Abstract
Circadian rhythmicity in mammals is under the control of a molecular pacemaker constituted of clock gene products organized in transcriptional autoregulatory loops. Phase resetting of the clock in response to light involves dynamic changes in the expression of several clock genes. The molecular pathways used by light to influence pacemaker-driven oscillation of clock genes remain poorly understood. We explored the functional integration of both light- and clock-responsive transcriptional regulation at the promoter level of the Period (Per) genes. Three Per genes exist in the mouse. Whereas mPer1 and mPer2 are light-inducible in clock neurons of the hypothalamic suprachiasmatic nucleus, mPer3 is not. We have studied the promoter structure of the three mPer genes and compared their regulation. All three mPer promoters contain E-boxes and respond to the CLOCK/brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like protein 1 (BMAL1) heterodimer. On the other hand, only mPer1 and mPer2 promoters contain bona fide cAMP-responsive elements (CREs) that bind CRE-binding protein (CREB) from suprachiasmatic nucleus protein extracts. The mPer1 promoter is responsive to synergistic activation of the cAMP and mitogen-activated protein kinase pathways, a physiological response that requires integrity of the CRE. In contrast, activation of mPer promoters by CLOCK/BMAL1 occurs regardless of an intact CRE. Altogether, these results constitute strong evidence that CREB acts as a pivotal endpoint of signaling pathways for the regulation of mPer genes. Our results reveal that signaling-dependent activation of mPer genes is distinct from the CLOCK/BMAL1-driven transcription required within the clock feedback loop.
Circadian rhythmicity is a conserved physiological feature of almost all organisms (1–3). Light is the most prominent stimulus that has contributed in shaping circadian physiology during evolution (4, 5). Through several photoreception systems, light is capable of synchronizing circadian oscillations to the environment (4, 6). In mammals the core pacemaker is located in the suprachiasmatic nucleus (SCN) of the hypothalamus, whose neurons receive photic input signals from the retina by way of the retinohypothalamic tract (7).
Although several nonphotic stimuli have also been shown to reset the mammalian circadian system (8–13), light is the major entraining signal and it delays the pacemaker if administered at early night and advances it at late night (6). The effect is intimately connected to the clock mechanism because light has no effect when applied during the subjective day. The process of synchronization involves the transcriptional activation of several genes. In mice, brief exposure to light during the subjective night causes rapid induction of immediate-early genes, such as c-fos (14), and of clock genes, such as the homologs of the Drosophila period gene (15–17). Three period genes exist in the mouse, and although mPer1 is induced by a light pulse within 15–30 min and mPer2 within 2 h (15–17), the mPer3 gene is not light-responsive (18, 19). Arousal (11) and serotonin receptor activation (20) induce acute down-regulation of mPer1 and mPer2 expression in the SCN, identifying them as common targets for both photic and nonphotic cues.
Whereas mPer1 and mPer2 seem to play a crucial role in the molecular organization of the pacemaker (21–25), mPer3 seem to operate on clock output pathways (26). Per genes are known to be positively regulated by other clock proteins belonging to the basic helix-loop-helix–period/aryl hydrocarbon receptor nuclear translocator/single-minded (PAS) class. These are CLOCK and brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like protein 1 (BMAL1) which, associated as heterodimers, bind to E-box enhancer elements (27–29). In addition, mPER proteins constitute multimeric complexes with the products of the Cryptochrome genes, mCRY1 and mCRY2, which in turn inhibit transcription mediated by CLOCK/BMAL1 (30, 31). The mPer genes exhibit circadian cycling expression in the SCN (15, 19) and in several peripheral tissues, e.g., liver and skeletal muscle (19, 32), and in cultured cell lines stimulated with a number of stimuli (33–37).
Several lines of evidence indicate that the mPer1 gene plays a central role in conveying the light-entraining information to the central clock. mPer1 is the only clock gene that has been convincingly shown to be induced very rapidly after light stimulation (15–17, 38). In addition, light-induced resetting of locomotor activity and glutamate-induced resetting of firing rhythms can be blocked by mPer1 antisense oligonucleotides (39). Finally, some reports on mPer1-deficient mice confirm this view (40).
Although resetting of the circadian system seems to involve changes in gene expression, little is known about the signal transduction pathways that initiate this transcriptional response. Signaling pathways for light-dependent clock resetting include glutamate release from retinohypothalamic tract terminations within the SCN, N-methyl-D-aspartate receptor activation, and Ca2+ influx (41, 42). Calcium influx may be linked to cAMP-responsive element (CRE)-mediated transcription activated by the extracellular signal-regulated kinase (ERK)-mitogen-activated protein kinase (MAPK) pathway (43). Indeed, a light pulse during the night activates the MAPK signaling cascade (44) and consequently induces CRE-binding protein (CREB) phosphorylation (45) in SCN neurons.
We have studied the regulation of mPer promoters by signaling stimuli. Our analysis has identified significant differences and similarities among the three promoters. We demonstrate that CREB acts as a major effector of converging signaling pathways to the mPer1 promoter and that this regulation is independent of CLOCK/BMAL1 action.
Materials and Methods
Plasmids.
The mPer1, mPer2, and mPer3 promoter regions were isolated and cloned in pGL3-Basic Vector (Promega). The mPer1 region spans from −1803 to +40 (+1 is the putative transcription start site), and the sequence is identical with that in GenBank accession no. AB030818 (46). The mPer2 and mPer3 regions are from −1670 to +53 and from −1594 to +128, respectively, and were deposited in GenBank (accession nos. AF491941 and AF491942). Mutation in the CREs within mPer1 and mPer2 promoters was generated by deletion of the central 4 nt (TGACGTCA → TGCA). Oligonucleotides corresponding to CRE-mPer1 (5′-tccgcttTGACGTCAcctccct-3′), CRE-mPer2 (5′-ccaccatTGACGTCAatgtaag-3′), or their mutated forms (5′-tccgctcTCACAAAAcctccct-3′ and 5′-ccaccgcTGACAAAatgtaag-3′) were inserted in the pGL3-Promoter Vector (Promega). Mouse Clock ORF was amplified by PCR from a cDNA kindly provided by J. Takahashi (Northwestern University, Chicago), cloned in the pSG5 vector (Stratagene) with a FLAG epitope sequences at the 5′ end. Mouse Bmal1 was amplified by reverse transcription–PCR from mouse brain RNA. This cDNA was cloned in pCS2+MTK, a derivative of pCS2 with five Myc-tag sequences at the 5′ end of the cDNA.
Cell Culture and Transient Transfections.
Human choriocarcinoma JEG3 cells were cultured in monolayers with Dulbecco's modified Eagle's medium (GIBCO/BRL) supplemented with 10% FCS. Cells were transfected by the calcium phosphate coprecipitation technique with 1 μg of reporter construct. Medium was replaced with 0.5% FCS medium 12–16 h before cells were treated with various agents: forskolin (10 μM), epidermal growth factor (EGF; 50 ng/ml), 4-bromo-calcium ionophore A23187 (1 μM), phorbol 12-tetradecanoate 13-acetate (TPA) (100 ng/ml), and/or 20% FCS for 6 h before harvest. In specific experiments inhibitors PD 98059 (30 μM; Calbiochem) and SB 203580 (10 μM; Calbiochem) were added to cells 1 h before treatments. Luciferase activity was measured as described (47). All experiments were performed in triplicate.
Nuclear Extract Preparation and DNA-Binding Assays.
Adult male Wistar rats were kept on a 12-h light:12-h dark cycle for 3 weeks before dissection. SCNs and underlying optic chiasms were isolated under the dissecting microscope. Blocks ≈1 × 1 × 1 mm were rapidly frozen in liquid nitrogen and stored at −80°C. SCN (48) and Rat-1 fibroblasts (49) nuclear extracts were prepared as described. Western analyses and immunostaining were as described (50) with anti-phospho-CREB or anti-CREB antibodies (New England Biolabs). For the gel retardation assays, CRE oligonucleotide probes or their mutated forms (same sequences as above) were used as described (51). In competition or in antibody-supershifting assays, unlabeled oligonucleotides or anti-CREB, anti-ATF1, anti-ATF2 antibodies (Santa Cruz Biotechnology) were added to the extracts 30 min before the labeled oligonucleotides. Recombinant CREB was produced as described (52).
Results
Similarities and Differences Between the mPer Promoters.
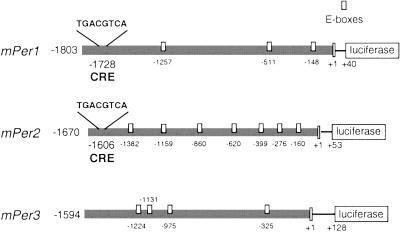
We have analyzed the sequence of the 5′ flanking regulatory regions of the three mouse Per genes. Three CACGTG E-boxes and one canonical CRE (TGACGTCA) are within 1,803 bp of the mPer1 gene promoter, centered at 1,728 bp upstream of the transcription start site (Fig. 1). Within the 1,670-bp promoter region of mPer2 and 1,594 bp of mPer3 genes, seven and four E-boxes (CANNTG) were found, respectively (Fig. 1). None of these E-boxes corresponds to the CACGTGA sequence, the strict consensus binding site for CLOCK/BMAL1 heterodimers (28). A canonical CRE is also present within mPer2 promoter, but not in the mPer3 promoter (Fig. 1). The CRE has a common location within both mPer1 and mPer2 promoters, being consistently upstream from the E-boxes. This reciprocal location of CREs and E-boxes is characteristic also in promoters of other genes, such as renin and transforming growth factor β2 (53, 54).
Figure 1.
Schematic representation of mPer1, mPer2, and mPer3 promoters. The upstream sequence of mPer1, mPer2, and mPer3 genes was fused to a luciferase reporter. On the left is indicated the size of the genomic fragment upstream of the transcription start site (+1) that is included in the construction. The numbers below the box representing the promoter sequence are the positions of the CRE (TGACGTCA) and E-boxes (small white boxes).
mPer1 and mPer2 CREs Bind CREB in SCN Nuclear Extract.
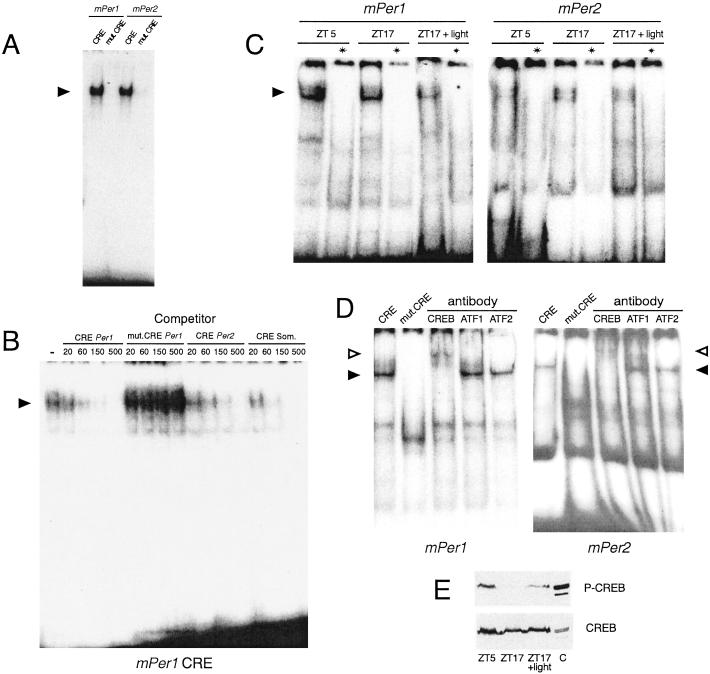
The CREs present in the promoters of the mPer1 and mPer2 genes efficiently bind recombinant CREB protein, whereas their mutated forms do not (Fig. 2A). A nuclear extract from Rat-1 fibroblasts was also used to assess binding to the mPer1 CRE (Fig. 2B). Specificity of binding was confirmed by competition with unlabeled CRE oligonucleotides. The canonical somatostatin CRE (55) and the mPer1 and mPer2 CREs successfully competed for binding, whereas a mutated mPer1 CRE was not an effective competitor. Analogous results were obtained with the mPer2 CRE (not shown). We also performed binding assays using nuclear extracts from rat SCN collected at Zeitgeber time (ZT) 5 or ZT17, or 1 h after a 30-min light pulse at ZT17 (Fig. 2C). SCN extracts display a robust CRE-binding activity. Complex formation for both mPer1 and mPer2 CREs is constant independent of circadian phase or photic stimulation. Binding specificity was assessed by competition with the somatostatin CRE. To identify the nature of the natural mPer CRE-binding activity supershift assays were performed with the SCN extracts. Whereas preincubation with anti-CREB antibodies drastically decreased complex formation on both mPer1 or mPer2 CREs, neither anti-ATF1 nor anti-ATF2 antibodies had any effect (Fig. 2D). These results show that CREB naturally binds the mPer promoter sequences in the SCN.
Figure 2.
Specific binding of CREB to wild-type CRE of mPer1 and mPer2 promoters. (A) Gel mobility-shift assay using wild-type or mutated mPer1 or mPer2 CRE oligonucleotides, plus bacterially expressed CREB protein. The specific complex is indicated by an arrowhead. (B) Competition gel mobility-shift assay using labeled mPer1 CRE oligonucleotide together with nuclear extract from Rat-1 fibroblasts. Competition is made by preincubating the labeled DNA with increasing amounts (20–500 ng) of unlabeled wild-type mPer1, mPer2, or somatostatin CRE or mutated mPer1 oligonucleotides. The specific complex is indicated by an arrowhead. (C) Gel mobility-shift assay using mPer1 or mPer2 CRE oligonucleotides along with nuclear extracts from the SCN of rats killed during the day (ZT5), and during the night (ZT17) either in the darkness or 1 h after the beginning of a 30-min light pulse. (D) Supershift assay on SCN nuclear extracts with mPer1 or mPer2 CRE after preincubation of SCN nuclear extract with anti-CREB, anti-ATF1, and anti-ATF2 antibodies. The specific complex is indicated by a closed arrowhead and the supershifted by an open arrowhead. The first lane is the control without antibody and the second lane is a complex with mutated oligonucleotide. (E) Immunoblotting of SCN protein extracts from rats killed as in C, with an anti-phosphorylated CREB (P-CREB) antibody. “C” is a control containing P-CREB (EGF-stimulated fibroblasts).
A light stimulus that phase-shifts circadian rhythms has been shown to induce phosphorylation of CREB at Ser-133, a critical event in transcriptional activation (45, 56). To link this notion to the activation of mPer genes, we analyzed CREB phosphorylation in our SCN nuclear extracts. We used antibodies raised against CREB phosphorylated at Ser-133 (45). CREB is phosphorylated at ZT5, but not at ZT17, and is induced by a light pulse at ZT17 (Fig. 2E). These results implicate CREB as a signaling-responsive switch for mPer1 transcriptional response in the SCN.
Distinct Effects of CLOCK/BMAL1 and Signaling Pathways on mPer Promoter Activation.
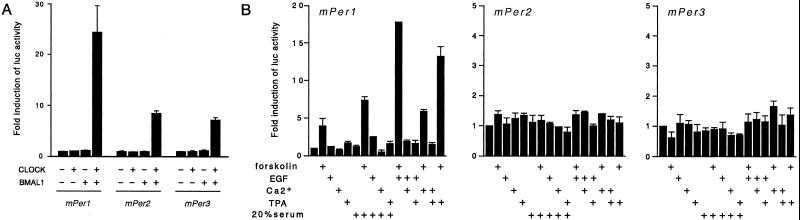
Cotransfection of JEG-3 cells with mPer1, mPer2, and mPer3 promoter reporters along with CLOCK and BMAL1 expression vectors resulted in transcriptional stimulation of all three mPer promoters (Fig. 3A), as reported for mPer1 (27). CLOCK/BMAL1-dependent activation of mPer1 is significantly higher than mPer2 and mPer3.
Figure 3.
Activation of mPer1, mPer2, and mPer3 promoters by CLOCK/BMAL1 and by the induction of different signaling pathways. (A) JEG-3 cells were transfected with a construct of the mPer promoter regions linked to the luciferase reporter gene and either CLOCK or BMAL1 alone or together. Cells were washed after 12 h and processed for luciferase assay 24 h later. (B) For stimulation, cells transfected with either of the mPer promoter constructs were treated with forskolin (10 μM), EGF (50 ng/ml), 4-bromo-calcium ionophore A23187 (1 μM), TPA (100 ng/ml), 20% fetal calf serum, or combinations. Six hours later, cells were harvested for luciferase assay. Data are expressed as fold increase over the value for unstimulated cells. Each bar represents the mean and SEM of three independent samples. The figures are representative from several independent experiments with similar results.
To explore the ability of mPer promoters to respond to stimulation of different signaling pathways, we treated transfected cells with adenylate cyclase activator forskolin, EGF, 4-bromo-calcium ionophore A23187, TPA, and 20% FCS. Only forskolin caused significant elevation of mPer1 promoter activity (Fig. 3B). However, remarkable up-regulation was observed when cells were concurrently treated with forskolin and EGF or TPA. These results indicate that maximal induction of mPer1 promoter activity requires a cooperative activation of both cAMP and MAPK pathways. No response was observed for either mPer2 or mPer3 after treatments (Fig. 3B), which is noteworthy considering the similarity in the organization of mPer1 and mPer2 promoters (Fig. 1) and the capacity of the mPer2 CRE to bind CREB (Fig. 2).
Essential Role of the mPer1 CRE in the Response to Signaling Inducers.
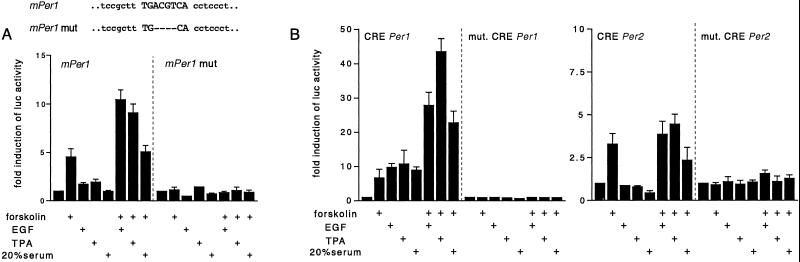
To assess the role played by the CRE in the remarkable responsiveness of the mPer1 promoter, we generated a deletion of the four internal nucleotides of the CRE within the context of the whole mPer1 promoter (Fig. 4A). This mutation of the CRE abolishes CREB binding (not shown) and fully blocks responsiveness of the mPer1 promoter (Fig. 4A). This result underscores the importance of the CRE in signaling response and demonstrates that the rest of the mPer1 promoter, including the E-boxes, is not sufficient to elicit inducibility.
Figure 4.
Mutation within CRE abolishes the response of the promoter to cell-signaling stimulation. JEG-3 cells were transfected with a luciferase construct with either wild-type or CRE-mutated mPer1 promoter (A), or with isolated wild-type or mutated CRE motifs of mPer1 and mPer2 promoters (B). Stimulation and luciferase assays were done as in Fig. 3.
The functional importance of the CRE was further verified by using the isolated mPer1 and mPer2 CRE sequences inserted in heterologous promoter reporters. The mPer1 CRE confers remarkable responsiveness on stimulation with forskolin, EGF, TPA, or serum, and synergistic activation after combined treatments, analogous to what was observed with the full mPer1 promoter. All responses were abolished when the CRE was mutated (Fig. 4B). Thus, the CRE alone is capable of conveying MAPK- and cAMP-inducible activation and could thereby constitute a link between light-stimulated MAPK pathway and light induction of the mPer1 gene. When isolated, the CRE of mPer2 promoter can also serve as a target site of activated signaling pathways, especially cAMP (Fig. 3B). This observation suggests that additional sequences within the mPer2 promoter may modulate the responsiveness of the CRE.
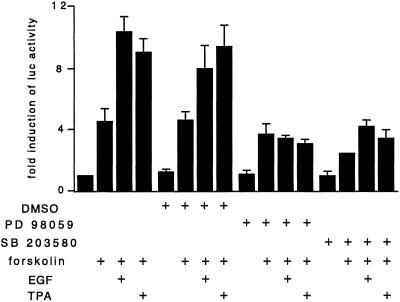
MAPK Cascade Inhibitors Block mPer1 EGF and TPA Inducibility.
Phosphorylation of CREB in the SCN in response to a light stimulus has been proposed to depend on the MAPK pathway (43). To examine the specificity of mPer1 stimulation by EGF and TPA, we treated cells transfected with the full-length mPer1 promoter with the specific MEK inhibitor PD 98059 1 h before stimulation (Fig. 5). Inhibition of MEKs blocks the synergistic activation of the mPer1 promoter by concurrent application of EGF or TPA and forskolin. However, forskolin-dependent activation is not affected, stressing the independence from MAPKs of the cAMP-inducible activation. These results suggest the convergence of multiple signaling routes whose effects are integrated at the level of the mPer1 promoter. In support of this interpretation, SB 203580, a specific inhibitor of the p38 MAPK cascade, also significantly inhibits EGF- and TPA-mediated activation of mPer1. As for the inhibitor PD 98059, cAMP inducibility of mPer1 is not affected by SB 203580.
Figure 5.
Inhibitors of MAPK cascades abolish synergistic activation of the mPer1 promoter by forskolin and EGF or TPA. Transfection, treatments, and luciferase analysis were conducted as in Fig. 3B, except that PD 98059 (30 μM) or SB 203580 (10 μM) or the vehicle was applied to the cells 1 h before the inducing treatments.
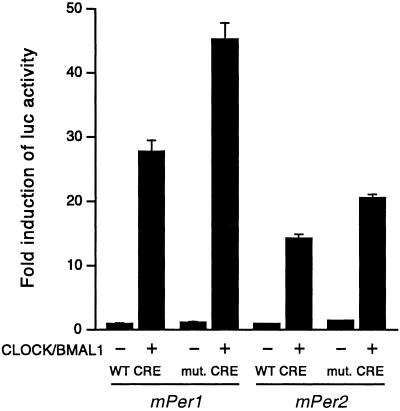
Activation by CLOCK/BMAL1 Is Independent of the CREs.
We wished to establish whether signaling-dependent induction through the CRE would influence the clock-controlled CLOCK/BMAL1 activation. We used the full-length mPer1 and mPer2 promoters containing either an intact or a mutated CRE in cotransfection experiments with CLOCK and BMAL1 expression vectors. The results clearly show that CLOCK/BMAL1 activation does not require the CRE (Fig. 6). This finding indicates that specific, independent sequences within the mPer1 promoter integrate signaling stimuli and clock-dependent regulation.
Figure 6.
Activation of mPer1 or mPer2 promoters by CLOCK/BMAL1 is unaffected by a CRE mutation. Cells were transfected with the wild-type or CRE-mutated mPer1 or mPer2 promoter constructs, and with CLOCK and BMAL1 expression plasmid or the empty vectors, and processed as in Fig. 3A.
Discussion
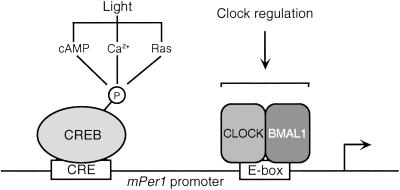
The pathways that light uses to impinge on the clock molecular mechanism are still poorly understood. We reasoned that studying how the expression of clock genes is regulated by intracellular signaling systems would provide useful information in this respect. The mPer genes have been shown to respond differentially to light in the SCN (15, 16, 18, 19, 38). mPer1 is induced quickly, with kinetics of an immediate-early gene; mPer2 responds more slowly, reaching peak values 2–3 h after the light stimulus; mPer3 is not light-responsive. Our results on the inducibility of the mPer promoters by various signaling agents establish an interesting parallel with light responsiveness. Whereas mPer1 and mPer2 contain a CRE, mPer3 does not (Fig. 1). The absence of a CRE in the mPer3 promoter is likely to be linked to its uninducibility by activation of the cAMP and MAPK pathways. Although the mPer2 regulatory region contains a canonical and functional CRE, the promoter is poorly responsive, in sharp contrast to mPer1 (Fig. 3). It seems that the mPer2 CRE is functional, but that in the context of the whole promoter it becomes inactivated. This different activation potential of mPer1 and mPer2 CREs could account for the diverse induction kinetics of the two genes in the SCN of light-stimulated animals. One possibility is that newly synthesized factors are needed for the induction of mPer2 in the SCN, which would explain its delayed kinetics of induction. These de novo synthesized proteins may be lacking in our cell-based system. Another aspect to take into account is the likely interactions that CREB may have with additional factors regulating the mPer promoters, including E-box-binding proteins, because these interactions may modulate responsiveness of the promoter. In this respect the combined presence of CREs and E-boxes in several promoters is noteworthy (53, 54). The CRE is often placed upstream from the E-boxes, suggesting a code of possible protein–protein interactions. Yet direct interactions of CREB with E-box-binding proteins have not been described. We have shown that CLOCK/BMAL1-dependent activation is independent of the CRE (Fig. 7).
Figure 7.
Schematic representation of the mPer1 promoter and its regulation. While the E- boxes (only one is shown here) are targets of the clock-controlled CLOCK/BMAL1 regulation, the CRE is essential for response to various signaling pathways.
The involvement of CREB in light-induced clock resetting is based on the observation that CREB becomes phosphorylated in vivo in response to photic stimuli (45), and in vitro after glutamate treatment (57). Furthermore, light exposure at night, when it causes phase shifts of overt rhythms, elicited a robust increase in CRE-mediated transcription in the retinorecipient part of the SCN (43). Here we have established a direct link between intracellular signaling, CRE-regulated transcription, and mPer gene expression. It is important to explore the role played by the CREB coactivator CBP (CREB-binding protein) in SCN neurons, because it has been shown that CBP itself is a target of signaling regulation (56).
CREB activation by phosphorylation at Ser-133 occurs in response to several signaling stimuli, including the ERK-MAPK pathway (58, 59). In the SCN, stimulation of these kinases occurs after a light pulse that would shift the clock (44) and contributes to activation of CREB by glutamate (43). A variety of treatments trigger expression of endogenous Per1 and other clock and clock-controlled genes in mammalian cell lines (33–37, 60). Our results identify the cAMP and MAPK pathways, and their synergistic combination, as essential for CRE-mediated induction of mPer1. Future studies will need to focus on the important issue of how these signaling pathways are modulated within SCN neurons. Phosphorylation and dephosphorylation of specific nuclear substrates, including CREB and histone H3 tail (45, 50, 56), are essential in the fine tuning of transcription regulation.
This study provides further support to the hypothesis that light-induced resetting of the clock proceeds by activation of Per genes, namely mPer1 (15, 16, 38, 39). It is still unclear how the same mPer1 activation gives rise to different responses depending on the time in the circadian cycle (16, 50, 61). The changing phase relationships of clock genes and relative protein abundance likely play an important role in this respect (62). The possibility that synergistic action of signaling routes, as well as their known cross-talks, may occur at restricted temporal windows will need to be taken into account to decipher the mechanisms of clock physiology.
Acknowledgments
We thank Estelle Heitz for her expert technical assistance, Mark Zylka and Xiaowei Jin for the cloning of Per promoter regions, Joseph Takahashi for the kind gift of the Clock cDNA, and all of the members of the Sassone-Corsi laboratory for help, reagents, and discussions. Z.T.-B. was supported by a European Molecular Biology Organization long-term fellowship. N.C. was supported by a Human Frontier Science Program long-term fellowship and a Canadian Institutes of Health Research postdoctoral fellowship. Work in our laboratory is supported by grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Régional Universitaire, Human Frontier Science Program, Organon Akzo/Nobel, Fondation pour la Recherche Médicale, and Association pour la Recherche sur le Cancer.
Abbreviations
- SCN
suprachiasmatic nucleus
- ARNT
aryl hydrocarbon receptor nuclear translocator
- BMAL1
brain and muscle ARNT-like protein 1
- ZT
Zeitgeber time
- ERK
extracellular signal-regulated kinase
- MAPK
mitogen-activated protein kinase
- CRE
cAMP-responsive element
- CREB
CRE-binding protein
- EGF
epidermal growth factor
- TPA
phorbol 12-tetradecanoate 13-acetate
Footnotes
References
- 1.Cermakian N, Sassone-Corsi P. Nat Rev Mol Cell Biol. 2000;1:59–67. doi: 10.1038/35036078. [DOI] [PubMed] [Google Scholar]
- 2.Dunlap J C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 3.Young M W, Kay S A. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 4.Foster R G. Neuron. 1998;20:829–832. doi: 10.1016/s0896-6273(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 5.Menaker M, Moreira L F, Tosini G. Braz J Med Biol Res. 1997;30:305–313. doi: 10.1590/s0100-879x1997000300003. [DOI] [PubMed] [Google Scholar]
- 6.Daan S, Pittendrigh C S. J Comp Physiol. 1976;106:253–266. [Google Scholar]
- 7.Moore R Y, Lenn N Y. J Comp Neurol. 1972;146:1–14. doi: 10.1002/cne.901460102. [DOI] [PubMed] [Google Scholar]
- 8.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humlova M, Illnerova H. Neuroendocrinology. 1990;52:196–199. doi: 10.1159/000125573. [DOI] [PubMed] [Google Scholar]
- 10.Krieger D T, Hauser H, Krey L C. Science. 1977;197:398–399. doi: 10.1126/science.877566. [DOI] [PubMed] [Google Scholar]
- 11.Maywood E S, Mrosovsky N, Field M D, Hastings M H. Proc Natl Acad Sci USA. 1999;96:15211–15216. doi: 10.1073/pnas.96.26.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mrosovsky N. Biol Rev Camb Philos Soc. 1996;71:343–372. doi: 10.1111/j.1469-185x.1996.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 13.Stokkan K A, Yamazaki S, Tei H, Sakaki Y, Menaker M. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 14.Kornhauser J M, Nelson D E, Mayo K E, Takahashi J S. Neuron. 1990;5:127–134. doi: 10.1016/0896-6273(90)90303-w. [DOI] [PubMed] [Google Scholar]
- 15.Albrecht U, Sun Z S, Eichele G, Lee C C. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 16.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F, Jr, Reppert S M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 17.Wilsbacher L D, Yamazaki S, Herzog E D, Song E J, Radcliffe L A, Abe M, Block G, Spitznagel E, Menaker M, Takahashi J S. Proc Natl Acad Sci USA. 2002;99:489–494. doi: 10.1073/pnas.012248599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, et al. EMBO J. 1998;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zylka M J, Shearman L P, Weaver D R, Reppert S M. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 20.Horikawa K, Yokota S, Fuji K, Akiyama M, Moriya T, Okamura H, Shibata S. J Neurosci. 2000;20:5867–5873. doi: 10.1523/JNEUROSCI.20-15-05867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bae K, Jin X, Maywood E S, Hastings M H, Reppert S M, Weaver D R. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 22.Cermakian N, Monaco L, Pando M P, Dierich A, Sassone-Corsi P. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shearman L P, Sriram S, Weaver D R, Maywood E S, Chaves I, Zheng B, Kume K, Lee C C, van der Horst G T, Hastings M H, Reppert S M. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 24.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun Z S, Eichele G, Bradley A, Lee C C. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 25.Zheng B, Larkin D W, Albrecht U, Sun Z S, Sage M, Eichele G, Lee C C, Bradley A. Nature (London) 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 26.Shearman L P, Jin X, Lee C, Reppert S M, Weaver D R. Mol Cell Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 28.Hogenesch J B, Gu Y Z, Jain S, Bradfield C A. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi S, Mitsui S, Miyake S, Yan L, Onishi H, Yagita K, Suzuki M, Shibata S, Kobayashi M, Okamura H. Curr Biol. 2000;10:873–876. doi: 10.1016/s0960-9822(00)00602-3. [DOI] [PubMed] [Google Scholar]
- 30.Griffin E A, Jr, Staknis D, Weitz C J. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 31.Kume K, Zylka M J, Sriram S, Shearman L P, Weaver D R, Jin X, Maywood E S, Hastings M H, Reppert S M. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 32.Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Biochem Biophys Res Commun. 1998;253:199–203. doi: 10.1006/bbrc.1998.9779. [DOI] [PubMed] [Google Scholar]
- 33.Akashi M, Nishida E. Genes Dev. 2000;14:645–649. [PMC free article] [PubMed] [Google Scholar]
- 34.Balsalobre A, Damiola F, Schibler U. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 35.Balsalobre A, Marcacci L, Schibler U. Curr Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 36.Yagita K, Okamura H. FEBS Lett. 2000;465:79–82. doi: 10.1016/s0014-5793(99)01724-x. [DOI] [PubMed] [Google Scholar]
- 37.Yagita K, Tamanini F, van Der Horst G T, Okamura H. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 38.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros J J, Dunlap J C, Okamura H. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 39.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albrecht U, Zheng B, Larkin D, Sun Z S, Lee C C. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 41.Colwell C S, Foster R G, Menaker M. Brain Res. 1991;554:105–110. doi: 10.1016/0006-8993(91)90177-w. [DOI] [PubMed] [Google Scholar]
- 42.Ding J M, Chen D, Weber E T, Faiman L E, Rea M A, Gillette M U. Science. 1994;266:1713–1717. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- 43.Obrietan K, Impey S, Smith D, Athos J, Storm D R. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- 44.Obrietan K, Impey S, Storm D R. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- 45.Ginty D D, Kornhauser J M, Thompson M A, Bading H, Mayo K E, Takahashi J S, Greenberg M E. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 46.Hida A, Koike N, Hirose M, Hattori M, Sakaki Y, Tei H. Genomics. 2000;65:224–233. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 47.Drust D S, Troccoli N M, Jameson J L. Mol Endocrinol. 1991;5:1541–1551. doi: 10.1210/mend-5-10-1541. [DOI] [PubMed] [Google Scholar]
- 48.Howard T, Balogh R, Overbeek P, Bernstein K E. Mol Cell Biol. 1993;13:18–27. doi: 10.1128/mcb.13.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crosio C, Cermakian N, Allis C D, Sassone-Corsi P. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- 51.Foulkes N S, Borrelli E, Sassone-Corsi P. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 52.Delmas V, Laoide B M, Masquilier D, de Groot R P, Foulkes N S, Sassone-Corsi P. Proc Natl Acad Sci USA. 1992;89:4226–4230. doi: 10.1073/pnas.89.10.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan L, Black T A, Shi Q, Jones C A, Petrovic N, Loudon J, Kane C, Sigmund C D, Gross K W. J Biol Chem. 2001;276:45530–45538. doi: 10.1074/jbc.M103010200. [DOI] [PubMed] [Google Scholar]
- 54.Scholtz B, Lamb K, Rosfjord E, Kingsley M, Rizzino A. Dev Biol. 1996;173:420–427. doi: 10.1006/dbio.1996.0037. [DOI] [PubMed] [Google Scholar]
- 55.Montminy M R, Bilezikjian L M. Nature (London) 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 56.De Cesare D, Fimia G M, Sassone-Corsi P. Trends Biochem Sci. 1999;24:281–285. doi: 10.1016/s0968-0004(99)01414-0. [DOI] [PubMed] [Google Scholar]
- 57.von Gall C, Duffield G E, Hastings M H, Kopp M D, Dehghani F, Korf H W, Stehle J H. J Neurosci. 1998;18:10389–10397. doi: 10.1523/JNEUROSCI.18-24-10389.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Impey S, Obrietan K, Wong S T, Poser S, Yano S, Wayman G, Deloulme J C, Chan G, Storm D R. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 59.Riccio A, Ahn S, Davenport C M, Blendy J A, Ginty D D. Science. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 60.Motzkus D, Maronde E, Grunenberg U, Lee C C, Forssmann W, Albrecht U. FEBS Lett. 2000;486:315–319. doi: 10.1016/s0014-5793(00)02315-2. [DOI] [PubMed] [Google Scholar]
- 61.Miyake S, Sumi Y, Yan L, Takekida S, Fukuyama T, Ishida Y, Yamaguchi S, Yagita K, Okamura H. Neurosci Lett. 2000;294:41–44. doi: 10.1016/s0304-3940(00)01545-7. [DOI] [PubMed] [Google Scholar]
- 62.Field M D, Maywood E S, O'Brien J A, Weaver D R, Reppert S M, Hastings M H. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]