isolates, which includes examples of the previously described EMRSA and GISA clones, and compare these to a collection of 553 methicillin-susceptible S. aureus (MSSAs). We demonstrate the limited number of major EMRSA genotypes and provide an unambiguous method for characterizing MRSA and GISA clones and a rational nomenclature. We also identify the ancestral MRSA clone and its MSSA ancestor and suggest the evolutionary pathways by which MRSA clones have repeatedly emerged from successful MSSA clones.
Materials and Methods
Bacterial Isolates.
A total of 359 MRSA isolates were collected between 1961 and 1999 from 20 countries. Isolates were confirmed as MRSAs in our laboratory by detecting the presence of the mecA gene with PCR (14). The collection contains members of previously described EMRSA clones, including the Iberian (15), Portuguese/Brazilian (16), Vienna (17), New York/Japan (18, 19), pediatric (20), Berlin (17), Hannover (17), South German (17), EMRSA-3, -15, and -16 (21), and six of the first GISA isolates (minimum inhibitory concentration ≥8 μg vancomycin ml−1) from Japan, the United States, and Scotland (6, 7). The allelic profiles of the MRSA isolates were compared with those of 553 MSSA isolates from disease and carriage; details of all isolates are available at the MLST database (http://www.mlst.net).
MLST.
MLST was performed as described (10). Alleles at the seven loci were assigned by comparing the sequences at each locus to those of the known alleles in the S. aureus MLST database. The allele numbers at each of the seven loci define the allelic profile of each isolate. An allelic profile is defined as a sequence type (ST) that provides a convenient and unambiguous descriptor for each S. aureus genotype. The allelic profiles of all 912 isolates were compared by using the program burst (Based Upon Related Sequence Types). The relatedness of lineages was displayed as a dendrogram constructed from the matrix of pairwise differences in allelic profiles by using the unweighted pairgroup method with arithmetic averages.
burst Analysis.
burst is an algorithm devised by E.J.F. that is used to discern groups of isolates among large MLST datasets that have some defined level of similarity in allelic profile (in this study, identity at five or more of the seven loci), and to predict the ancestral allelic profile (genotype) of each group, or clonal complex (CC), and the most parsimonious patterns of evolutionary descent of all isolates in the group from this putative ancestral allelic profile. A CC should include all of the isolates in the MLST dataset that have descended from the ancestral genotype, although it could include other isolates, for example, descendents of isolates related to the original ancestral genotype. As the CCs are observed within a very small sample of the total S. aureus population, the founding isolate is likely to have gained some adaptive (fitness) advantage such that it increased in frequency within the population. During this clonal expansion slight genetic diversification will occur so that descendents of the ancestral genotype that differ at one of the seven MLST loci will accumulate (single locus variants; SLVs), and the putative ancestral genotype within each CC is therefore defined as the allelic profile that has the largest number of SLVs. Further details are available at http://www.mlst.net/BURST/burst.htm.
mec Gene Analyses.
The SCCmec type was determined for 304 MRSA isolates by PCR detection of the ccr (cassette chromosome recombinase) and mec genes as described by Hiramatsu et al. (5).
Results
MLST of MRSA and GISA Isolates.
MLST revealed 162 STs among the 912 isolates and 38 different allelic profiles (STs) among the 359 MRSA isolates. Twenty five of the MRSA STs included only a single isolate, and there were only 12 STs that contained multiple MRSAs recovered from more than one country (Table 1). Several of these major STs included MRSAs differing in SCCmec type, which presumably have arisen by independent acquisitions of the mec genes. Defining MRSA clones as isolates with the same ST and the same SCCmec type, there were 11 major MRSA clones (more than 10 isolates; Table 1).
Table 1.
Details of STs and clones containing MRSA from more than one country
| EMRSA clone* | No. of isolates MRSA† | No. of isolates MSSA (I/C)‡ | Allelic profile§ | Geographic spread, MRSA¶ | Previous name(s) of EMRSA clone |
|---|---|---|---|---|---|
| ST5-MRSA-1 | 12 | 34 (14/20) | 1-4-1-4-12-1-10 | Pol,Slo,UK | EMRSA-3 |
| ST5-MRSA-II | 21 | 1-4-1-4-12-1-10 | Fin,Ire,Jap,UK,USA | New York/Japan/GISA | |
| ST5-GISA-II | 4 | 1-4-1-4-12-1-10 | Jap,USA | ||
| ST5-MRSA-III | 1 | 1-4-1-4-12-1-10 | Bel | ||
| ST5-MRSA-IV | 5 | 1-4-1-4-12-1-10 | Fra,Por,UK,USA | Pediatric | |
| ST8-MRSA-I | 1 | 38 (21/17) | 3-3-1-1-4-4-3 | Aus | |
| ST8-MRSA-II | 12 | 3-3-1-1-4-4-3 | Ire,UK,USA | Irish-1 | |
| ST8-MRSA-III | 1 | 3-3-1-1-4-4-4 | UK | EMRSA-7 | |
| ST8-MRSA-IV | 20 | 3-3-1-1-4-4-3 | Fin,Fra,Ger,Ire,Net,UK,USA | EMRSA-2,-6 | |
| ST22-MRSA-IV | 43 | 28 (9/19) | 7-6-1-5-8-8-6 | Ger,Ire,Swe,UK | EMRSA-15, Barnim |
| ST30-MRSA-IV | 3 | 85 (38/47) | 2-2-2-2-6-3-2 | Ger,Swe | |
| ST36-MRSA-II | 44 | 0 | 2-2-2-2-2-3-2 | Fin,UK | EMRSA-16 |
| ST45-MRSA-II | 1 | 27 (18/9) | 10-14-8-6-10-3-2 | USA | |
| ST45-MRSA-IV | 13 | 10-14-8-6-10-3-2 | Bel,Fin,Ger,Swe | Berlin | |
| ST228-MRSA-I | 13 | 0 | 1-4-1-4-12-24-29 | Ger,Slo | Southern Germany |
| ST239-MRSA-III | 21 | 0 | 2-3-1-1-4-4-3 | Fin,Ger,Gre,Ire,Net,Pol,Por,Slo,Swe,UK,USA | EMRSA-1, -4, -11 Por/Bra, Vienna |
| ST241-MRSA-III | 2 | 0 | 2-3-1-1-4-4-30 | Fin,UK | |
| ST247-MRSA-I | 39 | 0 | 3-3-1-12-4-4-16 | Bel,Fin,Fra,Ger,Por,Slo,Spa,Swe,UK,USA | EMRSA-5, -17, Iberian |
| ST250-MRSA-I | 21 | 0 | 3-3-1-1-4-4-16 | Den,Ger,Swi,Uga,UK | First MRSA |
| ST254-MRSA-I | 1 | 0 | 3-32-1-1-4-4-3 | UK | |
| ST254-MRSA-IV | 2 | 3-32-1-1-4-4-3 | Ger,UK | EMRSA-10, Hannover |
The 11 major MRSA clones among the 359 MRSA isolates are underlined.
Numbers of MRSA isolates differ from those in Fig. 2 as not all isolates could be SSCmec typed.
Number of MSSA isolates of ST; I = invasive disease isolate; C = carried isolate.
Alleles at the seven MLST loci in the order arcC-aroE-glpF-gmk-pta-tpi-yqiL.
Country codes: Aus-Australia, Bel-Belgium, Den-Denmark, Fra-France, Fin-Finland, Ger-Germany, Gre-Greece, Ire-Republic of Ireland, Jap-Japan, Net-Netherlands, Pol-Poland, Por-Portugal, Slo-Slovenia, Spa-Spain, Swe-Sweden, Swi-Switzerland, Tha-Thailand, Uga-Uganda, UK-United Kingdom, USA-United States.
Several EMRSA clones considered to be distinct through the use of pulsed-field gel electrophoresis and other molecular typing methods were indistinguishable with MLST (Table 1). For example, the clones EMRSA-1, -4, and -11, the Portuguese clone, the Brazilian clone, and the Vienna clone all were ST239. Similarly, EMRSA-2, -6, -7, -12, -13, and -14 and the Irish-1 clone were indistinguishable by MLST (ST8), as were EMRSA-5 and -17 and the Iberian clone (ST247), EMRSA-15 and the Barnim clone (ST22), and EMRSA-3, New York/Japan and the pediatric clone (ST5). Four GISAs from the United States and Japan (6, 7) were ST5, and of the other GISAs studied, one (from the United States) was a member of the ST5 CC. A Scottish GISA isolate belonged to ST235, which is unrelated to other STs when studied by burst, but shares four alleles in common with ST5. In some cases previously defined EMRSA clones with the same ST could be distinguished by the presence of different SCCmec types (Table 1).
Analysis of the mec Genes of MRSA Isolates.
The SCCmec types of 304 MRSA isolates were determined. It was not possible to type all MRSA isolates by using the published primers as ambiguous results were found in some cases, perhaps indicating the existence of a novel SCCmec class. Of those tested, 93 isolates (30.6%) had SCCmec I, 87 (28.6%) had SCCmec II, 28 (9.2%) had SCCmec III, and 96 (31.6%) had SCCmec IV.
The SCCmec types present within the 12 STs that included MRSA from more than one country are shown in Table 1. STs 5, 8, 45, and 254 included MRSA isolates with different SCCmec types, whereas the other STs were uniform in SCCmec type. Isolates with all four SCCmec types were found within both ST5 and ST8.
A Rational Nomenclature of MRSA Clones.
EMRSA clones described in different countries often have been given different names although they are indistinguishable in genotype with MLST. The allelic profile defined by MLST is unambiguous, and we argue that MRSA or EMRSA genotypes should be defined by their STs. Because some of the MRSA STs also include MSSA or GISA isolates, clones are further designated where necessary as ST5–MRSA, ST5–MSSA, or ST5–GISA. Any nomenclature has to take account of the fact that MRSA clones appear to have emerged on more than one occasion in the same genetic background, as MRSA isolates of the same ST may have different SCCmec types. We therefore propose that MRSA clones are defined by both their genotype (ST) and their SCCmec type. Thus, isolates of ST5 may be ST5–MSSA, ST5–MRSA-I, ST5–MRSA-II, etc., or ST5–GISA-II. The relationship between this ST–SCCmec nomenclature and the original arbitrary nomenclature of MRSA clones is shown in Table 1.
Identification of the Ancestral MRSA Genotype.
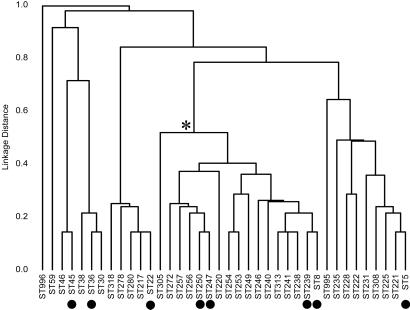
Fig. 1 shows the relatedness of the 38 STs that include MRSA isolates. It is apparent from the dendrogram that many MRSA isolates are only distantly related to each other, although there is one cluster of related MRSA lineages (marked by * in Fig. 1) that includes four of the major international EMRSA STs. All except one of the 35 early MRSA isolates (those from the 1960s) are within this cluster of related STs. The other early MRSA isolate (ST996) is distantly related to all of the isolates that we studied and its origins are unclear.
Figure 1.
Relatedness of MRSA isolates. The 359 MRSA isolates were resolved by MLST into 38 STs, and a dendrogram was constructed from the pairwise differences in their allelic profiles. * identifies a major cluster of related STs, which includes four of the major EMRSA STs (and five of the major EMRSA clones) and all but one of the MRSA isolates from the 1960s. ● denote the seven major international EMRSA STs and one additional EMRSA ST (ST36) that was very abundant in the dataset, but that was largely restricted to the United Kingdom (Table 1).
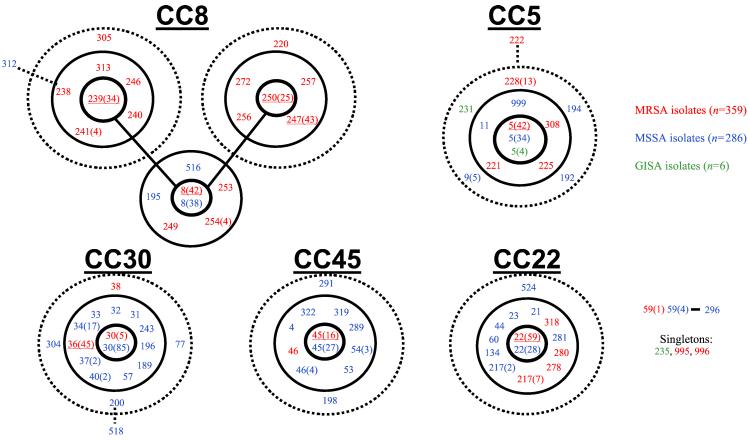
The relationships among similar genotypes are poorly represented by a dendrogram, and the algorithm burst was used to identify groups of related genotypes (CCs) and to predict the ancestral genotype of each group and the most parsimonious patterns of descent from this ancestor. The analysis was applied to all 912 S. aureus for which allelic profiles have been determined with MLST, including the 359 MRSAs and 553 MSSAs from disease and nasal carriage. Fig. 2 shows those CCs identified by burst that include at least one MRSA (or GISA) isolate. CCs that include only MSSA isolates are not shown.
Figure 2.
Analysis of S. aureus isolates with burst. The collection of 914 S. aureus isolates was analyzed by burst, and the five CCs that included MRSA isolates are shown. CCs are named after the ST predicted to be the ancestral genotype (e.g., CC8). The ST prefix is not shown (i.e., 250 corresponds to ST250). The ST of the predicted ancestral genotype is placed in the central circle, SLVs are within the second circle, and double locus variants within the outer dotted circle. Three isolates within CCs but that are not SLVs or double locus variants of the ancestral genotype are also shown (STs 222, 312, and 518). One further small group of isolates for which an ancestral genotype cannot be inferred and three STs that are not members of any CC (singletons) are also shown. The major EMRSA STs are underlined. MRSA STs are shown in red, MSSA STs in blue, and GISA STs in green. In some cases the same ST includes MRSA and MSSA isolates and, for ST5, also GISA isolates. Some MRSA STs include more than one MRSA clone, identified by the presence of different SCCmec types (Table 1). Where more than one, the number of isolates in each ST is shown in parentheses.
All except one of the 35 early MRSA isolates were within a single CC whose predicted ancestor is ST8 (CC8). All except one of these isolates belonged to a subgroup of CC8 that was identified by burst, and most (65%) were identical by MLST and had the allelic profile of the putative ancestral genotype (ST250) of the latter subgroup.
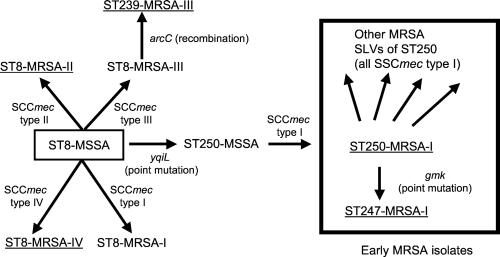
All isolates of the ST250 subgroup are MRSA and possess SCCmec type I; most are isolates from the 1960s. Because ST250 is the predicted ancestor of this subgroup, and this ST includes most of the early isolates, it is considered to be the original MRSA clone (ST250–MRSA-I in our nomenclature). ST250 is predicted to be derived from ST8. These STs differ only at yqiL and the distribution of the alleles at this locus supports the burst prediction that ST8 is the ancestor of the ST250 subgroup. ST8 possesses allele 3 of yqiL, which is found in several other distantly related lineages, whereas ST250, and all SLVs predicted by burst to be derived from it, possess allele 16. Allele 16 is found only in the ST250 subgroup and differs from allele 3 at a single nucleotide site, which results in a polymorphism that is not found in any of the other yqiL alleles, indicating that allele 16 arose from allele 3 by a point mutation, and thus that ST250 arose from ST8.
Although ST250–MRSA-I appears to have been derived from ST8, the latter includes no early MRSA isolates, making it unlikely that mec first appeared in ST8, which then diversified slightly to become ST250. It seems more likely that ST8 was a successful MSSA lineage and slight diversification occurred to produce the SLV, ST250–MSSA, and the latter acquired SCCmec type I and became the EMRSA clone that was first detected in 1961 (Fig. 3).
Figure 3.
Evolutionary origins and patterns of descent within CC8. ST8 is the predicted ancestor of the CC8 CC. Alterations in the allelic profile (and the locus that has changed), and the acquisition of the SCCmec types are shown. The five major EMRSA clones are underlined. All except one of the MRSA isolates from the 1960s are within the STs included in the box. For isolates outside the box not all SLVs are shown. ST250–MSSA isolates were not present in our collection but have been described (26).
Origins of Other Major EMRSA Clones Within CC8.
In addition to ST250–MRSA-I, there are three other major EMRSA STs within CC8. ST8 is a successful MSSA clone but there are also many MRSAs with this genotype. Two major EMRSA clones are apparent within ST8, one with SCCmec type II and one with SCCmec type IV, although single isolates possessed SCCmec types I and III. The ST8–MRSA clones probably emerged by multiple independent introductions of mec into the successful ST8–MSSA clone (Fig. 3), rather than from ST250 (see above), and this view is supported by the fact there are no early MRSA isolates corresponding to ST8.
ST250 now appears to be very uncommon (no ST250–MSSA and only one ST250–MRSA was found among the approximately 870 S. aureus isolates in our collection recovered since 1990), but a SLV of ST250 (ST247) has emerged as one of the most commonly encountered and internationally disseminated multidrug-resistant EMRSA clones (ST247–MRSA-I), and corresponds to the Iberian clone described by Sanches et al. (15). ST247 differs from ST250 at the gmk locus and is almost certainly derived from ST250, because the single nucleotide difference in the gmk allele of ST247 (allele 12) is not found in any other gmk allele and therefore is presumed to be a recent point mutation. Furthermore, ST247 has allele 16 at yqiL, which is restricted to the ST250 subgroup, and the same SCCmec type as ST250 (Fig. 3).
The fourth major EMRSA ST within CC8 is ST239 (ST239–MRSA-III), which corresponds to the Brazilian clone described by de Sousa et al. (16). ST239 is a SLV of ST8 and is assigned by burst as the putative ancestral genotype of another subgroup within CC8. All isolates of ST239 (and all of its associated SLVs and double locus variants) are MRSA, which suggests it was derived from an ancestor that was itself MRSA, and all isolates tested possess SCCmec type III, which within CC8 is otherwise only found in an MRSA isolate of ST8. ST239–MRSA-III therefore is probably derived from a ST8–MRSA isolate that contained SCCmec type III (Fig. 3).
Evolution of the Other Major EMRSA Clones.
Of the major EMRSA STs in Table 1, four are within the group of closely related genotypes that define CC8 (ST8, ST239, ST247, and ST250). However, the other three major international EMRSA STs (ST5, ST22, and ST45), and the prevalent but less widely distributed ST36, are very different in genotype, differing from each other and from the predicted ancestor of CC8 at six or all seven of the loci used in MLST (Fig. 1; Table 1). Similarly, two of the unique MRSA isolates (ST59 and ST996) are very different in genotype from all other MRSA isolates. The presence of mec in such widely divergent genotypes is almost certainly the consequence of the horizontal transfer of the mec region into distantly related S. aureus lineages.
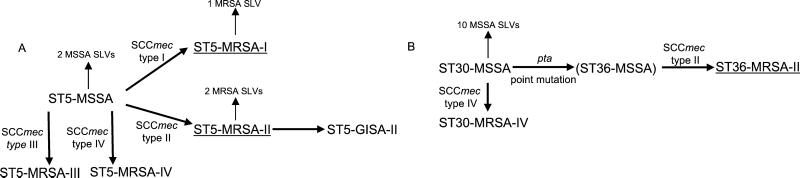
The major international EMRSA STs 5, 22, and 45 are identified by burst as the putative ancestral genotypes of their corresponding CCs (Fig. 2). In each case these STs also include many MSSA isolates, from which the MRSA clones have arisen by the acquisition of mec. There were single EMRSA clones within ST22 and ST45, although one isolate of ST45 possessed a different SCCmec type. ST5 isolates with all four SCCmec types were identified, and ST5–MRSA-I and ST5–MRSA-II were major EMRSA clones (Table 1). The first GISAs from Japan and the United States are ST5 and possess SCCmec type II and are derivatives of the ST5–MRSA-II clone (Fig. 4).
Figure 4.
Evolutionary origins and patterns of descent within CC5 and CC30. (A) The proposed pathways to the major EMRSA clones of ST5 (underlined) and the origins of GISA–ST5-II are shown. (B) The origin of MRSA–ST36-II (underlined) is shown. MSSA–ST36 is on the proposed evolutionary pathway from MSSA–ST30 to MRSA–ST36 but isolates with this genotype have not been observed so far. Other details are as in Fig. 3. Not all SLVs are shown.
ST36 is a major EMRSA genotype in the United Kingdom but in our collection there was only one isolate of ST36 from outside the United Kingdom. All isolates of ST36 were MRSA and appeared to be a single clone possessing SCCmec type II. ST36 is a SLV of a predominant MSSA clone (ST30) that was the predicted ancestor of the corresponding CC. ST36–MRSA-II presumably has emerged by the introduction of SCCmec type II into a SLV of ST30 (Fig. 4). Five of the 90 ST30 isolates were MRSA but possessed SCCmec type IV rather than SCCmec type II found in ST36–MRSA. These ST30–MRSA-IV isolates probably have emerged by a recent introduction of SCCmec type IV into the successful ST30–MSSA clone, rather than being ancestors of ST36–MRSA-II (or derived from ST36–MRSA).
Discussion
MLST provides an unambiguous method for characterizing MRSA isolates via the internet. The procedure demonstrates that many of the clones of EMRSA that were previously described as distinct with pulsed-field gel electrophoresis are indistinguishable in genotype with MLST. In one extreme case, isolates of the EMRSA-2, -6, -7, -12, -13, and -14 clones described from the United Kingdom all were indistinguishable by MLST (ST8). It could be argued that MLST lacks sufficient discriminatory ability or is grouping unrelated genotypes within the same ST. However, this notion is very unlikely given that there is an average of 42 alleles per locus, which provides the ability to resolve >200 billion STs, and the probability of two unrelated genotypes having the same ST is essentially zero. It is far more likely that the rapid accumulation of the genetic variation indexed by pulsed-field gel electrophoresis has led to considerable differences in SmaI DNA fragment patterns among the descendents of each of the ancestral genotypes of the EMRSA clones, resulting in some EMRSA clones being inappropriately subdivided.
Pulsed-field gel electrophoresis or other high-resolution typing methods such as fluorescent-amplified fragment-length polymorphism (22) may be appropriate for identifying clusters of related genotypes within a laboratory, but for comparing strains between laboratories, or for the unambiguous assignment of MRSA (or MSSA or GISA) isolates as known clones or new clones, we consider that MLST has very considerable advantages. MLST is already the gold standard for precisely assigning Neisseria meningitidis isolates to the known hypervirulent clones (8) and for defining antibiotic-resistant clones of pneumococci (23). MLST also provides a logical nomenclature for MRSA clones, as the ST precisely defines a strain as having a unique and unambiguous allelic profile and identifies those MRSA isolates that have descended from the same recent common ancestor. However, there is clear evidence that successful MSSA clones have become MRSA on more than one occasion, as evidenced by the presence of isolates with the same ST but with different SCCmec types. For example, MRSA isolates of both ST5 and ST8 are found with each of the four known SCCmec types and MRSA isolates of these STs have presumably arisen on at least four occasions (Table 1). The proposed rational nomenclature of MRSA clones takes this into account by including both the ST and the SCCmec type.
MLST combined with SCCmec typing establishes that there are relatively few major EMRSA clones. Only 38 STs contain MRSA of the 162 currently present in the MLST database, demonstrating that MRSA have evolved in relatively few lineages. Only 11 MRSA clones were represented by more than 10 isolates among the international collection of 359 MRSA isolates. Of these 11 major clones, five belonged to a single group of closely related S. aureus lineages (CC8) whereas the other six were distantly related to this CC and (excepting the two major MRSA clones within ST5) to each other. The presence of distantly related lineages of MRSA has been shown in several studies (12, 13, 24) and is evidence that MRSA isolates are not all descended over the last 40 years by diversification of a single original MRSA clone. Rather, horizontal transfer of mec into different lineages has been highly significant in MRSA evolution (12, 13).
The evolutionary origins of MRSA clones were explored with burst, which, together with an analysis of the distribution and nucleotide sequences of alleles within SLVs and their presumed ancestors, provides a powerful approach to discerning the likely evolutionary relationships among bacterial clones. By combining this approach with an analysis of the SCCmec types we have produced a putative evolutionary history for all of the major EMRSA clones. burst identified a large complex of related MSSA and MRSA genotypes (CC8) and resolved it into three subgroups. Significantly, almost all of the MRSA isolates from the 1960s were within a single subgroup of CC8 and most of these were in ST250, the predicted ancestor of this subgroup. Our data support the suggestion that methicillin resistance first arose within ST250 (25), but the results from the burst analysis, and the alleles at the yqiL locus, show that ST250–MSSA probably first evolved from a MSSA isolate of ST8 and then acquired the mec gene. Whereas ST250 isolates are not current causes of epidemic MRSA disease, a minor variant (ST247–MRSA-I; the Iberian clone) has evolved resistance to most antimicrobial agents and is one of the major EMRSA clones currently recovered from European hospitals (19). ST247–MRSA-I may have arisen relatively early from ST250–MRSA-I because there are six isolates of MRSA–ST247-I from the 1960s. ST8, the predicted ancestor of ST250 (and of the whole CC), has itself also developed into two major EMRSA clones (ST8–MRSA-II and ST8–MRSA-IV) although we believe this development happened independently and subsequent to the emergence of ST250–MRSA-I.
MRSA arose recently and in many cases should have retained the allelic profile of the MSSA isolate that acquired the mec determinant. In the collection of 912 S. aureus isolates there were no MSSAs with the allelic profile of the original MRSA clone, ST250–MRSA-I. ST250–MSSA has, however, recently been described among early isolates from Denmark, and ST250–MSSA was a successful clone within hospitals in the 1950s (24), but it appears that this MSSA ancestor of the first MRSA clone is no longer commonly encountered among MSSAs from disease or carriage. However, MSSA isolates with the same allelic profile as the major EMRSA STs 5, 8, 22, and 45 were common among the MSSA population that we studied, which were mostly recovered in Europe during the late 1990s. Similarly, ST36–MRSA-II is a SLV of a very successful MSSA clone (ST30–MSSA). MSSA isolates of the major EMRSA clones ST239–MRSA-III and ST247–MRSA-I were not observed, presumably because these clones were derived directly as SLVs of preexisting MRSA clones.
The major EMRSA clones have emerged either as descendants (SLVs) of preexisting EMRSA clones or by the horizontal transfer of the mec determinants into MSSA. In the latter cases, the mec genes are most likely to have been introduced into those S. aureus clones that were already common within hospitals. The fact that most of these EMRSA clones correspond to major MSSA clones, and the evidence that in several instances MRSA clones have arisen on multiple occasions from the same successful MSSA clone, supports this view. Interestingly, four of the six GISA isolates also have emerged within one of the major EMRSA clones (ST5–MRSA-II). This finding suggests a depressing evolutionary progression, with MSSA strains that are well adapted to transmission within hospitals repeatedly receiving the mec determinant after the introduction of methicillin to treat penicillinase-producing MSSA, and then becoming the successful EMRSA clones within hospitals. Now these same successful EMRSA clones are responding to the increasing use of vancomycin by becoming less susceptible to glycopeptides, resulting in EMRSA variants that are also GISAs.
The ccr and mec genes that are the basis of SCCmec typing are thought to have first been introduced into coagulase-negative staphylococci (4, 26, 27) from an unknown source, where deletion of the mec regulatory genes occurred, and then into S. aureus. It is unclear which staphylococcal species donated the four SCCmec types found among MRSAs, but the presence of four types suggests multiple introductions into S. aureus, and their presence in the same ST indicates that horizontal transfer of mec genes is relatively frequent within S. aureus.
Finally, we stress the value of burst that provides an objective evaluation of the relationships between closely related isolates within a CC. The burst analysis provides a hypothesis about ancestry and patterns of descent whose validity can be tested. For MRSA, analysis of the distribution of alleles at MLST loci, and of SCCmec types, and consideration of the genotypes of the early MRSA isolates, provides a consistent and compelling scenario for the evolutionary history of the major EMRSA clones.
Acknowledgments
We thank all donors of isolates. This work was funded by the Wellcome Trust. M.C.E. is a Royal Society University Research Fellow. B.G.S. is a Wellcome Trust Principal Research Fellow.
Abbreviations
- MRSA
methicillin-resistant Staphylococcus aureus
- EMRSA
epidemic MRSA
- MSSA
methicillin-susceptible S. aureus
- SCCmec
staphylococcal cassette chromosome mec
- GISA
glycopeptide intermediately susceptible S. aureus
- MLST
multilocus sequence typing
- ST
sequence type
- SLV
single locus variant
- CC
clonal complex
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jevons M P. Br Med J. 1961;1:124–125. [Google Scholar]
- 2.Hussain F M, Boyle-Vavra S, Bethel C D, Daum R S. Pediatr Infect Dis J. 2000;19:1163–1166. doi: 10.1097/00006454-200012000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Morbid Mortal Wkly Rep. 1999;48:707–710. [Google Scholar]
- 4.Hiramatsu K, Cui L, Kuroda M, Ito T. Trends Microbiol. 2001;9:486–493. doi: 10.1016/s0966-842x(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 5.Hiramatsu K, Kuroda M, Baba T, Okuma K. In: Diagnostic Molecular Microbiology. Persing D, editor. Washington, DC: Am. Soc. Microbiol.; 2002. , in press. [Google Scholar]
- 6.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Morbid Mortal Wkly Rep. 1997;46:765–766. [Google Scholar]
- 8.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, et al. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright M C, Spratt B G. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 10.Enright M C, Day N P, Davies C E, Peacock S J, Spratt B G. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 12.Musser J M, Kapur V. J Clin Microbiol. 1992;30:2058–2063. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald R F, Sturdevant D E, Mackie S M, Gill S R, Musser J M. Proc Natl Acad Sci USA. 2001;98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokue Y, Shoji S, Satoh K, Watanabe A, Motomiya M. Antimicrob Agents Chemother. 1992;36:6–9. doi: 10.1128/aac.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanches I S, Ramirez M, Troni H, Abecassis M, Padua M, Tomasz A, de Lencastre H. J Clin Microbiol. 1995;33:1243–1246. doi: 10.1128/jcm.33.5.1243-1246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Sousa M A, Sanches I S, Ferro M L, Vaz M J, Saraiva Z, Tendeiro T, Serra J, de Lencastre H. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witte W. J Antimicrob Chemother. 1999;44:1–9. doi: 10.1093/jac/44.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- 18.Ito T, Katayama Y, Hiramatsu K. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira D C, Tomasz A, de Lencastre H. Microb Drug Resist. 2001;7:349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 20.Sa-Leao R, Sanches I S, Dias D, Peres I, Barros R M, de Lencastre H. J Clin Microbiol. 1999;37:1913–1920. doi: 10.1128/jcm.37.6.1913-1920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anonymous. Commun Dis Rep Weekly. 1997;7:1. [PubMed] [Google Scholar]
- 22.Grady R, Desai M, O'Neill G, Cookson B, Stanley J. J Clin Microbiol. 1999;37:3198–3203. doi: 10.1128/jcm.37.10.3198-3203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGee L, McDougal L, Zhou J, Spratt B G, Tenover F C, George R, Hackenbeck R, Hryniewicz W, Lefevre J C, Tomasz A, et al. J Clin Microbiol. 2001;39:2565–2571. doi: 10.1128/JCM.39.7.2565-2571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crisostomo M I, Westh H, Tomasz A, Chung M, Oliveira D C, de Lencastre H. Proc Natl Acad Sci USA. 2001;98:9865–9870. doi: 10.1073/pnas.161272898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. Antimicrob Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki E, Kuwahara-Arai K, Richardson J F, Hiramatsu K. Antimicrob Agents Chemother. 1993;37:1219–1226. doi: 10.1128/aac.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]