Abstract
We showed previously that rrn P1 promoters require unusually high concentrations of the initiating nucleoside triphosphates (ATP or GTP, depending on the promoter) for maximal transcription in vitro. We proposed that this requirement for high initiating NTP concentrations contributes to control of the rrn P1 promoters from the seven Escherichia coli rRNA operons. However, the previous studies did not prove that variation in NTP concentration affects rrn P1 promoter activity directly in vivo. Here, we create conditions in vivo in which ATP and GTP concentrations are altered in opposite directions relative to one another, and we show that transcription from rrn P1 promoters that initiate with either ATP or GTP follows the concentration of the initiating NTP for that promoter. These results demonstrate that the effect of initiating NTP concentration on rrn P1 promoter activity in vivo is direct. As predicted by a model in which homeostatic control of rRNA transcription results, at least in part, from sensing of NTP concentrations by rrn P1 promoters, we show that inhibition of protein synthesis results in an increase in ATP concentration and a corresponding increase in transcription from rrnB P1. We conclude that translation is a major consumer of purine NTPs, and that NTP-sensing by rrn P1 promoters serves as a direct regulatory link between translation and ribosome synthesis.
Because overexpression of ribosomes would be energetically costly, whereas underexpression would prevent the cell from taking full advantage of its nutritional environment, ribosome synthesis is regulated with the demand for protein synthesis. rRNA transcription is the rate-limiting step in ribosome synthesis in Escherichia coli and is controlled by several regulatory mechanisms acting at the level of transcription initiation (1, 2). In addition, an antitermination system ensures efficient rRNA transcription elongation (3).
Each of the seven rRNA (rrn) operons in E. coli has two promoters, P1 and P2. The P1 promoters are responsible for the majority of rRNA transcription at moderate to fast growth rates and have been characterized extensively. Much of the intrinsic strength of the rrn P1 promoters results from A+T-rich sequences (UP elements) upstream of the core promoters that recruit RNA polymerase (RNAP) to the promoter through specific interactions with the RNAP α-subunit (4–6). At least two trans-acting proteins affect rRNA transcription. Fis activates transcription from each of the 7 rrn P1 promoters by binding to sites upstream of −60 relative to the transcription start site, +1 (4, 7), whereas H-NS contributes to repression of rrn P1 promoters during stationary phase (8).
Although UP elements and Fis sites are required for maximal strength, rrn P1 promoters lacking these sequences (core promoters) are still regulated in response to the cell's nutritional environment (9, 10). Consistent with this finding, cells lacking the fis gene regulate transcription from rrn P1 promoters similarly to wild-type strains, because feedback systems compensate for the loss of Fis (7).
rrn P1 promoters form open complexes with much shorter half-lives than those formed by most E. coli promoters (11), making them subject to regulation in vivo by factors that do not directly regulate longer-lived promoters (reviewed in ref. 12). For example, the high levels of ppGpp that are produced during a stringent response severely inhibit rRNA synthesis (reviewed in ref. 13). ppGpp shortens the half-lives of open complexes formed at all promoters, but it only inhibits transcription from those promoters (such as rrn P1) where this step is rate-limiting (11). Strains that cannot make ppGpp exhibit relatively normal rRNA transcription during steady-state growth, in contrast to the situation during a stringent response (9, 11, 14).
The short half-life of rrn P1 open complexes also results in a requirement in vitro for concentrations of initiating nucleoside triphosphates that are much higher than for other promoters (15). Consistent with this observation, when purine NTP concentrations are elevated in vivo by limitation for pyrimidines, rrnB P1 promoter activity increases in parallel with the ATP concentration (15). RNAP variants and rrnB P1 promoter mutations that affect NTP requirements for transcription initiation in vitro also affect promoter regulation in vivo (16, 17). Furthermore, because all seven rrn P1 promoters initiate with purine NTPs (6 with ATP and rrnD P1 with GTP), and because translation consumes both ATP and GTP, it was proposed that free purine NTP concentrations could serve as a homeostatic regulatory link between translation and ribosome synthesis (15). In this “NTP-sensing” model, fluctuations in NTP pools (resulting from changes in NTP production and/or consumption) would lead to adjustments in rRNA production in response to changes in the demand for protein synthesis.
Previous results did not distinguish whether variation of NTP concentrations in vivo directly vs. indirectly affects rrn P1 promoter activity. That is, variation in initiating NTP concentrations could have affected other factors that subsequently affected rRNA promoters. Here, we demonstrate that rrn P1 promoter activities vary in concert with the concentrations of their respective initiating NTPs, even when these NTP concentrations change in opposite directions in vivo. Furthermore, we find that inhibition of translation results in elevated purine NTP concentrations and in increased rrn P1 promoter activity. The results support the model that protein synthesis is a major consumer of purine NTPs, and that the NTP-sensing mechanism directly links rRNA transcription to the level of translation.
Materials and Methods
Strain Construction.
Promoter constructs were generated by PCR by using oligonucleotides with EcoRI sites upstream and HindIII sites downstream of the promoter sequence for insertion into bacteriophage λ “system I” (5) or plasmid pRLG770 (7). λ monolysogens carrying promoter-lacZ fusions were constructed in VH1000 (MG1655 lacZ, lacI, pyrE+ (15). Strains or plasmids are listed in the appropriate figure legends. rrn P1 promoter variants were generated by site-directed oligonucleotide-mediated mutagenesis with standard methods and confirmed by DNA sequencing. The promoter constructs all contained their natural UP elements, ensuring high signal-to-noise ratios both in vitro and in vivo (see figure legends for promoter endpoints).
Mutations affecting purine synthesis were transduced into lysogens by using P1vir (18). The purE mutation is a Tn10 insertion [purE79, CAG12171 (19)] conferring tetracycline-resistance. Auxotrophy was verified by plating on M9 agar (18) containing glucose and casamino acids at 30°C in the absence of exogenous purines. A point mutation in guaB from SO1784 (resulting in partial function) was obtained from K. F. Jensen (Univ. of Copenhagen, Denmark; ref. 20) and was moved into lysogens by cotransduction of a linked Tn10 cassette (zff208, CAG18481; ref. 19). Tetr colonies were screened for slow growth on M9 agar with glucose and casamino acids in the absence of purines at 30°C. Two fresh transductants were used for each experiment to minimize the potential for occurrence of suppressor mutations.
In Vitro Transcription.
Transcription reactions were performed essentially as described (6) at 30°C and were started by the addition of 4 nM RNAP [Eσ70), a generous gift from R. Landick (Univ. of Wisconsin, Madison, WI), purified as described; ref. 21]. Reactions (25 μl) contained 0.6 nM supercoiled plasmid templates (pRLG770 derivatives, see Fig. 1 for list of plasmids) in 40 mM Tris⋅HCl, pH 8.0/10 mM MgCl2/1 mM DTT/0.1 μg/μl BSA/170 mM KCl/10–1600 μM ATP (or GTP)/500 μM GTP (or ATP)/10 μM CTP and UTP/5 μCi α-[32P]UTP.
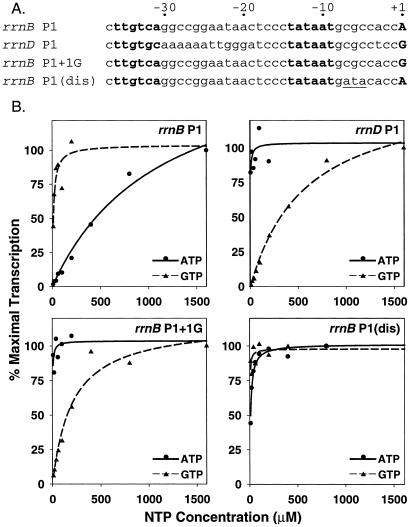
Figure 1.
rrn P1 promoter activity depends on the concentration of the initiating NTP in vitro. (A) Core promoter sequences: −10 and −35 recognition hexamers for RNAP are in bold, initiating NTP is uppercase in bold, and the 3-bp substitution at positions −5 to −7 in the rrnB P1(dis) promoter is underlined. (B) In vitro transcription from supercoiled plasmids carrying the indicated promoters at different initiating NTP concentrations. In each panel, the solid line was derived from varying ATP, and the dashed line was derived from varying GTP. The invariant purine NTP was 500 μM, and CTP and UTP were 10 μM (see Materials and Methods). Each data point is the average of two identical reactions; variation was less than 10%. Lines are best-fit nonlinear regressions. Templates (promoter sequence endpoints and plasmid names in parentheses): rrnB P1 (−66 to +50, pRLG6214); rrnD P1 (−60 to +10, pRLG3426); rrnB P1 + 1G (−66 to +50, pRLG6215); rrnB P1(dis) (−66 to +9, pRLG6120). We note that solution parameters such as anion and cation concentration, temperature, template topology, etc. dramatically affect open complex half-life and, thereby, the absolute concentration of initiating NTP required for transcription (15–17). Therefore, absolute NTP concentrations required for transcription in vitro should not be extrapolated to concentrations required in vivo.
NTP Measurements.
Cultures were grown in media described in the figure legends to an OD600 of ≈0.4. Promoter activities (see below) and NTP concentrations were determined from the same cultures. In all experiments, NTPs were extracted by using two methods: formaldehyde fixation followed by alkaline extraction (22) or direct formic acid extraction (23). Extracts were filtered (0.22 μm pore size), stored at −80°C, thawed on ice, and fractionated either by C18 reverse-phase HPLC with a Supelco LC-18T column and a Beckman System Gold 125 HPLC or by TLC (23). Peaks were identified by comparison with commercial standards (Amersham Pharmacia), quantified by integration of the peak area and comparison with standard curves, and normalized to the OD600 of the culture at the time of extraction. Reported values represent the averages of duplicate extractions from at least two different cultures. Although formic acid extraction resulted in higher NTP yields than those obtained by the formaldehyde/alkaline extraction method, relative changes in NTP levels (between strains or between the same strain grown under the different conditions used here) were virtually identical with both extraction methods. Values from the formic acid extraction are reported in the figures. We note that these values may reflect the total rather than the free cellular NTP content. The NTP concentrations we reported previously (using the formaldehyde/alkaline extraction method; ref. 15) were systematically inflated 10-fold from an error in decimal point placement.
Promoter Activity in Vivo.
λ monolysogens containing promoter-lacZ fusions were grown in the media described in the figure legends for 3–4 generations to an OD600 of ≈0.4. Cultures were placed on ice for >30 min, lysed by sonication (16), and β-galactosidase activity (in Miller units) was measured (18). Where indicated, direct measurement of promoter activity was performed by primer extension. RNA was extracted from cultures with the Bio-Rad Aqua Pure extraction kit. RNAs were measured by primer extension of an unstable mRNA made from either rrnB P1 or rrnB P1(dis) single-copy lacZ-fusions, as described in ref. 10, except that hybridization of the labeled primer was performed in M-MLV buffer (Promega) at 48°C, subsequent precipitation steps were eliminated, and extension reactions were stopped by the addition of formamide loading buffer. The mRNAs made from these constructs are identical.
RNA:Protein Ratios.
Wild-type and mutant strains (lysogens and nonlysogens) were grown at different growth rates in the media indicated in the figure legends, harvested, and lysed; RNA and protein levels were quantified as described (11).
Results
Rationale.
We showed previously (15) that both rrnB P1 promoter activity and ATP concentration are elevated relative to wild type in a carA strain limited for pyrimidines. Although this result suggested that the initiating NTP concentration affects rrn P1 promoter activity in vivo, it did not distinguish whether the observed effects were direct or indirect. Changes in nucleotide concentrations could potentially have affected other regulatory factor(s). For example, ppGpp concentrations decrease during pyrimidine limitation (24), and this decrease could have been responsible for the observed increase in rrnB P1 activity.
In wild-type cells, rrn P1 promoters (initiating with either ATP or GTP) are regulated in parallel (refs. 4, 15, 25, 26, and data not shown) either by responding together to the same factor and/or to different factors that change in parallel. To determine whether changes in purine NTP concentrations affect rRNA transcription directly in vivo, we examined the activities of rrn P1 promoters initiating with either ATP or GTP in purE and guaB mutant strains where the normal parallel regulation of ATP and GTP concentrations was disrupted. If the activities of rrn P1 promoters paralleled the concentrations of their respective initiating nucleotides, even when those NTP concentrations changed in opposite directions, the simplest interpretation would be that NTP concentrations directly affect rrn P1 promoter activity in vivo. Alternatively, if the activities of rrn P1 promoters initiating with ATP or with GTP were affected in parallel although though the ATP and GTP concentrations diverged, the simplest interpretation would be that changing NTP concentrations affect rrn P1 promoters indirectly by acting through one or more other regulators.
To test the effects of diverging ATP and GTP concentrations on transcription in vivo, we used the rrnB P1 and rrnD P1 promoters, which initiate with ATP and GTP, respectively. Because rrnB P1 and rrnD P1 differ at other positions besides +1 (Fig. 1A), and because these differences could potentially affect regulation by factors other than the initiating NTP concentration, we also constructed an rrnB P1 promoter derivative that initiates with GTP rather than ATP (rrnB P1 + 1G; Fig. 1A). rrnB P1 required high concentrations of ATP and not GTP for maximal activity in vitro (see also ref. 15), whereas both rrnD P1 and rrnB P1 + 1G required high concentrations of GTP and not ATP for maximal activity in vitro (Fig. 1B). As a control, we also used a 3-bp variant of rrnB P1 that makes a longer-lived open complex and is, therefore, no longer regulated in vivo [rrnB P1(dis); Fig. 1A; refs. 10, 11, 17]. The rrnB P1(dis) promoter required neither high ATP nor high GTP for maximal activity in vitro (Fig. 1B).
rrn P1 Promoter Activities Correlate with Initiating NTP Concentrations in Vivo in purE Mutants.
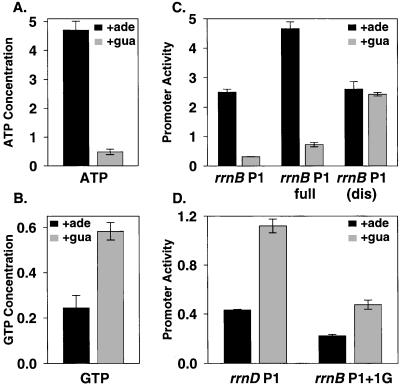
The purE gene encodes an enzyme involved in the early steps of purine metabolism, and disruption of purE leads to purine auxotrophy. However, purE mutants will grow in the presence of exogenous adenine or guanine. In a purE strain grown in adenine, ATP concentrations (measured either by formic acid extraction or by formaldehyde fixation followed by alkaline extraction; see Discussion) were ≈6-fold higher than in the same strain grown in guanine (Fig. 2A), although the growth rates in adenine and guanine were similar. In contrast, GTP concentrations were ≈2-fold lower in a purE strain grown in adenine than when grown in guanine (Fig. 2B). These results agree qualitatively with observations reported previously for Salmonella enterica (27).
Figure 2.
rrn P1 promoter activity depends on the concentration of the initiating NTP in purE mutants in vivo. (A and B) NTP concentrations from purE strains grown in M9 medium (18) supplemented with 0.2% glucose/0.2% casamino acids/10 μg/ml thiamine and with either adenine or guanine (0.1 mM), as indicated. The NTP concentrations [(pmol/ml/OD600) × 103] are from extractions performed with the formic acid method and were quantified by reverse-phase HPLC (see Materials and Methods). (C and D) Promoter activities (β-galactosidase Miller units × 103) from single-copy promoter-lacZ fusions. Bars represent averages of at least three measurements from at least two independent cultures. SDs are indicated. The purE strain had a growth rate of 0.58 doublings per hour in the presence of adenine and 0.49 doublings per hour in guanine. Promoter-lacZ fusions (promoter sequence endpoints and strain numbers in parentheses): rrnB P1 (−66 to +50, RLG6210); rrnB P1 full (−152 to +50, RLG6222); rrnB P1(dis) (−66 to +9, RLG6224); rrnD P1 (−60 to +10, RLG6223); rrnB P1 +1G (−66 to +50, RLG6213).
rrn P1 promoter activities were monitored in purE mutants by using lacZ fusions (rrnB P1, lacking the sites for the transcriptional activator Fis; rrnB P1 full, containing the Fis sites; rrnB P1(dis), rrnD P1, and rrnB P1 + 1G). Transcription from rrnB P1, which starts with ATP, was ≈6-fold higher in the purE strain grown in adenine than in the same strain grown in guanine (Fig. 2C), paralleling the ≈6-fold higher ATP concentrations. rrnB P1 full responded similarly to the construct that lacked Fis sites (Fig. 2C). In contrast, transcription from the unregulated rrnB P1(dis) promoter was approximately the same in purE cultures grown in either adenine or guanine, consistent with this promoter's requirement for lower concentrations of the initiating NTP than rrnB P1 in vitro.
In contrast to the ATP-responsive rrnB P1 promoters, the promoters initiating with GTP (rrnD P1 and rrnB P1 + 1G) had lower activities in purE cultures grown in adenine than in guanine (2.6- and 2.1-fold, respectively; Fig. 2D), in concert with the cellular GTP concentrations and not with the ATP concentrations. The simplest interpretation of these results is that changes in initiating NTP concentrations directly affect rrn P1 promoter activity in exponentially growing cells, in accord with the results obtained in vitro. Similar results also were obtained in purE mutant strains lacking ppGpp (purE ΔrelA ΔspoT mutants; data not shown), indicating that ppGpp is not required for NTP-sensing in vivo.
rrn P1 Promoter Activities Correlate with Initiating NTP Concentrations in Vivo in guaB Mutants.
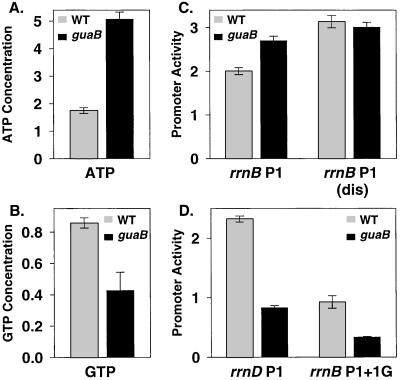
We also examined initiating NTP concentrations and promoter activities in a strain with a partial disruption of GuaB function. The guaB product catalyzes the first step in the guanine side of the de novo purine synthesis pathway, and partial disruption of guaB in S. enterica has been reported to reduce GTP concentrations in the absence of exogenous guanine while increasing ATP concentrations (27). We observed similar results in E. coli: ATP concentrations were ≈2.5-fold higher in the guaB strain than in the wild-type strain grown under the same conditions (Fig. 3A), whereas GTP concentrations were ≈2-fold lower in the guaB strain than in the wild type (Fig. 3B).
Figure 3.
rrn P1 promoter activity depends on the concentration of the initiating NTP in guaB mutants in vivo. (A and B) NTP concentrations and (C and D) rrn P1 promoter activities from wild-type and guaB strains grown in the same medium as in Fig. 2 but in the absence of exogenous purines; NTP concentrations and promoter activities were measured as in Fig. 2. The growth rates of the wild-type and guaB strains were 0.98 and 0.45 doublings per hour, respectively. Promoter-lacZ fusions (promoter sequence endpoints and strain numbers in parentheses): rrnB P1 (−66 to +50, RLG6209 in guaB background, RLG6208 in wild-type background); rrnB P1(dis) (−66 to +9, RLG6205 in guaB, RLG5651 in wild type); rrnD P1 (−60 to +10, RLG4591 in guaB, RLG6200 in wild type); rrnB P1 + 1G (−66 to +50, RLG6212 in guaB, RLG6211 in wild type).
Transcription from the promoter initiating with ATP, rrnB P1, was higher in the guaB strain than in the wild-type strain (Fig. 3C), in parallel with the increased ATP concentrations. This increase is especially significant because the guaB mutant grew >2-fold slower than the wild-type strain, and in a wild-type strain, rrnB P1 promoter activity decreases with decreasing growth rate (see Fig. 4). Thus, the guaB strain has an altered relationship between growth rate and rrnB P1 promoter activity that correlates with the elevated ATP concentration. In contrast, the promoter variant with the longer-lived open complex, rrnB P1(dis), showed no appreciable difference in activity in the guaB and wild-type strains. Transcription from the promoters initiating with GTP, rrnD P1 and rrnB P1 + 1G, decreased 2- to 3-fold in the guaB mutant relative to the wild-type strain, in parallel with the decreased GTP concentrations (and slower growth rate). Thus, rrn P1 promoter activity in the guaB strain depends on the concentration and identity of the initiating NTP, supporting the conclusion that rrn P1 promoters respond directly to changes in their initiating NTP concentrations in vivo. Furthermore, because the rrn P1 promoter activities increased when initiating NTP concentrations increased, the results suggest that the NTP concentrations present in vivo are not saturating for rrn P1 promoter activity (see Discussion).
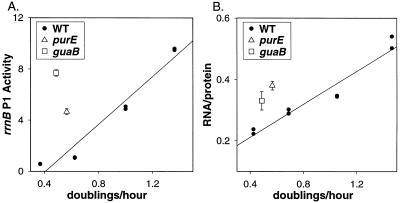
Figure 4.
rrn P1 promoter activity in purE and guaB mutants is disproportionate for the growth rate and leads to overproduction of rRNA. Wild-type cells were grown in different media (M9 with 0.2% glycerol, 0.2% glucose, 0.2% glucose, and 0.2% casamino acids, or LB). purE and guaB strains were grown in the media described in Figs. 2 and 3. (A) Promoter activity from an rrnB P1 full-lacZ fusion (β-galactosidase Miller units × 103) in a wild-type strain (●, RLG4747). A linear regression was drawn through points from at least two experiments with SIGMAPLOT V.5.0. The activity of the same promoter was measured from a purE strain (▵, RLG6222) or a guaB strain (□, RLG6202). (B) RNA:protein ratios from the same strains under the same conditions as in A.
Increased ATP Concentration Leads to Overproduction of rRNA.
Normally, rRNA promoter activity is proportional to the steady-state growth rate (growth rate-dependent control; refs. 26 and 28). Consistent with previous results, expression from the rrnB P1-lacZ fusion containing all three Fis sites (rrnB P1 full) increased with growth rate in a wild-type strain (Fig. 4A). The growth rates of the purE strain grown in adenine and of the guaB strain grown in the absence of exogenous guanine were relatively slow, but rrnB P1 promoter activity was disproportionately high in the mutant strains for their growth rates (Fig. 4A), in accord with their unusually high ATP concentrations.
Because rRNA accounts for most of the RNA present in cells (primarily because rRNAs are large and long-lived), the ratio of total RNA to total protein provides an estimate of rRNA synthesis. Like the rrn P1 promoter activities, RNA:protein ratios increase with the growth rate in the wild-type strain (Fig. 4B). To determine whether rRNA synthesis from the chromosomal rrn operons is uncoupled from the growth rate in the purine mutant strains, RNA:protein ratios in the purE and guaB strains were compared with RNA:protein ratios in wild-type strains at the same growth rate (Fig. 4B). The RNA:protein ratios in the purine mutant strains were unusually high for their respective growth rates, consistent with the elevated rrnB P1 promoter activity observed in these strains (Fig. 4A). We conclude that other mechanisms that regulate rRNA synthesis cannot fully compensate for the effect of the increased ATP levels in the mutant strains.
NTP-Sensing Plays a Role in Homeostatic Regulation of rrn P1 Promoter Activity.
Previous work indicated that translational activity regulates rRNA expression through a negative feedback mechanism whose molecular basis was unclear (29, 30). We hypothesized that because translation consumes purine NTPs, the initiating NTP concentration might be a feedback signal linking rRNA transcription initiation to translational activity (15). This model predicts that if translation were inhibited, ATP and GTP consumption would decrease, thereby causing an increase in intracellular ATP and GTP concentrations and a corresponding increase in rRNA transcription. To test the prediction that initiating NTP concentration might be responsible, at least in part, for feedback control of ribosome synthesis, we examined ATP levels and rrnB P1 promoter activity after translation inhibition by antibiotics in vivo.
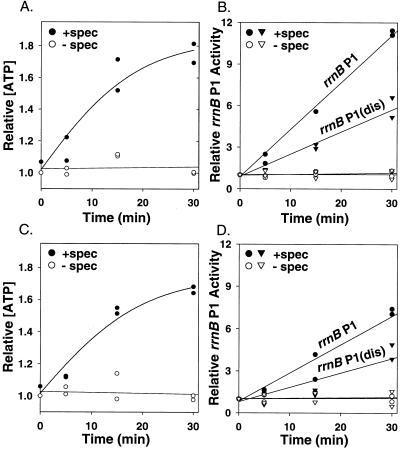
After the addition of 100 μg/ml spectinomycin (Fig. 5A) or chloramphenicol (data not shown) to log-phase cultures, the ATP concentration increased ≈2-fold within 30 min relative to the ATP level in an untreated culture (see also ref. 31). The increase in ATP concentration was accompanied by an increase in transcription from rrnB P1, as measured by primer extension (Fig. 5B), consistent with previous reports that rRNA accumulates under these conditions (32, 33). The increase in transcription from the wild-type rrnB P1 promoter was ≈2-fold greater than that from rrnB P1(dis) (Fig. 5B), consistent with the latter promoter's longer-lived open complex and lower initiating NTP concentration requirement. [We attribute the larger absolute increase observed in the experiment shown in Fig. 5 to effects of the antibiotic unrelated to the effect of increased ATP concentration on open complex lifetime. rrnB P1(dis) controls for these promoter nonspecific effects, because it makes exactly the same transcript as rrnB P1.] These findings indicate that protein synthesis consumes enough ATP to influence the cellular ATP concentration, and that this increase in ATP concentration is sufficient to serve as a feedback signal to rRNA promoters.
Figure 5.
Inhibition of protein synthesis leads to increased ATP concentration and increased rrn P1 promoter activity. Cells were grown in Mops medium (40) with 0.2% glucose/0.2% casamino acids/10 μg/ml thiamine. Filled symbols, 100 μg/ml spectinomycin in water added at time 0; open symbols, water only (control). (A) Relative ATP concentrations in wild-type strains. (B) Promoter activities from the same strains as in A, activities measured by primer extension. Circles, rrnB P1 promoter (−66 to +9, RLG3739); triangles, rrnB P1(dis) promoter (−66 to +9, RLG5651). (C and D) Same as A and B, but from a ΔrelA ΔspoT strain [circles, rrnB P1–66 to +9, RLG6218; triangles, rrnB P1(dis) −66 to +9, RLG6219].
Although these experiments were performed at high growth rates where ppGpp concentrations are very low, it was conceivable that the increased rrnB P1 promoter activity observed after spectinomycin addition resulted from a potential decrease in the ppGpp concentration rather than from the increase in ATP concentration. Therefore, we repeated the experiment in a strain deleted for the genes responsible for synthesizing ppGpp (ΔrelA ΔspoT). As in the wild-type strain, both ATP concentration and rrnB P1 promoter activity increased after addition of the antibiotic, whereas rrnB P1(dis) promoter activity increased much less (Fig. 5 C–D). We conclude that feedback by initiating NTP concentration contributes to homeostatic control of rRNA synthesis in the presence or absence of ppGpp.
Discussion
Initiating NTP Concentration Directly Affects rrn P1 Promoter Activity.
Previous studies implicated changing NTP concentrations in the control of rRNA synthesis, but the potential for indirect effects in vivo was not excluded. The results reported here strongly support the model that the effect of the initiating NTP concentration on rrn P1 promoter activity in vivo is direct, although of course more complex models are possible. Specific responses to initiating NTP concentration in vivo were limited to promoters that form short-lived open complexes. Transcription from promoters that form longer-lived open complexes [e.g., rrnB P1 (dis), Figs. 2, 3, and 5; lacUV5, data not shown] was much less affected by changes in NTP concentration in vivo.
rrn P1 Promoters Are Not Saturated for NTPs in Vivo.
It has been proposed that rrn P1 promoters are saturated for their initiating NTPs during exponential growth in vivo, rendering regulation by changing ATP and GTP concentrations unlikely (34). However, when ATP concentrations were increased over the wild-type level by growth of a guaB mutant in guanine (Fig. 3), or by spectinomycin or chloramphenicol treatment of wild-type cells grown in minimal or complex medium (Fig. 5 and data not shown), there was always a corresponding increase in rrn P1 promoter activity. These results indicate that rrn P1 promoter open complexes are not saturated for the initiating NTP in vivo.
The Role of NTP-Sensing in Homeostatic Regulation of rRNA Synthesis.
In wild-type cells, the rRNA synthesis rate is finely tuned to the cell's nutritional environment, yet remains remarkably constant following most genetic manipulations that might be expected to perturb it. For example, when the rRNA gene dose was altered by adding rRNA operons on plasmids or by inactivating chromosomal rRNA operons (35–37), when rRNA transcription initiation was altered by deletion of the fis gene or by mutation of the gene coding for the RNAP α-subunit (6, 7), or when rRNA transcription elongation was compromised by mutation of genes coding for Nus factors (38), rRNA core promoter activity changed to keep the overall rRNA synthesis rate appropriate for the growth rate.
The high ATP concentrations produced in the purine mutants resulted both in elevated rrn P1 promoter activity and in elevated rRNA levels (Fig. 4). Thus, unlike the situations just described, high ATP concentrations overwhelmed the mechanism(s) that potentially could have prevented rRNA overexpression. However, the increase in rRNA levels was not as large as that observed in rrn P1 promoter activity (Fig. 4). It is not surprising that the increase in rrn P1 promoter activity overestimates the increase in rRNA expression in the purine mutant strains. Some of the overproduced rRNA in the purine mutants might get degraded, because it might not get incorporated into ribosomes (see also ref. 36). Furthermore, at the moderate growth rates achieved by the purine mutant strains, much of the cell's rRNA transcription originates from the rrn P2 promoters, which initiate primarily with CTP and are therefore not affected by increased ATP concentration (H. D. Murray and R.L.G., unpublished work).
It has been argued that NTP pools are unlikely to function as feedback signals, informing rRNA operons about the translational state of the cell (39). However, the translation inhibition studies reported here (Fig. 5) strongly suggest that translation is a major consumer of ATP and GTP in vivo. Furthermore, as rrnB P1 activity increased in parallel with the increased NTP concentrations that resulted from shut-off of protein synthesis, these studies support the model that the NTP concentration serves as a feedback signal for homeostatic control of rRNA synthesis.
The increase in rrnB P1 activity in the ΔrelA ΔspoT strain after spectinomycin treatment was slightly less than that observed in the wild-type strain (Fig. 5). Because ppGpp concentrations have been reported to decrease after antibiotic addition (32), it is possible that both an increase in ATP concentration and a decrease in ppGpp concentration could contribute to stimulation of rrn P1 promoter activity in wild-type strains after spectinomycin addition. However, our studies clearly indicate that ppGpp is not essential for this feedback response.
When Does NTP-Sensing Affect rrn P1 Promoter Activity in Wild-Type Strains in Vivo?
We have demonstrated that rrn P1 promoters respond directly to changes in initiating NTP concentrations created by genetic manipulation or by protein synthesis inhibitors in vivo. Naturally occurring conditions in which NTP concentrations change and affect rRNA transcription have recently been defined (H. D. Murray, D.A.S., and R.L.G., unpublished results).
Previously, we and others (15, 31) have reported that ATP and GTP concentrations increase with the increases in growth rate achieved by varying the carbon source. This correlation between steady-state growth rate and initiating NTP concentrations suggested that NTP-sensing might contribute to the phenomenon referred to as growth rate-dependent control of rRNA synthesis (15). In contrast, another study reported recently that NTP concentrations do not change with growth rate and, therefore, that NTP-sensing could not be responsible for growth rate-dependent regulation of rRNA synthesis (39).
We compared the NTP extraction protocols used in these studies (formaldehyde fixation to inactivate ATPases followed by extraction with alkali, as in ref. 15, vs. formic acid extraction without formaldehyde treatment, as in ref. 39), and found that they account for the differences in the reported results (data not shown). Both extraction methods yielded highly reproducible results independent of strain background, but for reasons that remain unclear, NTP concentrations seem to be proportional to growth rate when extracted by the formaldehyde/alkali method from cells growing in different media, whereas NTP concentrations seem to be higher and relatively constant when extracted by the formic acid method from those same media. Although the extraction methods led to different conclusions with respect to growth-rate dependence of NTP concentrations, the two methods led to identical conclusions with respect to the ratios of NTP concentrations in the purE strain grown in the same medium with adenine vs. guanine and with respect to the ratios of NTPs in the same medium in the wild-type vs. the guaB strain. (We report here the NTP concentrations from formic acid extraction.)
Because there is no compelling reason to believe that one extraction method more accurately reflects the concentrations of free NTPs available to RNAP, further studies will be needed to evaluate the role of the NTP-sensing mechanism in growth-rate dependence of rRNA transcription. One possibility is that formic acid extraction liberates total NTP pools, whereas formaldehyde fixation followed by extraction with alkali results in detection of pools of NTPs not associated with protein. The relative efficiencies of extraction by the two methods could vary in media of different composition. In any case, the results reported here indicate that when initiating NTP concentrations change, rrn P1 promoters respond directly to these changes in vivo.
Acknowledgments
We thank Wilma Ross, Melanie Barker, Heath Murray, and Brian Paul for helpful comments and Kaj Frank Jensen for constructive criticism and strains. This work was supported by National Institutes of Health Grant GM37048 (to R.L.G.) and by predoctoral fellowships from the National Science Foundation and the Wisconsin Alumni Research Foundation (to D.A.S.).
Abbreviation
- RNAP
RNA polymerase
References
- 1.Gourse R L, Gaal T, Bartlett M S, Appleman J A, Ross W. Annu Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 2.Keener J, Nomura M. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1417–1431. [Google Scholar]
- 3.Squires C L, Zaporojets D. Annu Rev Microbiol. 2000;54:775–798. doi: 10.1146/annurev.micro.54.1.775. [DOI] [PubMed] [Google Scholar]
- 4.Hirvonen C A, Ross W, Wozniak C E, Marasco E, Anthony J R, Aiyar S E, Newburn V, Gourse R L. J Bacteriol. 2001;183:6305–6314. doi: 10.1128/JB.183.21.6305-6314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P J, Record M T, Jr, Gourse R L. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 6.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 7.Ross W, Thompson J F, Newlands J T, Gourse R L. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afflerbach H, Schroder O, Wagner R. Mol Microbiol. 1998;28:641–653. doi: 10.1046/j.1365-2958.1998.00829.x. [DOI] [PubMed] [Google Scholar]
- 9.Bartlett M S, Gourse R L. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josaitis C A, Gaal T, Gourse R L. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barker M M, Gaal T, Josaitis C A, Gourse R L. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 12.Gourse R L, Gaal T, Aiyar S E, Barker M M, Estrem S T, Hirvonen C A, Ross W. Cold Spring Harbor Symp Quant Biol. 1998;63:131–139. doi: 10.1101/sqb.1998.63.131. [DOI] [PubMed] [Google Scholar]
- 13.Cashel M, Gentry D R, Hernandez V H, Vinella D. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1458–1496. [Google Scholar]
- 14.Gaal T, Gourse R L. Proc Natl Acad Sci USA. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 16.Bartlett M S, Gaal T, Ross W, Gourse R L. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 17.Barker M M, Gourse R L. J Bacteriol. 2001;183:6315–6323. doi: 10.1128/JB.183.21.6315-6323.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 19.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poulsen P, Jensen K F. Mol Gen Genet. 1987;208:152–158. doi: 10.1007/BF00330436. [DOI] [PubMed] [Google Scholar]
- 21.Burgess R R, Jendrisak J J. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 22.Little R, Bremer H. Anal Biochem. 1982;126:381–388. doi: 10.1016/0003-2697(82)90531-0. [DOI] [PubMed] [Google Scholar]
- 23.Jensen K F, Houlberg U, Nygaard P. Anal Biochem. 1979;98:254–263. doi: 10.1016/0003-2697(79)90138-6. [DOI] [PubMed] [Google Scholar]
- 24.Vogel U, Pedersen S, Jensen K F. J Bacteriol. 1991;173:1168–1174. doi: 10.1128/jb.173.3.1168-1174.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gourse R L, de Boer H A, Nomura M. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 26.Miura A, Krueger J H, Itoh S, deBoer H A, Nomura M. Cell. 1981;25:773–782. doi: 10.1016/0092-8674(81)90185-9. [DOI] [PubMed] [Google Scholar]
- 27.Jensen K F. J Gen Microbiol. 1989;135:805–815. doi: 10.1099/00221287-135-4-805. [DOI] [PubMed] [Google Scholar]
- 28.Maaloe O, Kjeldgaard N O. Control of Macromolecular Synthesis: A Study of DNA, RNA, and Protein Synthesis in Bacteria. New York: Benjamin; 1966. [Google Scholar]
- 29.Yamagishi M, de Boer H A, Nomura M. J Mol Biol. 1987;198:547–550. doi: 10.1016/0022-2836(87)90299-3. [DOI] [PubMed] [Google Scholar]
- 30.Cole J R, Olsson C L, Hershey J W, Grunberg-Manago M, Nomura M. J Mol Biol. 1987;198:383–392. doi: 10.1016/0022-2836(87)90288-9. [DOI] [PubMed] [Google Scholar]
- 31.Bagnara A S, Finch L R. Eur J Biochem. 1973;36:422–427. doi: 10.1111/j.1432-1033.1973.tb02927.x. [DOI] [PubMed] [Google Scholar]
- 32.Lund E, Kjeldgaard N O. Eur J Biochem. 1972;28:316–326. doi: 10.1111/j.1432-1033.1972.tb01916.x. [DOI] [PubMed] [Google Scholar]
- 33.Shen V, Bremer H. J Bacteriol. 1977;130:1098–1108. doi: 10.1128/jb.130.3.1098-1108.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang S, Bipatnath M, Xu Y, Chen S, Dennis P, Ehrenberg M, Bremer H. J Mol Biol. 1999;292:19–37. doi: 10.1006/jmbi.1999.3056. [DOI] [PubMed] [Google Scholar]
- 35.Jinks-Robertson S, Gourse R L, Nomura M. Cell. 1983;33:865–876. doi: 10.1016/0092-8674(83)90029-6. [DOI] [PubMed] [Google Scholar]
- 36.Gourse R L, Takebe Y, Sharrock R A, Nomura M. Proc Natl Acad Sci USA. 1985;82:1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Condon C, French S, Squires C, Squires C L. EMBO J. 1993;12:4305–4315. doi: 10.1002/j.1460-2075.1993.tb06115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharrock R A, Gourse R L, Nomura M. Proc Natl Acad Sci USA. 1985;82:5275–5279. doi: 10.1073/pnas.82.16.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen C, Moller L B. J Biol Chem. 2000;275:3931–3935. doi: 10.1074/jbc.275.6.3931. [DOI] [PubMed] [Google Scholar]
- 40.Neidhardt F C, Bloch P L, Smith D F. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]