Abstract
Background
Liver transplantation (LT) for primary liver cancers achieves excellent patient outcomes, but a minority recur with poor prognosis. Survival may be improved by earlier recurrence detection. This study aims to evaluate the feasibility and performance of a personalized tumor-informed assay utilizing circulating tumor DNA (ctDNA) from peripheral blood for surveillance after LT in patients with hepatocellular carcinoma (HCC) or cholangiocarcinoma (CCA).
Methods
Here, we test whether a personalized tumor-informed assay utilizing ctDNA from peripheral blood informs post-LT surveillance. Personalized ctDNA assays were employed for surveillance in 38 LT recipients, alongside standard-of-care imaging and peripheral tumor biomarkers [alpha-fetoprotein (AFP), carbohydrate antigen 19-9 (CA19-9)].
Results
Recurrence was detected radiologically in 6 patients, with positive ctDNA assays in 3 and negative/insufficient to process (ITP) in 3. Nine ITP ctDNA assays were due to insufficient primary tumor tissue. In 31 patients without ITP ctDNA tests, the sensitivity and specificity of ctDNA were 75% [95% confidence interval (CI): 19–99%] and 100% (95% CI: 87–100%). Standard-of-care tumor biomarkers had sensitivity and specificity of 75% (95% CI: 19–99%) and 93% (95% CI: 76–99%), respectively (P>0.99 and P=0.16; McNemar χ2). Only 1 patient had ctDNA positive prior to imaging-based diagnosis.
Conclusions
This study corroborates the feasibility of ctDNA assays for recurrence surveillance in LT recipients. The results imply that ctDNA assays show promise in confirming recurrence and minimizing the need for invasive biopsy. However, additional prospective studies are needed to confirm ctDNA test utility in surveillance protocols.
Keywords: Transplant oncology, liver transplantation (LT), hepatocellular carcinoma (HCC), cholangiocarcinoma (CCA), circulating tumor DNA (ctDNA), liquid biopsy
Highlight box.
Key findings
• This study evaluated a personalized tumor-informed circulating tumor DNA (ctDNA) assay for surveillance after liver transplantation (LT) in 38 patients with hepatocellular carcinoma (HCC) and cholangiocarcinoma (CCA). Recurrence was detected in 6 patients, with ctDNA positive in 3 and negative or insufficient to process (ITP) in 3. Sensitivity of ctDNA was 75% and specificity was 100%, compared to 75% sensitivity and 93% specificity for standard biomarkers like alpha-fetoprotein (AFP) and carbohydrate antigen 19-9 (CA19-9). Only one patient had ctDNA positivity before imaging confirmed recurrence.
What is known and what is new?
• Post-LT surveillance for HCC and CCA typically relies on imaging and tumor markers like AFP and CA19-9, which have limited sensitivity and specificity. ctDNA, a non-invasive biomarker, has shown promise in other cancers but lacks robust data in LT settings.
• This study is the first to prospectively assess serial ctDNA testing post-LT, demonstrating its feasibility and high specificity, though with comparable sensitivity to existing methods.
What is the implication, and what should change now?
• The high specificity of ctDNA suggests it could confirm recurrence, potentially reducing invasive biopsies when imaging or biomarkers are inconclusive. However, its limited sensitivity and infrequent early detection indicate it should complement, not replace, current surveillance. Larger prospective studies are needed to refine its role, optimize testing frequency, and address ITP rates, particularly in patients with prior locoregional therapy.
Introduction
Primary liver malignancies have rising incidence and mortality rates in the United States and globally (1,2). Hepatocellular carcinoma (HCC) and cholangiocarcinoma (CCA) are the two most common types. With best palliative management alone, these have 5-year overall survival (OS) probability of under 5% (3,4). Multi-modal therapy including liver transplantation (LT) offers excellent treatment for both localized HCC (5) and CCA (6,7), as per the National Comprehensive Cancer Network (NCCN) and American Association for the Study of Liver Diseases (AASLD) guidelines (8-11), with 5-year OS probabilities of greater than 50%. However, post-LT 5-year recurrence rates are reported at greater than 15% for HCC and 40% for CCA patients. Recurrence is associated with poor prognosis and is the most common cause of death in these patients. Patients who recur have a median post-recurrence survival of under 1 year, due in part to limited treatment options in immunosuppressed LT recipients (12-15).
There is limited evidence on the role, modality, and frequency of surveillance post-LT for HCC and CCA recurrence. The NCCN, AASLD, and International Liver Transplantation Society (ILTS) (16,17) guidelines recommend circulating tumor markers, alpha-fetoprotein (AFP) and carbohydrate antigen 19-9 (CA19-9), and multiphasic cross-sectional imaging [i.e., computed tomography (CT) or magnetic resonance imaging (MRI)] every 3–6 months for 2 years then every 6 months for up to 5 years. One retrospective, multicenter study reported that greater use of surveillance imaging was associated with improved post-recurrence survival, though the threshold for “high frequency” was only 3 imaging studies in the first 24 months post-LT (17-19). More so, currently recommended circulatory markers, including AFP and CA19-9, have well-noted limitations in both sensitivity and specificity (20-22).
Circulating tumor DNA (ctDNA) in peripheral blood has been studied as a non-invasive biomarker for recurrence post-curative intent therapy in multiple cancers, especially breast (23) and colorectal (24-27). Further, ctDNA detection may inform the use of adjuvant and palliative systemic therapies (25,28), but not local therapies that require confirmatory imaging (29). In HCC and CCA, the current evidence for ctDNA is limited to retrospective series which only suggest a role in informing prognosis (30). Thus, integration of ctDNA testing into peri-LT care is not currently recommended by guidelines for primary liver cancers (8-11,31,32). The purpose of this study was to evaluate the feasibility of a personalized, tumor-informed ctDNA assay for post-LT recurrence surveillance in HCC and CCA patients, and to evaluate its performance relative to standard-of-care modalities. We present this article in accordance with the STARD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-791/rc).
Methods
Patient population
Patients undergoing LT between April 2018 and February 2023 at a single center were identified prospectively. The inclusion criterion was histologic diagnosis of primary liver cancer. Clinical criteria including demographics, pathology, treatment dates, time to recurrence, and time to last follow-up were extracted from the electronic medical records. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the Institutional Review Board of the Houston Methodist Hospital (No. PRO00035386). Informed consent was waived due to the use of de-identified data collected as part of routine clinical care.
Standard of care surveillance testing
Multiphasic cross-sectional imaging, AFP, and CA19-9 were obtained every 3 months for 2 years then every 6 months up to 5 years post-LT.
Tumor-informed ctDNA testing
Tumor-informed ctDNA testing was ordered post-transplant at the discretion of each patient’s treatment teams. The intention was to obtain these tests every 3 months, analogous to standard-of-care testing.
The cancer diagnosis was confirmed by histopathological examination of the explanted liver. Representative formalin-fixed, paraffin-embedded (FFPE) tumor tissue from the explanted liver underwent whole exome sequencing. From this, a set of up to 16 somatic, clonal single nucleotide variants (SNVs) were selected per patient to design a personalized, tumor-informed ctDNA assay [SignateraTM, bespoke multiplex (m) polymerase chain reaction (PCR)-next-generation sequencing], as previously described (24). Peripheral blood samples were collected from patients following LT. Peripheral blood samples with ≥2 SNVs detected were defined as ctDNA-positive. All biological samples were processed at Natera, Inc. (Austin, Texas, USA), following Clinical Laboratory Improvement Amendments (CLIA)-validated standard operating procedures. Timing of blood draws was intended to be performed every 3 months for 2 years then every 6 months for up to 5 years post-LT.
Outcomes
True recurrence was defined as any new lesion on CT or MRI with appropriate features, as reported by a board-certified clinical radiologist, and subsequently confirmed clinically by a multi-disciplinary team, with or without tissue confirmation. Positive AFP and CA19-9 were defined as an abnormally elevated level above 8.3 and 35 ng/mL, respectively, in the absence of other possible causes (e.g., biliary obstruction). AFP and CA19-9 values were tracked serially post-transplant. For patients with positive ctDNA tests, the post-transplant AFP and CA19-9 values up to the date of the positive ctDNA blood draw were considered. For patients with serially negative ctDNA tests, post-transplant values were considered up to the latest date of ctDNA blood draw. Positive ctDNA assay results were defined by the manufacturer. Test outcomes were categorized as successful (negative or positive) and insufficient to process (ITP). Primary outcomes were test parameters for ctDNA, AFP, and/or CA19-9. Secondary outcomes included observed ctDNA testing frequency, turnaround time, and ITP assay rate.
Statistical analysis
Patient characteristics were summarized according to test outcomes. All data were presented as median and interquartile range (IQR) for continuous variables, and number and % for categorical variables. Fisher’s exact test for categorical variables and Kruskal-Wallis test for continuous variables were used to compare between the test successful and ITP. Sensitivity, specificity, positive and negative predictive values were calculated and presented as percentage [95% confidence interval (CI)] (33). Statistical significance of differences in sensitivity and specificity between AFP, CA19-9, and ctDNA were determined by McNemar’s test. All analyses were performed in R (R Foundation for Statistical Computing, Vienna, Austria). Statistical significance was defined as two-tailed P<0.05 for all tests.
Results
Cohort
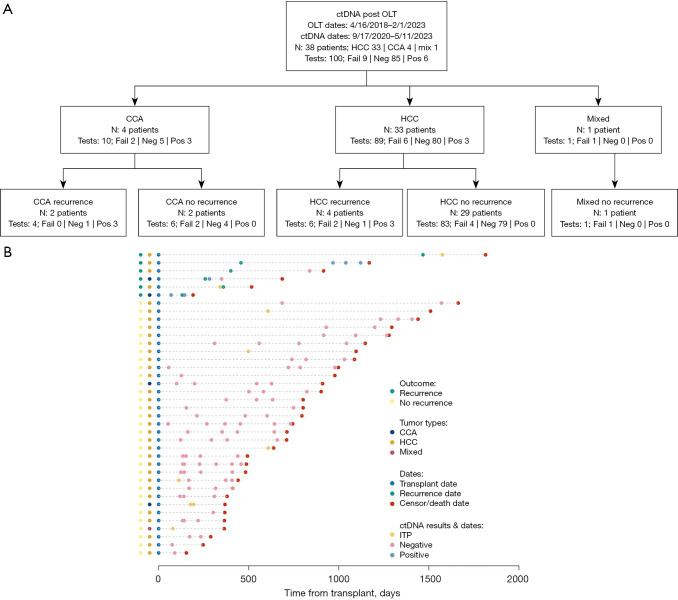
During the study period, 38 patients with primary liver cancer underwent LT followed by surveillance with ctDNA at a single institution (Table 1, Figure 1). No patients were excluded from downstream analyses. At the time of transplant, the median age was 63 years (IQR, 58–66 years), body mass index (BMI) 28.2 kg/m2 (IQR, 25.6–35.6 kg/m2), and laboratory Model for End-Stage Liver Disease (MELD) score of 14 (IQR, 9–30). Most patients were male (n=28, 73.7%), White (19, 50%), had chronic viral infections (12, 31.6%) or metabolic dysfunction-associated steatohepatitis (MASH) (12, 31.6%), and presented from home for LT (27, 71.1%). The median waitlist time was 284 days (IQR, 126–465 days). Thirty-one (81.6%) patients underwent pre-LT therapy, including loco-regional (29, 76.3%) and/or systemic (9, 23.7%). One (2.6%) had pre-LT hepatectomy. All patients underwent LT, with 3 (7.9%) receiving simultaneous kidney transplants. Eight (21.1%) patients received adjuvant systemic therapy. This included 4 patients with CCA and 1 with mixed tumor who received 5FU or gemcitabine for eight cycles, and 3 with HCC who received sorafenib or lenvatinib for 6 months. This center’s practice is to give adjuvant therapy to all CCA and mixed tumor patients with 5-fluorouracil (5-FU) or gemcitabine, informed by the BILCAP trial (34), and to HCC patients with findings on final pathology concerning for high recurrence risk, including greater than 3 nodules, maximum size greater than 5 cm, and lympho-vascular invasion. Median recurrence-free survival (RFS) was 583 days (IQR, 368–931 days), and OS was 728 days (IQR, 411–1,087 days) (Figure 2A,2B). Six (15.8%) patients had recurrences. Sites of initial recurrence included the liver allograft (n=3), lung (n=2), and bone (n=2). Three (7.9%) patients died, 2 of malignancy and 1 of respiratory failure.
Table 1. Clinical, pathologic, and outcome variables of patients undergoing ctDNA surveillance for primary liver cancer recurrence after liver transplantation.
| Variable | n | Total | Test outcome | P | |
|---|---|---|---|---|---|
| ITP (n=7) | Successful (n=31) | ||||
| Recipient clinical variables | |||||
| Age at transplant (years) | 38 | 63.0 (58.0, 66.0) | 66.0 (57.5, 66.0) | 63.0 (58.0, 65.0) | 0.61 |
| Sex | 38 | 0.75 | |||
| Female | 10 (26.3) | 1 (14.3) | 9 (29.0) | ||
| Male | 28 (73.7) | 6 (85.7) | 22 (71.0) | ||
| Race/ethnicity | 38 | 0.13 | |||
| White | 19 (50.0) | 4 (57.1) | 15 (48.4) | ||
| Hispanic | 15 (39.5) | 1 (14.3) | 14 (45.2) | ||
| Black | 4 (10.5) | 2 (28.6) | 2 (6.5) | ||
| BMI at transplant (kg/m2) | 37 | 28.2 (25.6, 35.6) | 29.9 (24.3, 32.2) | 28.2 (26.3, 35.7) | 0.61 |
| Laboratory MELD at transplant | 38 | 14.0 (9.0, 30.0) | 9.0 (8.5, 10.0) | 18.0 (9.0, 33.5) | 0.04 |
| Primary tumor type | 38 | 0.09 | |||
| Cholangiocarcinoma | 4 (10.5) | 1 (14.3) | 3 (9.7) | ||
| Hepatocellular carcinoma | 33 (86.8) | 5 (71.4) | 28 (90.3) | ||
| Hepatocellular-cholangiocarcinoma | 1 (2.6) | 1 (14.3) | 0 (0.0) | ||
| Etiology of liver disease | |||||
| Alcohol-associated liver disease | 38 | 7 (18.4) | 0 (0.0) | 7 (22.6) | 0.33 |
| MASH | 12 (31.6) | 2 (28.6) | 10 (32.3) | ||
| Viral (HCV, HBV) | 12 (31.6) | 4 (57.1) | 8 (25.8) | ||
| Other | 7 (18.4) | 1 (14.3) | 6 (19.4) | ||
| Time on waitlist (days) | 38 | 284.0 (126.0, 465.0) | 293.0 (206.0, 565.0) | 278.0 (72.0, 464.5) | 0.30 |
| Medical condition at transplant | 38 | 0.17 | |||
| Home | 27 (71.1) | 7 (100.0) | 20 (64.5) | ||
| Hospital, not in ICU | 4 (10.5) | 0 (0.0) | 4 (12.9) | ||
| In ICU | 7 (18.4) | 0 (0.0) | 7 (22.6) | ||
| Simultaneous liver-kidney transplant | 38 | 3 (7.9) | 0 (0.0) | 3 (9.7) | 0.94 |
| AFP at transplant (ng/mL) | 27 | 3.2 (2.5, 8.4) | 2.2 (2.2, 2.5) | 4.1 (2.5, 10.1) | NA |
| CA19-9 at transplant (U/mL) | 7 | 53.0 (32.0, 287.5) | 487.0 (487.0, 487.0) | 52.5 (12.0, 88.0) | NA |
| CEA at transplant (ng/mL) | 10 | 5.1 (1.8, 12.2) | 1.5 (1.5, 1.5) | 5.1 (4.1, 12.2) | NA |
| Adjunctive therapy | |||||
| Pre-transplant hepatectomy | 38 | 1 (2.6) | 1 (14.3) | 0 (0.0) | 0.41 |
| Pre-transplant LRT | 38 | 29 (76.3) | 7 (100.0) | 22 (71.0) | 0.03* |
| Pre-transplant systemic therapy | 38 | 9 (23.7) | 2 (28.6) | 7 (22.6) | >0.99 |
| Peri-transplant Whipple procedure | 38 | 1 (2.6) | 1 (14.3) | 0 (0.0) | 0.41 |
| Adjuvant systemic therapy | 38 | 8 (21.1) | 3 (42.9) | 5 (16.1) | 0.29 |
| Palliative systemic therapy | 38 | 2 (5.3) | 1 (14.3) | 1 (3.2) | 0.81 |
| Explant pathology | |||||
| Tumor focality | 38 | 0.22 | |||
| Solitary | 22 (57.9) | 6 (85.7) | 16 (51.6) | ||
| Multifocal | 16 (42.1) | 1 (14.3) | 15 (48.4) | ||
| Number of tumor nodules | 38 | 1.0 (1.0, 3.0) | 1.0 (1.0, 1.0) | 2.0 (1.0, 3.5) | 0.19 |
| Maximum tumor size (cm) | 37 | 2.9 (1.8, 3.6) | 2.8 (2.5, 3.6) | 2.9 (1.8, 3.5) | 0.89 |
| Lympho-vascular invasion present | 38 | 8 (21.1) | 2 (28.6) | 6 (19.4) | 0.98 |
| Perineural invasion present | 38 | 5 (13.2) | 1 (14.3) | 4 (12.9) | >0.99 |
| R1 margins | 38 | 2 (5.3) | 1 (14.3) | 1 (3.2) | 0.81 |
| Response to neoadjuvant therapy (percentage necrosis) | 25 | 65.0 (30.0, 85.0) | 99.0 (35.0, 100.0) | 55.0 (30.0, 75.0) | 0.21 |
| Cirrhosis present | 38 | 36 (94.7) | 6 (85.7) | 30 (96.8) | 0.81 |
| Outcomes | |||||
| Hospital length of stay (days) | 37 | 15.0 (10.0, 19.0) | 10.0 (9.5, 16.5) | 15.0 (11.0, 20.0) | 0.38 |
| Experienced recurrence | 38 | 6 (15.8) | 2 (28.6) | 4 (12.9) | 0.65 |
| Recurrence-free survival (days) | 38 | 583.0 (368.0, 931.0) | 638.0 (366.0, 1,259.0) | 577.0 (390.0, 920.0) | 0.63 |
| Location of recurrence | |||||
| Local | 38 | 3 (7.9) | 1 (14.3) | 2 (6.5) | >0.99 |
| Regional | 38 | 1 (2.6) | 0 (0.0) | 1 (3.2) | >0.99 |
| Distant | 38 | 3 (7.9) | 1 (14.3) | 2 (6.5) | >0.99 |
| Recurrence site | |||||
| Liver allograft | 38 | 3 (7.9) | 1 (14.3) | 2 (6.5) | >0.99 |
| Lung | 38 | 2 (5.3) | 1 (14.3) | 1 (3.2) | 0.81 |
| Bone | 38 | 2 (5.3) | 0 (0.0) | 2 (6.5) | >0.99 |
| Patient status | 38 | >0.99 | |||
| Alive | 34 (89.5) | 6 (85.7) | 28 (90.3) | ||
| Dead | 4 (10.5) | 1 (14.3) | 3 (9.7) | ||
| Patient survival (days) | 38 | 728.0 (411.0, 1,087.0) | 638.0 (442.0, 1,302.5) | 744.0 (426.0, 988.0) | 0.68 |
| Cause of death | 3 | >0.99 | |||
| Cancer | 2 (66.7) | 1 (100.0) | 1 (50.0) | ||
| Respiratory | 1 (33.3) | 0 (0.0) | 1 (50.0) | ||
Data are presented as median (IQR) or n (%). *, P<0.05. AFP, alpha fetoprotein; BMI, body mass index; CA19-9, carbohydrate antigen 19-9; CEA, carcinoembryonic antigen; ctDNA, circulating tumor DNA; HBV, hepatitis B virus; HCV, hepatitis C virus; ICU, intensive care unit; IQR, interquartile range; ITP, insufficient to process; LRT, locoregional therapy; MASH, metabolic dysfunction-associated steatohepatitis; MELD, Model for End-Stage Liver Disease; NA, not available.
Figure 1.
Overview of study cohort. (A) Study cohort including 100 ctDNA assays performed on 38 primary liver cancer patients post-liver transplantation. (B) Point-and-segment plot depicting the natural history of each patient, including recurrence status (far left), tumor type (middle left) and dates of liver transplant, recurrence, death/censor, and ctDNA reporting dates, normalized to transplant date. CCA, cholangiocarcinoma; ctDNA, circulating tumor DNA; Fail, failed; HCC, hepatocellular carcinoma; ITP, insufficient to process; Neg, negative; OLT, orthotopic liver allotransplantation; Pos, positive.
Figure 2.
Post-transplant survival. (A) Overall and (B) recurrence-free survival for the entire cohort. The dotted lines indicate the 95% confidence interval for the survival curves.
Tumor types included 33 HCC, 4 CCA, and 1 mixed HCC-CCA. At transplant, the median AFP was 2.8 ng/mL (IQR, 2.4–8.4 ng/mL) in HCC patients, and the median CA19-9 was 287.5 U/mL (IQR, 70–611 U/mL) in CCA patients. On explant pathology, the median number of tumor nodules was 1 (IQR, 1–3) and the maximum tumor size was 2.9 cm (IQR, 1.8–3.7 cm). There were 3 HCC (9.1% of 33) beyond Milan criteria (35). Lymphovascular invasion (LVI) was present in 8 (21.1%), perineural invasion (PNI) in 5 (13.2%), and background liver cirrhosis in 36 (94.7%). One (25% of 4) patient with CCA had lymph node metastases and 2 (50%) had R1 margins, requiring either margin revision or Whipple surgery. The median treatment effect, measured as histologic percent tumor necrosis, was 62.5% (IQR, 30–87.5%). Recipient and donor features stratified by tumor type are available in Table S1.
ctDNA test features
During post-LT surveillance, 100 ctDNA assays were ordered between 9/17/2020 and 5/11/2023 (Figure 3). The median time from LT to first test ordering was 117.5 days (IQR, 78–467 days), to first blood collection 148 days (IQR, 88–467 days), and first reporting 175 days (IQR, 122–607 days). The median time between test orders was 135.6 days (IQR, 78–189.3 days). The median number of tests per patient was 3 (range, 1–7). The time from ordering to collection increased significantly with each serial assay (P<0.001, Kruskal-Wallis), while the time from collection to reporting significantly decreased from the first to subsequent assays (P<0.001, Kruskal-Wallis), resulting in no significant difference in serial time from ordering to reporting (P=0.06, Kruskal-Wallis).
Figure 3.
Circulating tumor DNA assay metrics. (A) Barplot depicting time from first order to first blood collection (dark blue) and first ctDNA report (light blue) per patient, normalized to first order date. (B) Boxplots of times from ctDNA test order to blood collection for first and subsequent assays. (C) Boxplots of times from blood collection to ctDNA result report for first and subsequent assays. (D) Boxplots of times from ctDNA test order to result report for first and subsequent assays. ctDNA, circulating tumor DNA.
ctDNA assay logistics
Of the 100 ctDNA assays, there were 6 positive results, 85 negative, and 9 ITP. The number of positive tests per patient ranged from 0–3, negative tests 0–7, and ITP tests 0–2. ITP test assay was significantly associated with patients who received neoadjuvant loco-regional therapy and whose explanted tumors were of lower T stage and poorer differentiation (Tables 1,2; all P<0.05). The causes for ITP assays included insufficient tumor tissue (n=8, 21.1%) and improper blood handling/storage (n=1, 2.6%). The 9 tests with ITP results were performed on 8 patients. Of those 8 patients, 6 had only 1 test (i.e., they were not retested) and 2 were retested. Of the 2 retested patients, 1 had a second “ITP” test result, and 1 had 3 subsequent “negative” tests. Seven patients did not receive test results, only ITP assays (Table 2).
Table 2. ctDNA test features for the pooled cohort.
| Variable | N | Values |
|---|---|---|
| Test results per patient | 38 | |
| Positive | 3 (7.9) | |
| Negative | 28 (73.7) | |
| ITP | 7 (18.4) | |
| Reason for test ITP | 38 | |
| Insufficient tumor tissue | 8 (21.1) | |
| Improper blood storage/handling | 1 (2.6) | |
| Number of ctDNA tests per patient | 38 | 3.0 (1.0, 3.8) |
| Number of positive ctDNA tests per patient | 38 | 1.0 (1.0, 1.0) |
| Number of negative ctDNA tests per patient | 38 | 3.5 (2.0, 4.0) |
| Time between test orders (days) | 26 | 135.6 (78.0, 189.3) |
| Time between blood collections (days) | 26 | 148.8 (96.3, 193.0) |
| First ctDNA tests | ||
| Time from transplant to test order (days) | 38 | 117.5 (78.0, 467.0) |
| Time from transplant to blood collection (days) | 38 | 148.0 (88.0, 467.0) |
| Time from transplant to first test report (days) | 38 | 175.0 (122.0, 607.0) |
| Time from test order to blood collection, (days) | 38 | 0.0 (0.0, 21.0) |
| Time from blood collection to report (days) | 38 | 33.5 (22.0, 41.0) |
| Time from test order to report (days) | 38 | 41.0 (34.0, 64.0) |
| Subsequent ctDNA tests | ||
| Time from test order to blood collection (days) | 26 | 18.5 (10.0, 48.0) |
| Time from blood collection to report (days) | 26 | 9.0 (8.0, 10.0) |
| Time from test order to report (days) | 26 | 31.8 (19.0, 57.0) |
Data are presented as median (IQR) or n (%). ctDNA, circulating tumor DNA; IQR, interquartile range; ITP, insufficient to process.
ctDNA test sensitivity, specificity, and predictive values
In the 29 HCC patients without recurrence, there were 79 negative and 4 ITP assays. In the 4 HCC patients with recurrence, there were 3 positive tests in 1 patient, 1 negative test in 1 patient, and 2 ITP tests in 2 patients. In the 2 CCA patients without recurrence, there were 4 negative and 2 ITP tests. In the 2 CCA patients with recurrence, there was 1 positive test prior to and 1 positive test post-imaging-based recurrence in 1 patient, and 1 positive test and 1 negative test post-recurrence in another patient. In the 1 mixed tumor patient, there was 1 ITP test.
Ignoring the ITP tests (n=31 patients, Table 3), ctDNA test parameters include sensitivity 75% (95% CI: 19–99%), specificity 100% (95% CI: 87–100%), negative predictive value 96.4% (95% CI: 82–100%) and positive predictive value 100% (95% CI: 29–100%). For standard-of-care biomarkers in the same 31 patients, namely AFP for HCC or CA19-9 for CA19-9 patients, test parameters included sensitivity 75% (95% CI: 19–99%), specificity 93% (95% CI: 76–99%), negative predictive value 96% (95% CI: 80–100%) and positive predictive value 60% (95% CI: 15–95%). There was no significant difference in either sensitivity or specificity between standard and ctDNA biomarkers (P>0.99, P=0.16, respectively), though ctDNA specificity was non-significantly greater. When considering ITP tests as negative results (n=38 patients; Table 3), statistical significances of sensitivity and specificity differences remained the same. When considering HCC patients alone, the test features and statistical significances of sensitivity and specificity differences also remained the same (Table S2). As the most positive ctDNA tests (5/6) were ordered after imaging-based tumor recurrence, the time from assay positivity to clinical diagnosis could not be estimated.
Table 3. Biomarker test parameters for circulating tumor DNA versus traditional primary liver cancer biomarkers.
| Variables | Detection method, % [95% CI] | P value† | |||||
|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | Sensitivity | Specificity | ||
| Cholangiocarcinoma only | |||||||
| Excluding patients with non-complete ctDNA tests | 0.32 | 0.32 | |||||
| ctDNA | 100 [16–100] | 100 [3–100] | 100 [3–100] | 100 [16–100] | |||
| CA19-9 | 50 [1–99] | 0 [0–97] | 0 [0–97] | 50 [1–99] | |||
| Non-complete tests = negative results | 0.32 | 0.32 | |||||
| ctDNA | 100 [16–100] | 100 [16–100] | 100 [16–100] | 100 [16–100] | |||
| CA19-9 | 50 [1–99] | 50 [1–99] | 50 [1–99] | 50 [1–99] | |||
| Hepatocellular carcinoma only | |||||||
| Excluding patients with non-complete ctDNA tests | 0.32 | 0.32 | |||||
| ctDNA | 50 [1–99] | 100 [87–100] | 96 [81–100] | 100 [3–100] | |||
| AFP | 100 [16–100] | 96 [80–100] | 100 [86–100] | 67 [9–99] | |||
| Non-complete tests = negative results | 0.08 | 0.32 | |||||
| ctDNA | 25 [1–81] | 100 [88–100] | 91 [75–98] | 100 [3–100] | |||
| AFP | 100 [40–100] | 97 [82–100] | 100 [88–100] | 80 [28–99] | |||
| All tumor types | |||||||
| Excluding patients with non-complete ctDNA tests | >0.99 | 0.16 | |||||
| ctDNA | 75 [19–99] | 100 [87–100] | 100 [29–100] | 96 [82–100] | |||
| AFP/CA19-9 | 75 [19–99] | 93 [76–99] | 60 [15–95] | 96 [80–100] | |||
| Non-complete tests = negative results | 0.32 | 0.16 | |||||
| ctDNA | 50 [12–88] | 100 [89–100] | 100 [29–100] | 91 [77–98] | |||
| AFP/CA19-9 | 83 [36–100] | 94 [79–99] | 71 [29–96] | 97 [83–100] | |||
| Including patients with non-complete ctDNA tests | |||||||
| ctDNA and AFP/CA19-9 | 100 [54–100] | 94 [79–99] | 75 [35–97] | 100 [88–100] | |||
†, P values reflect differences between ctDNA and AFP/CA-19-9 tests. AFP, alpha-fetoprotein; CA19-9, carbohydrate antigen 19-9; CI, confidence interval; ctDNA, circulating tumor DNA; NPV, negative predictive value; PPV, positive predictive value.
When results of ctDNA, AFP, and CA19-9 were considered jointly, test parameters included sensitivity 100% (95% CI: 54–100%), specificity 94% (95% CI: 79–99%), positive predictive value 75% (95% CI: 35–97%), and negative predictive value 100% (95% CI: 88–100%).
Discussion
This study presents a clinical cohort of 38 patients with primary liver cancers prospectively undergoing post-LT tumor-informed ctDNA testing in serial peripheral blood samples. Approximately 10% of ctDNA assays were ITP, often due to insufficient solid tumor tissue availability, which was associated with receipt of pre-LT locoregional therapy. Specificity and positive predictive value were greater than sensitivity and negative predictive value. The specificity of ctDNA was non-significantly improved relative to conventional peripheral blood biomarkers (AFP, CA19-9). To our knowledge, this is the first study to implement serial ctDNA prospectively post-LT for cancer recurrence surveillance.
Data on the utility of peripheral blood ctDNA assays includes numerous studies on post-resection prognosis for primary liver cancers and recurrence surveillance in other solid tumors, including breast and colorectal cancers. However, most studies are retrospective and use differing methodologies. There is very little data specific to primary liver cancers and LT (30). Two abstracts from China presented at the 2022 American Society for Clinical Oncology (ASCO) Annual Meeting retrospectively explored the utility of ctDNA testing peri-LT for HCC (36,37). The first by Huang et al. evaluated pre- and post-LT ctDNA in 74 HCC patients (36). The second study by Jiang et al., in 45 patients, included only pre-LT ctDNA (37). In both, ctDNA detection was associated with recurrence but not with OS. Thus, the clinical translation of ctDNA assays in post-LT cancer surveillance remains in question. We did not identify a difference in sensitivity and specificity of the ctDNA assay over standard-of-care methods of detecting recurrence. Often (5/6 patients), recurrences were detected sooner by imaging than by ctDNA. Even in the 1 patient where ctDNA positivity preceded other tests, it led to confirmatory imaging rather than treatment. Given its non-significantly greater specificity than standard biomarkers, ctDNA may better serve as a confirmatory test for situations of diagnostic uncertainty in standard-of-care surveillance testing results. This would fulfill the terminology of “liquid biopsy”, potentially usurping the role of invasive solid tissue biopsies. To develop its potential utility in this role, trials allowing diagnosis based on ctDNA-positivity are needed (29).
This study demonstrates the feasibility of using longitudinal ctDNA testing post-LT in patients with HCC or CCA. Testing frequency was approximately every 4 months, in line with recommendations for standard screening every 3–6 months (8). The observed test ITP rate is higher than that reported in other solid tumors, reflecting the high frequency and effectiveness of pre-LT locoregional therapies in this clinical context. The increasing time interval from ordering to blood collection with serial testing observed here also raises concerns for patient compliance, which would ultimately limit its role as a surveillance test over the recommended 5-year post-LT period. Interestingly, high failure rate and low compliance were not reported in a prospective series of peripheral blood multi-analyte testing as pan-cancer screening, suggesting a different adoption of ctDNA in pre- versus post-treatment patients (38).
This study was limited by its relatively small sample size, low frequency of recurrences, short follow-up, and single-center nature. The sample size was inherently limited by the various histologies of primary liver cancers and their different peri-operative adjunctive therapies. This study focused on determining ctDNA test parameters for post-transplant recurrence in the pooled cohort. Factors including test timing relative to transplant and number of tests performed per patient could not be considered given sample size limitations. Analyses considering these factors would require a larger sample size. Nonetheless, we feel that our results are meaningful to clinicians practicing in this space, where single center patient sizes are typically on the scale included here or smaller. The infrequent recurrences prevented the association of ctDNA positivity with the anatomic site of recurrence. Primary liver cancers can recur after our 2-year follow-up period when the frequency of standard-of-care surveillance decreases. Paradoxically, less frequent imaging may accentuate the utility of ctDNA as a screening modality by introducing lead-time bias (27). Nonetheless, this is the first prospective study of peripheral blood tumor-informed ctDNA testing post-LT and can inform future studies.
Conclusions
Our study informs the application of peripheral blood tumor-informed ctDNA assays in surveillance post-LT for primary liver cancer. The high specificity and low sensitivity suggest a confirmatory or diagnostic role may supplant invasive solid tissue biopsy when abnormal conventional modalities suggest recurrence and show the need for more investigations in the surveillance setting. Additional studies are required to determine the best way to utilize these tests before clinical adoption in this context.
Supplementary
The article’s supplementary files as
Acknowledgments
We extend our sincere gratitude to Drs. Charuta C. Palsuledesai, Vasily N. Aushev, Chris M. Brewer, Meenakshi Malhotra, Adham Jurdi, and Minetta C. Liu from Natera, Inc., who provided technical assistance with the Signatera ctDNA tests.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the Institutional Review Board of the Houston Methodist Hospital (No. PRO00035386). Informed consent was waived due to the use of de-identified data collected as part of routine clinical care.
Footnotes
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-791/rc
Funding: This study was supported by the 2023 American Society of Transplant Surgeons Faculty Development Award (to A.A.C.).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-791/coif). R.M.G. has served as a consultant for Sanofi within the past 3 years; received an honorarium from Sanofi for lectures; participated on the Advisory Board for Sirtex, TransMedics (not currently an Advisory Board member), and LyGenesis; held leadership roles in the International Liver Transplantation Society and the American Society of Transplant Surgeons; owned but sold all stock in TransMedics; and owned stock in Eli Lilly. D.W.V. reports on owning common stock in Pfizer and Bristol Myers Squibb (BMS) and participating in a single advisory board meeting with Eisai. The other authors have no conflicts of interest to declare.
Data Sharing Statement
Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-791/dss
References
- 1.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913-21. 10.1158/0008-5472.CAN-14-0155 [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 3.Brar G, Greten TF, Graubard BI, et al. Hepatocellular Carcinoma Survival by Etiology: A SEER-Medicare Database Analysis. Hepatol Commun 2020;4:1541-51. 10.1002/hep4.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebata T, Mizuno T, Yokoyama Y, et al. Surgical resection for Bismuth type IV perihilar cholangiocarcinoma. Br J Surg 2018;105:829-38. 10.1002/bjs.10556 [DOI] [PubMed] [Google Scholar]
- 5.Koh JH, Tan DJH, Ong Y, et al. Liver resection versus liver transplantation for hepatocellular carcinoma within Milan criteria: a meta-analysis of 18,421 patients. Hepatobiliary Surg Nutr 2022;11:78-93. 10.21037/hbsn-21-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cambridge WA, Fairfield C, Powell JJ, et al. Meta-analysis and Meta-regression of Survival After Liver Transplantation for Unresectable Perihilar Cholangiocarcinoma. Ann Surg 2021;273:240-50. 10.1097/SLA.0000000000003801 [DOI] [PubMed] [Google Scholar]
- 7.Ziogas IA, Giannis D, Economopoulos KP, et al. Liver Transplantation for Intrahepatic Cholangiocarcinoma: A Meta-analysis and Meta-regression of Survival Rates. Transplantation 2021;105:2263-71. 10.1097/TP.0000000000003539 [DOI] [PubMed] [Google Scholar]
- 8.Singal AG, Llovet JM, Yarchoan M, et al. AASLD Practice Guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology 2023;78:1922-65. 10.1097/HEP.0000000000000466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowlus CL, Arrivé L, Bergquist A, et al. AASLD practice guidance on primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2023;77:659-702. 10.1002/hep.32771 [DOI] [PubMed] [Google Scholar]
- 10.Benson AB, D'Angelica MI, Abrams T, et al. NCCN Guidelines® Insights: Biliary Tract Cancers, Version 2.2023. J Natl Compr Canc Netw 2023;21:694-704. 10.6004/jnccn.2023.0035 [DOI] [PubMed] [Google Scholar]
- 11.Corbett V, Li D, Chauhan A. Review of practice informing data and current state of NCCN consensus guidelines in hepatobiliary cancers. Hepatobiliary Surg Nutr 2023;12:798-803. 10.21037/hbsn-23-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajendran L, Ivanics T, Claasen MP, et al. The management of post-transplantation recurrence of hepatocellular carcinoma. Clin Mol Hepatol 2022;28:1-16. 10.3350/cmh.2021.0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama I, Carr B, Saitsu H, et al. Accelerated growth rates of recurrent hepatocellular carcinoma after liver transplantation. Cancer 1991;68:2095-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gül-Klein S, Schmitz P, Schöning W, et al. The Role of Immunosuppression for Recurrent Cholangiocellular Carcinoma after Liver Transplantation. Cancers (Basel) 2022;14:2890. 10.3390/cancers14122890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodzin AS, Lunsford KE, Markovic D, et al. Predicting Mortality in Patients Developing Recurrent Hepatocellular Carcinoma After Liver Transplantation: Impact of Treatment Modality and Recurrence Characteristics. Ann Surg 2017;266:118-25. 10.1097/SLA.0000000000001894 [DOI] [PubMed] [Google Scholar]
- 16.Ho CM, Lee CH, Lee MC, et al. Survival After Treatable Hepatocellular Carcinoma Recurrence in Liver Recipients: A Nationwide Cohort Analysis. Front Oncol 2020;10:616094. 10.3389/fonc.2020.616094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berenguer M, Burra P, Ghobrial M, et al. Posttransplant Management of Recipients Undergoing Liver Transplantation for Hepatocellular Carcinoma. Working Group Report From the ILTS Transplant Oncology Consensus Conference. Transplantation 2020;104:1143-9. 10.1097/TP.0000000000003196 [DOI] [PubMed] [Google Scholar]
- 18.Sapisochin G, Goldaracena N, Astete S, et al. Benefit of Treating Hepatocellular Carcinoma Recurrence after Liver Transplantation and Analysis of Prognostic Factors for Survival in a Large Euro-American Series. Ann Surg Oncol 2015;22:2286-94. 10.1245/s10434-014-4273-6 [DOI] [PubMed] [Google Scholar]
- 19.Lee DD, Sapisochin G, Mehta N, et al. Surveillance for HCC After Liver Transplantation: Increased Monitoring May Yield Aggressive Treatment Options and Improved Postrecurrence Survival. Transplantation 2020;104:2105-12. 10.1097/TP.0000000000003117 [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Lai SL, Chen J, et al. Validated preoperative computed tomography risk estimation for postoperative hepatocellular carcinoma recurrence. World J Gastroenterol 2017;23:6467-73. 10.3748/wjg.v23.i35.6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe H, Midorikawa Y, Higaki T, et al. Magnetic resonance elastography-based prediction of hepatocellular carcinoma recurrence after curative resection. Surgery 2021;170:167-72. 10.1016/j.surg.2021.02.027 [DOI] [PubMed] [Google Scholar]
- 22.Benson AB, D'Angelica MI, Abbott DE, et al. Hepatobiliary Cancers, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2021;19:541-65. 10.6004/jnccn.2021.0022 [DOI] [PubMed] [Google Scholar]
- 23.Magbanua MJM, Swigart LB, Wu HT, et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann Oncol 2021;32:229-39. 10.1016/j.annonc.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol 2019;5:1124-31. 10.1001/jamaoncol.2019.0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tie J, Cohen JD, Lahouel K, et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med 2022;386:2261-72. 10.1056/NEJMoa2200075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tie J, Cohen JD, Wang Y, et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol 2019;5:1710-7. 10.1001/jamaoncol.2019.3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fakih M, Sandhu J, Wang C, et al. Evaluation of Comparative Surveillance Strategies of Circulating Tumor DNA, Imaging, and Carcinoembryonic Antigen Levels in Patients With Resected Colorectal Cancer. JAMA Netw Open 2022;5:e221093. 10.1001/jamanetworkopen.2022.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sartore-Bianchi A, Pietrantonio F, Lonardi S, et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: the phase 2 CHRONOS trial. Nat Med 2022;28:1612-8. 10.1038/s41591-022-01886-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venook AP. Colorectal Cancer Surveillance With Circulating Tumor DNA Assay. JAMA Netw Open 2022;5:e221100. 10.1001/jamanetworkopen.2022.1100 [DOI] [PubMed] [Google Scholar]
- 30.Johnson P, Zhou Q, Dao DY, et al. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2022;19:670-81. 10.1038/s41575-022-00620-y [DOI] [PubMed] [Google Scholar]
- 31.Abdelrahim M, Esmail A, Abudayyeh A, et al. Transplant Oncology: An Emerging Discipline of Cancer Treatment. Cancers (Basel) 2023;15:5337. 10.3390/cancers15225337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy T, Esmail A, Chang JC, et al. Utility of Cell-Free DNA Detection in Transplant Oncology. Cancers (Basel) 2022;14:743. 10.3390/cancers14030743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trevethan R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front Public Health 2017;5:307. 10.3389/fpubh.2017.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bridgewater J, Fletcher P, Palmer DH, et al. Long-Term Outcomes and Exploratory Analyses of the Randomized Phase III BILCAP Study. J Clin Oncol 2022;40:2048-57. 10.1200/JCO.21.02568 [DOI] [PubMed] [Google Scholar]
- 35.Mazzaferro V, Regalia E, Doci R, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med 1996;334:693-9. 10.1056/NEJM199603143341104 [DOI] [PubMed] [Google Scholar]
- 36.Huang A, Guo DZ, Zhang X, et al. Application of circulating tumor DNA for prediction and surveillance of tumor recurrence after liver transplantation: A pilot study. J Clin Oncol 2022;40:abstr e16149.
- 37.Jiang N, Zeng X, Tang J, et al. Circulating tumor DNA is a potential prognostic risk factor of recurrence in patients with hepatocellular carcinoma treated by liver transplantation. J Clin Oncol 2022;40:abstr e16196.
- 38.Lennon AM, Buchanan AH, Kinde I, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 2020;369:eabb9601. 10.1126/science.abb9601 [DOI] [PMC free article] [PubMed] [Google Scholar]