Abstract
P201 is a short (eight-residue) nonacidic peptide that comprises a strong transcriptional activating region when tethered to DNA in yeast. Here we identify the mediator protein Gal11 as an essential target of P201. Deletion of Gal11, which modestly decreases activation elicited by the typical acidic yeast activator, abolishes activation by DNA-tethered P201. A point mutation in Gal11, which has no effect on other Gal11 functions, also greatly diminishes activation by DNA-tethered P201. P201 binds to a fragment of Gal11 in vivo and in vitro, and the interaction is diminished by mutations in either component that decrease activation in vivo. P201, unlike the typical yeast acidic activating region, does not work in mammalian cells, which is consistent with the notion that the unique target of P201 (Gal11) is absent from mammalian cells.
The typical yeast transcriptional activator (e.g., Gal4) bears two functions: a DNA-binding region and an activating region. The activator works by recruitment; the DNA-binding domain tethers the activating region near a gene, and the activating region, an “adhesive” peptide, binds and recruits the transcriptional machinery to the nearby promoter (1, 2). The transcriptional machinery comprises some 50 proteins, and it has proved difficult to specify the direct targets of activating regions. Gal4's activating region, for example, has been reported to interact with many proteins in the machinery (e.g., components found in complexes including the mediator, TFIID, and SAGA; refs. 3–11), but it is not clear which of these is relevant in vivo. In particular we do not know whether at a given promoter contact with a single target will suffice or whether contacts with multiple targets is required.
Similar to many other natural activating regions, that found on Gal4 bears an excess of acidic residues in addition to crucial hydrophobic ones (12–14). Previously uncharacterized acidic activating regions have been found encoded in random bits of Escherichia coli DNA, and a peptide designed to form an amphipathic helix, with acidic residues along one surface, has been described (15–17). All these acidic activating regions, where tested, work in cells of higher eukaryotes as well as in yeast (18–22).
Short activating regions, rich in hydrophobic residues but lacking charged residues, have been described also (23). These regions were produced by creating a library of genes encoding a DNA-binding fragment of Gal4 attached to short, randomly generated peptides and screening the expressed library for strong activators in yeast. One of those recovered, called P201, bears the 8-amino acid peptide YLLPTCIP attached to Gal4(1–100). This protein activates as efficiently in yeast as does Gal4 itself. Replacing any one of the first seven residues in P201 with Arg (or in some cases with Ala) virtually abolishes activity (23).
Here we define the protein Gal11 as a crucial, perhaps unique, target of P201. Elimination of the interaction (by point mutation of Gal11 or deletion of that protein) greatly decreases activation by P201. The point mutation in Gal11 has no effect on activation by certain natural yeast activators, and the deletion of Gal11 decreases their activities only modestly. Moreover, as assayed in vitro and in vivo, interaction of wild-type and mutant forms of Gal11 and P201 correlates with activation. We also confirm an earlier suggestion (23) that DNA-tethered P201 does not work in mammalian cells, a fact we attribute to the absence of its target from those cells.
Materials and Methods
Genetic Analysis.
Saccharomyces cerevisiae strains used in this study are listed in Table 1. Yeast media were prepared as described (24) except the ethidium bromide-galactose medium, which was prepared as described by Suzuki et al. (25). Yeast transformations were done by using standard methods (24). The transformants were assayed for β-galactosidase activity as described (26).
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| AYW3-1B | Matα ade2 his3 leu2 ura3 |
| DY150 | Mata ade2 can1 his3 leu2 trp1 ura3 |
| DY1702 | Mata ade2 can1 his3 leu2 trp1 ura3 sin4∷TRP1 |
| JPY7 | Mata his3Δ200 leu2Δ1 trp1Δ63 ura3-52 gal11∷TRP1 |
| JPY9 | Matα his3Δ200 leu2Δ1 trp1Δ63 ura3-52 lys2Δ385 gal4Δ11 |
| JPY10 | Matα his3Δ200 leu2Δ1 trp1Δ63 ura3-52 lys2Δ385 gal4Δ11 gal11∷TRP1 |
| JPY52 | Matα his3Δ200 leu2Δ1 trp1Δ63 ura3-52 lys2Δ385 gal4Δ11 gal11∷LYS2 |
| MG106 | Mata ade2-1 can1-100 his3-11 15 leu2-3 112 trp11-1 ura3-1 |
| MG107 | Mata ade2-1 can1-100 his3-11 15 leu2-3 112 trp11-1 ura3-1 med2Δ1∷TRP1 |
| SSAB-2C | Matα ade2 his3 leu2 ura3 hrs1Δ∷LEU2 |
Mutagenesis and Screening.
Gal11 was mutagenized as described by Lehming et al. (27). Briefly, a DNA fragment encoding residues 144–574 of Gal11 was mutagenized by PCR. This mutagenized PCR fragment was cotransformed into yeast (JPY7, which lacks Gal11) along with a linearized plasmid that expresses full-length Gal11 under the control of its own promoter. In linearizing the plasmid we removed residues 158–551 of Gal11. Thus, the plasmid and the PCR fragment share homologous sequences of Gal11 (residues 144–158 at the N terminus and residues 551–574 at the C terminus). This degree of homology is sufficient for the gap-repair system to recombine and incorporate the PCR fragment efficiently into the linearized plasmid. The transformants were selected for growth on ethidium bromide-galactose plates for 3 days; these conditions eliminate mutants that do not express Gal11 (25, 28).
In Vivo Protein Interaction.
The split-ubiquitin system was used to test interactions in vivo as described (29). Gal4(2–100)+P201 (wild type or mutant) was fused to the Nub plasmid, and Gal11(186–617) (wild type or mutant) was fused to the Cub plasmid. Nub and Cub fusions were cotransformed into JPY9 cells. An equal number of cells, determined by cell counting, was spotted on Ura− and Ura+ plates.
Protein Purification.
Plasmids encoding glutathione S-transferase (GST)-Gal11 (wild type and mutants) were transformed into BL21 (DE3) pLysE cells from Stratagene. Cells were grown to an OD of 0.8, and protein expression was induced for 8 h with 0.75 mM isopropyl β-D-thiogalactoside at 16°C. Cells were pelleted, washed, and resuspended in buffer A [10 mM phosphate buffer (pH 7.4)/27 mM KCl/137 mM NaCl (PBS)/10% glycerol/0.1% Nonidet P-40]. Cells were lysed in buffer A containing a protease inhibitor mixture (1 mM PMSF/1 μl/ml 2M chymostatin/2 mM pepstatin/2 mM benzamadine), and the debris was pelleted by centrifugation at 11,000 × g for 15 min. The supernatant was incubated for 45 min with glutathione-Sepharose beads that were preequilibrated in buffer A (Amersham Pharmacia). The beads were washed six times, and the equal volumes of beads were mixed with SDS-containing sample buffer, resolved on a 6% SDS-Tricine gel, and visualized by Coomassie staining.
In Vitro Binding Assay.
Radiolabeled activator proteins were synthesized by using the Promega, rabbit reticulocyte transcription and translation (TNT) per manufacturer instructions.
Equal amounts of glutathione-Sepharose-bound GST-fusion proteins (as estimated by Bio-Rad assay of eluted proteins as well as the approximate amount of full-length product on the gel) were used for coprecipitation (pull-down) assays. Roughly 20 μl of beads were incubated for 30 min with 2 μl of radiolabeled activators (TNT mix) in 600 μl of binding buffer. The “relaxed” binding buffer comprises buffer A, 100 μg/ml acetylated BSA, and 0.1% Triton. “Stringent” binding buffer comprises 20 mM Tris, 200 mM NaCl, 1 mM EDTA, 100 μg/ml tRNA, 100 μg/ml acetylated BSA, 0.1% Triton, and 75 mg/ml salmon sperm DNA.
CAT Assay.
HeLa cells were cotransfected with plasmids carrying the Gal4-responsive chloramphenicol acetyltransferase (CAT) gene and Gal4 derivatives by using the Lipofectamine method suggested by the manufacture (Promega). A plasmid expressing β-galactosidase also was included in transfection to normalize the transfection efficiency. Cell extracts were prepared ≈40 h after transfection. CAT and β-galactosidase activities were determined as described (30). Transcriptional activity of the Gal4 derivative was indicated as relative CAT activity normalized to β-galactosidase activity.
Results
Deletion of Gal11.
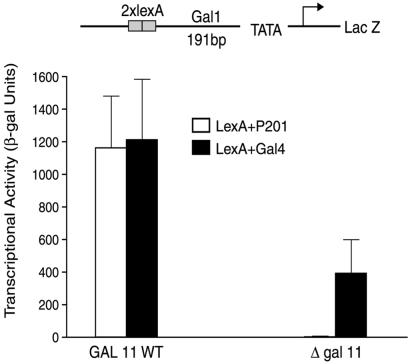
To search for a possible target of P201 we deleted, singly, various components of the mediator and measured the effect on activation by LexA+Gal4 and by LexA+P201. “LexA+Gal4” bears the LexA DNA-binding domain fused to the activating region of Gal4, and “LexA+P201” bears the LexA DNA-binding domain and a bit of the Gal4 dimerization domain fused to P201 (see Lu et al., ref. 23). Fig. 1 shows that the deletion of Gal11, while decreasing activation by LexA+Gal4 some 3-fold (consistent with previous results), abolished activation by LexA+P201. This effect was independent of the DNA-binding domain used to tether the activating peptide. Thus deletion of Gal11 also abolished the activity of “Gal4+P201” [which comprises the Gal4 DNA-binding domain (residues 1–100) fused to P201; data not shown].
Figure 1.
Effect of deletion of Gal11 on the activity of P201. The reporter shown at the top was introduced into two yeast strains, one wild type (WT), and the other deleted for Gal11. Both strains were transformed with plasmids expressing, separately, LexA+P201 and LexA+Gal4. β-Galactosidase (β-gal) levels produced by cultures grown to mid-log phase are shown. In all the yeast experiments reported here the reporters were integrated at the Ura3 locus.
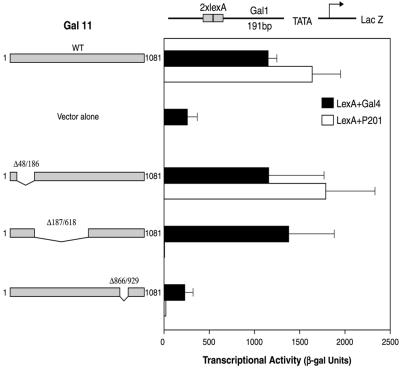
Deletion of Segments of Gal11.
Fig. 2 shows that deletion of residues 187–618 of Gal11 abolished activation by LexA+P201 but had little or no effect on activation by LexA+Gal4 (compare lines 1 and 2 with lines 7 and 8). In contrast, the deletion of Gal11 residues 48–186 had little effect on either LexA+P201 or LexA+Gal4 (lines 5 and 6). Deletion of Gal11 residues 866–929 (lines 9 and 10) had an effect equivalent to total deletion of Gal11 (lines 3 and 4). This part of Gal11 (i.e., residues 866–929) is required for whatever general role it plays in the mediator (31). These results, taken together, suggest that LexA+P201 contacts Gal11 somewhere in the segment encompassed by residues 187–618.
Figure 2.
Identification of a Gal11 fragment essential for the activity of P201. A yeast strain bearing the indicated reporter was deleted for Gal11. Plasmids expressing no Gal11, Gal11 wild type (WT), or one of the three depicted Gal11 deletion derivatives were introduced into these cells. A second plasmid, expressing either LexA+Gal4 or LexA+P201, was introduced, and the cells were assayed for β-galactosidase (β-gal) as in the experiment of Fig. 1.
Isolation of Gal11 Mutants.
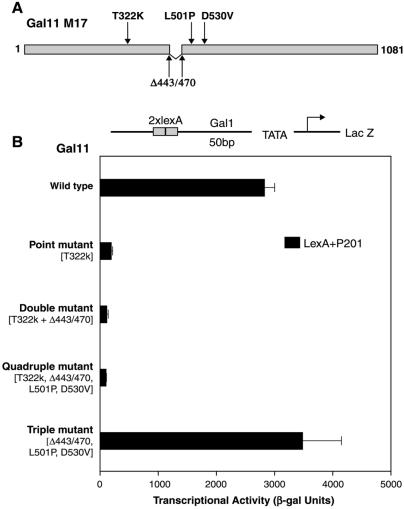
We used PCR to mutagenize the region encompassed by residues 187–618 in the context of intact Gal11 and introduced the mutagenized Gal11 into a strain deleted for wild-type Gal11. The strain bore a Gal1-LacZ reporter gene, transcription of which was activated by LexA+P201. We isolated mutant white colonies (i.e., those not expressing LacZ) from among the population of mostly blue colonies (i.e., those expressing LacZ). These were tested further for functional Gal11 by growing cells on plates bearing galactose as the sole sugar and in the presence of ethidium bromide, a medium on which cells deleted for Gal11 do not grow (25, 28). One of the mutants that retained Gal11 function in this assay, called Gal11M17, was analyzed further.
Gal11M17 bears the four changes shown in Fig. 3A: three point mutations and a short deletion. Fig. 3B shows that the single base-pair change T322K reduced more than 10-fold the response to LexA+P201 (line 2). When T322K was combined with the deletion 443–470, activation was reduced by approximately another 2-fold (line 3), and addition of the other two mutations from Gal11M17 had no further effect (line 4). Moreover, the three mutations found in Gal11M17 other than T322K, when present without that crucial point mutation, had little effect (line 5). The single mutation Gal11 T322K had virtually no effect on activation by LexA+Gal4 (see Table 2). The reporter gene used in this experiment bears two LexA binding sites positioned 50 base pairs upstream from the Gal1 promoter TATA box. When the activator binding sites were positioned 191 base pairs upstream, as in the experiment of Fig. 1, the point mutation T322K virtually abolished activation by LexA+P201 but had little or no effect on activation by LexA+Gal4 (data not shown).
Figure 3.
Changes in Gal11 mutant 17 (M17) and their effects on activation. (A) Gal11M17 bears the four changes indicated: the point mutations T322K, L501P, and D530V and the short deletion Δ443/470. (B) Mutations from Gal11M17 were introduced separately and in the indicated combinations into Gal11. The mutant Gal11 proteins, expressed from plasmids, were introduced into cells otherwise deleted for Gal11 and assayed as in the experiment of Fig. 1. β-gal, β-galactosidase.
Table 2.
The effect of Gal11 on different activators (activity compared to wild type)
| Δgal11, % | Gal11(T322K), % | |
|---|---|---|
| P201 | 3 | 5 |
| P64 | 30 | ND |
| Gal4 | 28 | 87 |
| Vp16 | 23 | 91 |
| Gcn4 | 74 | ND |
| Gal11P | Gal11P + T322K | |
| Gal4(1–100) | 100 | 109 |
In Gal11 wild-type strains, the β-galactosidase activity for each activator was: P201, 3,770; P64, 1,710; Gal4, 4,770; Vp16, 2,600; Gcn4, 125. Gal4(1–100) activity in Gal11P cell is 1,180. The reporter bore two activator-binding sites 50 bp upstream of the Gal1-LacZ gene. Standard deviation is within 25% in each case. ND, not determined.
Table 2 also shows that the T322K mutation had little effect on activation by the acidic activator, LexA+VP16 (second column, row 4). The deletion of Gal11 had a slight effect on activation by the acidic activator LexA+Gcn4 and a somewhat greater effect on activation by LexA+Vp16, an effect about equivalent to that seen with LexA+Gal4. P64, another peptide isolated in the screen that yielded P201, depended to a similarly modest extent on Gal11 (row 2).
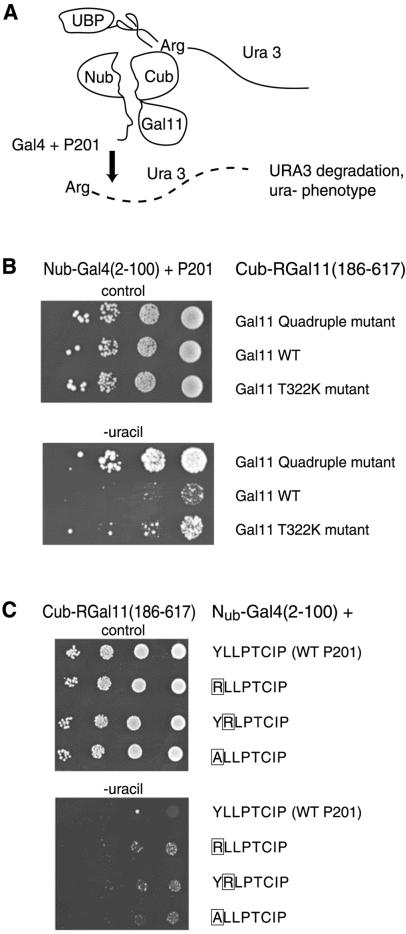
Binding Assayed in Vivo.
We measured binding of Gal4+P201 to a Gal11 fragment (residues 186–617) in vivo using the split-ubiquitin assay (29, 32). The fusion proteins used for the assay are diagrammed in Fig. 4A: interaction between the attached peptides apposes the two halves of ubiquitin, an interaction that leads to degradation of the fused Ura3 protein and therefore an inability to grow on medium lacking uracil.
Figure 4.
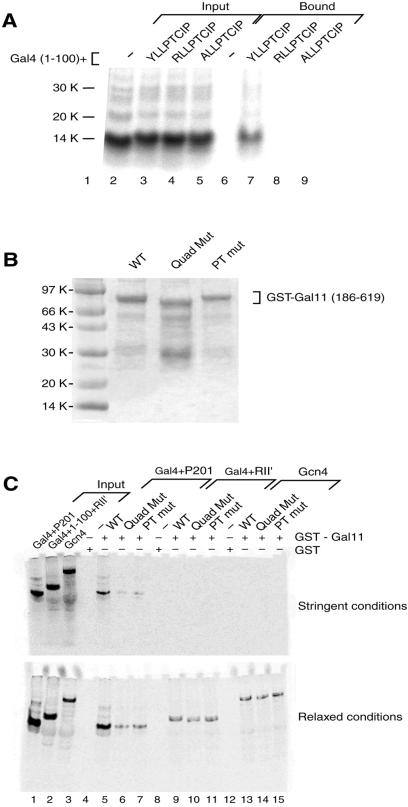
Interaction between P201 and Gal11 in vivo. (A) The split-ubiquitin two-hybrid system. Nub, an amino fragment of ubiquitin, was fused to Gal4+P201, and Cub, a carboxyl fragment, was fused to Gal11(186–617). Cub also bore a fused Ura3 gene. Interaction of the added peptides brings Nub together with Cub. The reconstituted ubiquitin is recognized by UBPs (ubiquitin C-terminal hydrolase), which in turn clip off the Ura3 protein. Because an Arg residue was placed at the cut site, the Ura3 protein is degraded rapidly. (B) Mutations in Gal11 weaken the interaction observed with the split-ubiquitin two-hybrid system. Cells were spotted in 10-fold dilutions, starting at the same concentration, onto plates lacking uracil (rows 4–6) and, as a control, on plates containing uracil (rows 1–3). Colony growth indicates an interaction between the wild-type (WT) Gal11 frag- ment and P201 (scant growth in row 5), an interaction that was decreased by introducing the single mutation T322K (row 6) or the quadruple mutation of Fig. 3 (row 4). (C) Mutations of P201 weaken the interaction observed with the split-ubiquitin two-hybrid system. Assays were performed as described for B. Colony growth indicates that the interaction (revealed by scant colony growth of cells bearing wild-type P201, row5) was decreased significantly by each of the indicated point mutants of P201.
Fig. 4B shows that, as measured in this assay, wild-type Gal4+P201 binds to Gal11(186–617). Thus, cells grew very poorly on plates lacking uracil if the Gal11 fragment was wild type (line 5). Introduction of the point mutation T332K into the Gal11 fragment greatly decreased the interaction (line 6), and introduction of the quadruple mutation of Fig. 3A decreased the interaction further still (line 4).
Fig. 4C shows that each of three point mutations in P201 also decreased interaction with the Gal11 fragment in the split-ubiquitin assay (compare lines 6–8 with line 5). Each of these mutations abolishes activation by Gal4+P201 (23).
Binding Assayed in Vitro.
Radiolabeled Gal4+P201 bound to GST-Gal11 (186–619) as shown in Fig. 5A (lane 7). For this experiment, the GST-Gal11 fragment was immobilized on beads, and the P201 derivatives were generated by TNT. No interaction was observed when either of the two point mutants of Gal4+P201 were used (lanes 8 and 9) or when Gal4(1–100) alone (Lane 6) was used. The point mutants of P201 used here were two of the three such mutants used in the in vivo assay of Fig. 4C, both of which decreased the interaction in vivo.
Figure 5.
Interaction between P201 and Gal11 in vitro. (A) Mutations in P201 abolish the interaction with Gal11(186–619) observed in a GST pull-down assay. Equal amounts of radiolabeled Gal4(1–100) and Gal4+P201 as well as two mutant derivatives of the latter, were tested for interaction with Gal11(186–619) in an in vitro pull-down assay. The Gal11 fragment was isolated as a GST-fusion protein, attached to beads, and incubated with radiolabeled Gal4 derivatives. Input lanes (2–5) contain 20% of the labeled activators incubated with the immobilized GST-Gal11. Bound lanes (6–9) show that only the derivative bearing wild-type P201 bound to Gal11 under these (stringent) conditions (see Materials and Methods). (B) Preparation of GST-Gal11 derivatives. Coomassie-stained gel showing the expression of three forms of GST-Gal11(186–619). The first lane shows the wild-type (WT) protein, the second lane shows the quadruple mutant (Quad Mut) of Fig. 3, and the third lane shows the T322K mutant (PT mut) form of the Gal11 fragment. As expected, the quadruple mutant, which includes the short deletion (residues 443–470), migrates slightly faster than the other two forms. (C) Mutations in Gal11 abolish the interaction with P201 observed in a GST pull-down assay. This experiment was performed as in A, except that various mutant versions of gal11 were tested for interaction with three activators. Input lanes (1–3) contain 20% of three radiolabeled activators used in the GST pull-down assay. Gal4(1– 100)+(840–881) is a weak acidic activator, and Gcn4 is the full-length form of another acidic yeast activator. Under stringent conditions (see Materials and Methods) the only activator that bound wild-type GST-Gal11(186–619) was Gal4+P201 (lane 5). That interaction was abolished by mutation of Gal11 (lanes 6 and 7). Under relaxed conditions, in addition to the strong interaction observed between wild-type Gal11 and Gal4+P201 (lane 5), a weaker interaction with either of the two mutant forms of Gal11 was observed (lanes 6 and 7). A weak interaction between Gal11 and the two other activators was observed also, but those interactions were unaffected by mutation of Gal11.
Fig. 5C shows that in a similar assay the Gal11 mutations that abolish activation by Gal4+P201 also greatly decrease interaction in vitro. GST-Gal11(186–619) in three forms was purified: wild type, the quadruple mutant of Fig. 3A, and the point mutant T322K (Fig. 5B lanes 2–4). The wild-type form, but neither mutant of Gal11, bound wild-type Gal4+P201 under stringent conditions. Under these conditions, no binding was observed to either of two acidic yeast activating regions: one from Gal4 RII′ and the other from the activator Gcn4. Under relaxed conditions these two acidic activating regions interacted weakly with Gal11, and these interactions were unaffected by mutation of Gal11.
Other Functions of Gal11.
We mentioned above that the mutants of Gal11 that did not support activation by LexA+P201 were normal for Gal11 function as determined by growth on galactose medium containing ethidium bromide and by response to activation by Gal4. Here we show that Gal11T322K (which is almost totally unable to support activation by P201) also is normal for another function of Gal11.
The change N342V in Gal11 is a gain-of-function mutation: in the presence of this mutated form of Gal11, called Gal11P (potentiator), the otherwise inert DNA-binding fragment of Gal4(1–100) works as a powerful activator (33, 34). The mutation has been shown to create an interaction between the dimerization region of Gal4(58–97) and Gal11 (35). As shown in Table 2, the T322K mutation has little effect on the ability of Gal11P to support activation by Gal4(1–100).
Activation in Mammalian Cells.
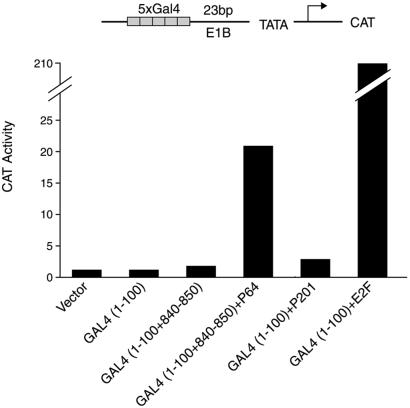
We mentioned in the Introduction that many acidic yeast activating regions, both natural and synthetic, also work in mammalian cells. The experiment of Fig. 6 shows that, in contrast, Gal4+P201 had very little (if any) activity in a mammalian cell as assayed in a transient transfection assay. The figure also shows two acidic activators that do work in these cells, Gal4+E2F and Gal4+P64. The former bears the acidic activating region from the mammalian activator E2F fused to Gal4(1–100). The latter bears, fused to Gal4(1–100), a short synthetic peptide with two acidic residues (in addition to hydrophobic ones) attached to an 11-residue bit of Gal4(840–850) that includes another acidic residue (23). Gal4+E2F works ≈10-fold more efficiently than does Gal4+P64. In these experiments, all the activators were expressed at similar levels (data not shown).
Figure 6.
Activation in mammalian cells. HeLa cells were transiently transfected with the indicated reporter and with plasmids expressing one or another of the activators listed at the bottom. After 40 h the cells were assayed for CAT as described by Lehming et al. (30).
Discussion
Our results define a specific amino acid change (T322K) in the mediator protein Gal11 that virtually abolishes transcription elicited by an activator bearing the synthetic activating region P201. As assayed both in vitro and in vivo, the mutation also drastically reduces binding of P201 to Gal11. In contrast, the mutation has virtually no effect on transcription elicited by a range of other activators. P201, similar to natural activators, bears hydrophobic residues, but unlike most natural yeast activators, it lacks acidic residues. Gal11 has been identified previously as an essential site of interaction with a DNA-tethered activating peptide (33, 34, 36). In that case the interaction occurs only if Gal11 bears a mutation at position 342 that changes an Asn to a hydrophobic residue. In the presence of this mutated Gal11 (called Gal11P), an otherwise inactive fragment of Gal4(1–100) works as a strong activator. Mutant studies revealed that in the Gal11P case, binding of the “activator” to a fragment of Gal11P was correlated with activation. Hidalgo et al. (35) showed that the region of Gal4 that interacts with Gal11P presents a hydrophobic surface. The Gal11 mutation T322K, which abolishes activation by P201, has no effect on activation by Gal4(1–100) if the Gal11 also bears the P mutation.
Our results are consistent with the idea that a single contact between a DNA-tethered peptide and Gal11 suffices for strong activation. Whether this restricted target choice limits the class of yeast genes that can be activated by DNA-bound P201 is not known. Of the four promoters we have tested, Gal1, His3, Pho5, and Cyc1, all are activated efficiently by activators bearing the P201 activating region (data not shown). Gal11 also has been proposed to be a target for various acidic activators (37–39). If so, that contact is not essential, because deletion of Gal11 has only a modest (2–5-fold) effect on activation in those cases.
How binding to Gal11 alone can elicit such high levels of activity (as with DNA-tethered P201) is unclear. Gal11 has been found in two complexes that contact RNA polymerase II (40–42). In one case it is part of the so-called Gal11 module of the mediator (43). The integrity of that module is not required for activation by P201, however; deletion of either of two other components of that module (Hrs1 and Sin4) has only modest deleterious effects, and deletion of another component (Med2) of mediator has a modest positive effect (data not shown).
One difference between P201 and typical acidic activators (Gal4, for example) is that whereas the latter work in mammalian cells (as well as in other metazoan cells), the former does not (23). This species-specific restriction could reflect the requirement for a specific target for P201 that is absent from mammalian cells. Consistent with this view, no clear Gal11 homologue has been found in higher eukaryotes. One weakly homologous protein, CA150 (44), lacks the sequence we have found to interact with P201. Perhaps acidic activating regions have a range that P201 lacks by virtue of their abilities to interact with an array of targets.
Acknowledgments
We thank Drs. A. Aguilera, S. Bjorklund, R. Kornberg, N. Lehming, H. Sakurai, and D. Stillman for yeast strains and plasmids. We also thank C. Heid and D. Spagna for technical assistance and members of the Ptashne lab for helpful discussions and G. Gill and A. Gann for comments on the manuscript. This work is supported by National Institutes of Health Grant GM32308 (to M.P.), the Ludwig professor of molecular biology, Ludwig Institute for Cancer Research.
Abbreviation
- GST
glutathione S-transferase
References
- 1.Ptashne M, Gann A. Genes and Signals. Plainview, NY: Cold Spring Harbor Lab. Press; 2002. [Google Scholar]
- 2.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 3.Bhaumik S R, Green M R. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larschan E, Winston F. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melcher K, Johnston S A. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Reece R J, Ptashne M. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 7.Brown C E, Howe L, Sousa K, Alley S C, Carrozza M J, Tan S, Workman J L. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 8.Koh S S, Ansari A Z, Ptashne M, Young R A. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 9.Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 10.Yudkovsky N, Logie C, Hahn S, Peterson C L. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emili A, Kobayashi R, Ingles C J. Proc Natl Acad Sci USA. 1998;95:11122–11127. doi: 10.1073/pnas.95.19.11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drysdale C M, Duenas E, Jackson B M, Reusser U, Braus G H, Hinnebusch A G. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan S M, Horn P J, Olson V A, Koop A H, Niu W, Ebright R H, Triezenberg S J. Nucleic Acids Res. 1998;26:4487–4496. doi: 10.1093/nar/26.19.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cress W D, Triezenberg S J. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 15.Giniger E, Ptashne M. Nature (London) 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 16.Ma J, Ptashne M. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 17.Ruden D M, Ma J, Li Y, Wood K, Ptashne M. Nature (London) 1991;350:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- 18.Fischer J A, Giniger E, Maniatis T, Ptashne M. Nature (London) 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- 19.Kakidani H, Ptashne M. Cell. 1988;52:161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- 20.Webster N, Jin J R, Green S, Hollis M, Chambon P. Cell. 1988;52:169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 21.Ma J, Przibilla E, Hu J, Bogorad L, Ptashne M. Nature (London) 1988;334:631–633. doi: 10.1038/334631a0. [DOI] [PubMed] [Google Scholar]
- 22.Ptashne M. A Genetic Switch: Phage Lambda and Higher Organisms. Cambridge, MA: Cell Press and Blackwell Scientific; 1992. [Google Scholar]
- 23.Lu X, Ansari A Z, Ptashne M. Proc Natl Acad Sci USA. 2000;97:1988–1992. doi: 10.1073/pnas.040573197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. [Google Scholar]
- 25.Suzuki Y, Nogi Y, Abe A, Fukasawa T. Mol Cell Biol. 1988;8:4991–4999. doi: 10.1128/mcb.8.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant G O, Struhl K, Ptashne M. Proc Natl Acad Sci USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehming N, McGuire S, Brickman J, M, Ptashne M. Proc Natl Acad Sci USA. 1995;92:10242–10246. doi: 10.1073/pnas.92.22.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nogi Y, Fukasawa T. Curr Genet. 1980;2:115–120. doi: 10.1007/BF00420623. [DOI] [PubMed] [Google Scholar]
- 29.Laser H, Bongards C, Schuller J, Heck S, Johnsson N, Lehming N. Proc Natl Acad Sci USA. 2000;97:13732–13737. doi: 10.1073/pnas.250400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehming N, Thanos D, Brickman J, M, Ma J, Maniatis T, Ptashne M. Nature (London) 1994;371:175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai H, Kim Y J, Ohishi T, Kornberg R D, Fukasawa T. Proc Natl Acad Sci USA. 1996;93:9488–9492. doi: 10.1073/pnas.93.18.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnsson N, Varshavsky A. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 34.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 35.Hidalgo P, Ansari A Z, Schmidt P, Hare B, Simkovich N, Farrell S, Shin E J, Ptashne M, Wagner G. Genes Dev. 2001;15:1007–1020. doi: 10.1101/gad.873901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Himmelfarb H J, Pearlberg J, Last D H, Ptashne M. Cell. 1990;63:1299–1309. doi: 10.1016/0092-8674(90)90425-e. [DOI] [PubMed] [Google Scholar]
- 37.Han S J, Lee Y C, Gim B S, Ryu G H, Park S J, Lane W S, Kim Y J. Mol Cell Biol. 1999;19:979–988. doi: 10.1128/mcb.19.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J M, Kim H S, Han S J, Hwang M S, Lee Y C, Kim Y J. Mol Cell Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong C J, Yang S H, Xie Y, Zhang L, Johnston S A, Kodadek T. Biochemistry. 2001;40:9421–9427. doi: 10.1021/bi010011k. [DOI] [PubMed] [Google Scholar]
- 40.Chang M, Jaehning J A. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y C, Park J M, Min S, Han S J, Kim Y J. Mol Cell Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sune C, Hayashi T, Liu Y, Lane W S, Young R A, Garcia-Blanco M A. Mol Cell Biol. 1997;17:6029–6039. doi: 10.1128/mcb.17.10.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]