Summary
Macrophages play a crucial role in mediating antibody-dependent cellular phagocytosis (ADCP), a process that enhances the effectiveness of several cancer immunotherapies. In this protocol, we outline detailed steps for isolating, differentiating, and polarizing macrophages from both mouse and human sources. Additionally, we describe an optimized technique to assess ADCP using in vitro co-culture studies followed by flow cytometry analysis. This protocol offers a reliable approach to evaluate the efficiency of macrophage-mediated phagocytosis of cancer cells during treatment with antibody-based therapies.
For complete details on the use and execution of this protocol, please refer to Osorio et al.1
Subject areas: Flow Cytometry, Cancer, Immunology, Antibody
Graphical abstract

Highlights
-
•
Isolation, differentiation, and polarization of mouse BMDMs and human MDMs in vitro
-
•
Phagocytosis of live tumor cells by antibody-treated primary mouse or human macrophages
-
•
Flow cytometry analysis of antibody-dependent cellular phagocytosis of cancer cells
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Macrophages play a crucial role in mediating antibody-dependent cellular phagocytosis (ADCP), a process that enhances the effectiveness of several cancer immunotherapies. In this protocol, we outline detailed steps for isolating, differentiating, and polarizing macrophages from both mouse and human sources. Additionally, we describe an optimized technique to assess ADCP using in vitro co-culture studies followed by flow cytometry analysis. This protocol offers a reliable approach to evaluate the efficiency of macrophage-mediated phagocytosis of cancer cells during treatment with antibody-based therapies.
Before you begin
The protocol outlines the procedures for the following steps: 1) Isolation, differentiation, and polarization of bone marrow-derived macrophages (BMDMs) from C57BL/6 mice and monocyte-derived macrophages (MDMs) from human peripheral blood. 2) Co-culture experiments of primary macrophages with either mouse or human cancer cells, during treatment with different antibody therapies. 3) Detection of macrophage phagocytosis of live tumor cells by flow cytometry.
Institutional permissions
Animal studies must be approved by and performed in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines. The ethics statement must be approved by the ethics committee reviewing research involving human cells in the institute. The blood of healthy donors is collected with their consent (e.g., by signing a consent form).
Preparation of materials
Timing: 1 week before ADCP assay
-
1.Mouse: Bone marrow-derived macrophages (BMDMs)
-
a.Prepare materials as buffers for mouse protocol as indicated under “materials and equipment”.
-
b.Determine the number of mice required for biological replicates.
-
c.Prepare and print a schematic outline of the 96-well plate indicating treatment conditions as well as technical and biological replicates (Figure 1).
-
d.Prepare and autoclave all surgical tools.
-
e.Prepare a dissection board, a 6-well plate, and collection tubes.
-
i.Fill wells of the 6-well plate with cold and sterile PBS (1X, Gibco). Keep on ice.
-
i.
-
f.Pierce the bottom of the 1.5 mL tube (Eppendorf) with a syringe needle (26G) and place it inside a 1.7 mL microcentrifuge tube (Thomas Scientific). Prepare two sets of tubes per mouse (Figures 2A–2C).
-
a.
-
2.Human: monocyte-derived macrophages (MDMs).
-
a.Prepare materials as buffers for human protocol as indicated under “materials and equipment”.
-
b.Determine the number of donors/participants required for biological replicates.
-
c.Prepare and print a schematic outline of the 96-well plate indicating treatment conditions as well as technical and biological replicates (Figure 1).
-
d.Prepare four 50 mL falcon tubes per donor (two for blood, two for density gradient medium [i.e., lymphoprep or Ficoll]).
-
e.Bring lymphoprep or preferred density gradient medium to a sterile hood.
-
f.Determine the number of human CD14 MicroBeads (Miltenyi Biotec) required per donor.
-
a.
Figure 1.
Schematic outline of treatment conditions
Scheme of cancer cells and macrophages with different antibody treatment conditions in a 96-well plate. Wells containing unstained controls, live/dead controls, and stained cells are included.
Figure 2.
Isolation of bone marrow-derived macrophages
(A–C) Pierce the bottom of a 1.5 mL microtube and place it inside a 1.7 mL Eppendorf tube.
(D) Collect tibias and femurs, remove tissue, and place in cold PBS.
(E and F) Cut the ends of the bones to expose bone marrow.
(G) Place two cut bones inside the pierced microtube. Spin down at 600 x g for 2 min. Bone marrow flows through the bottom of the microtube and collects in the Eppendorf tube.
Macrophage isolation (mouse)
Timing: 1 week before ADCP assay
-
3.
Euthanize mice by approved institutional guidelines.
-
4.
Remove tibias and femurs by cutting the bone, and muscles just below the hip joint and above the ankles.
-
5.
Remove the skin, muscle, and tissue from bones and place them in the 6-well plate containing ice-cold PBS (Figure 2D).
-
6.
Move to a sterile environment under a laminar flow/Biosafety hood.
Note: Consider additional trimming and removal of surrounding tissues using scissors and forceps under sterile conditions.
Note: Consider washing the bones with additional cold sterile PBS to remove the remaining hanging ligaments and tissue.
-
7.
Remove bones from the PBS and place them on a paper towel to dry.
-
8.
Expose the bone marrow cavity by trimming 1 mm of bone at the upper and lower end of the femurs and tibias using a razor blade (Figure 2E).
Note: Make sure the bone marrow is visible at both ends (Figure 2F).
-
9.
Place two bones (one femur, one tibia) inside the prepared pierced 1.5 mL tube within the 1.7 mL microcentrifuge tube (Figure 2C).
-
10.
Close the lid of the 1.5 mL tube and leave the 1.7 mL tube lids open.
-
11.
Centrifuge 1.5 mL/1.7 mL microcentrifuge tubes at 600 x g for 2 min.
Note: This will allow bone marrow to come out of the bone, pass through the pierced microtube, and collect at the bottom of the microcentrifuge tube (Figure 2G).
-
12.
Dispose of the 1.5 mL tube containing bones and spin down the microcentrifuge tube containing marrow at 300 x g for 5 min.
-
13.
Resuspend pellets in 1 mL of filter-sterilized RBC lysis buffer (1X) (Millipore Sigma) and incubate for 10 min at 20°C–25°C.
-
14.
Transfer the resuspended pellet into a 15 mL falcon tube and add 10 mL of PBS.
-
15.
Manually count the cells with a hemocytometer and centrifuge at 300 x g for 5 min at 4°C.
-
16.
Resuspend pellets in complete DMEM media (See materials) at 5 x 105 cells/mL.
-
17.
Add 50 ng/mL of recombinant murine M-CSF (PeproTech).
-
18.
Proceed to differentiation and polarization protocol for mouse.
Macrophage isolation (human)
Timing: 1 week before ADCP assay
-
19.
Obtain human leukocyte units (Leucopaks) or non-leukodepleted blood per institutional guidelines.
-
20.
In a sterile hood, cut the leukocyte packet with a razor blade and pour 20 mL of blood into a 50 mL falcon tube.
Note: Use as many falcon tubes as needed per donor.
-
21.
Dilute and mix leukocytes with cold PBS in a 1:1 ratio.
-
22.
In a separate falcon tube, add 20 mL of Lymphoprep (STEMCELL Technologies).
Alternatives: Using other density gradient media such as Ficoll (Millipore Sigma) is also acceptable.
-
23.
Use a pipette controller with an adjustable flow rate to take 25 mL of the 1:1 PBS/leukocyte mix.
-
24.
Adjust the pipette controller to slow speed flow and tilt the falcon tube containing the Lymphoprep to a 45-degree angle.
-
25.
Gently layer 1:1 PBS/leukocyte mix on top of the Lymphoprep layer.
Note: To do so, point the pipette on the side of the angled falcon tube and slowly pour the 25 mL of 1:1 PBS/leukocyte mix (Figure 3A). Use as many Lymphoprep falcon tubes as needed per donor.
Figure 3.
Isolation of peripheral blood mononuclear cells
(A) Tilt the falcon tube at a 45-degree angle and slowly pipette the PBS/leukocyte mixture along the wall of the tube to gently layer on top of the density gradient media.
(B) Centrifugation separates the mixture into four layers. Black arrows indicate the buffy coat layer.
(C) Gently collect the buffy coat layer with a pipette.
CRITICAL: Avoid agitation or high-speed flow through the pipette in this step to prevent mixing the two layers.
Note: After pipetting the mix on top, there must be two layers: a lower Lymphoprep layer and a top leukocyte layer (Figure 3B, left).
-
26.
Centrifuge at 750 x g with slow acceleration (=1), no break, for 20 min at 4°C.
CRITICAL: It is important to have these settings for the centrifuge to not disrupt the layers of blood.
Note: After centrifuging, there should be four distinct layers: 1) a red layer at the bottom containing RBCs and granulocytes; 2) a pink transparent layer of displaced Lymphoprep; 3) a thin white layer containing leukocytes (also known as the buffy coat); and 4) a top yellow layer of plasma (Figure 3B, right).
-
27.
Use a 1 mL transfer pipette to collect the buffy coat layer (leukocytes) which is between the top (yellow) layer and Lymphoprep (pink, clear) layer (Figure 3C).
Note: Consider removing the top yellow layer first before collecting the buffy coat layer to improve yield.
-
28.
Transfer all the buffy coats from one donor into a single 50 mL falcon tube.
-
29.
Add 20 mL PBS and spin at 350 x g for 4 min at 4°C.
-
30.
Repeat the wash from step 29 twice and discard the supernatant.
-
31.
Resuspend the cells in 15 mL of RBC lysis buffer (1X) (Millipore Sigma) and incubate for 10 min at 20°C–25°C.
-
32.
Add 35 mL PBS and count the cells.
-
33.
Centrifuge at 350 x g for 4 min and discard supernatant.
-
34.
Resuspend cells in 80 μL FACS buffer (See materials) per 107 total cells. Proceed to CD14+ monocyte isolation.
-
35.
CD14+ monocyte isolation: Add 20 μL of human CD14 MicroBeads (Miltenyi Biotec) per 107 total cells.
-
36.
Mix well and incubate for 15 min at 4°C.
-
37.
Wash cells by adding 1 mL of FACS buffer per 107 cells and centrifuge at 300 x g for 10 min.
-
38.
Aspirate supernatant, then resuspend cells in 500 μL FACS buffer per 108 cells.
-
39.
Place LS columns (Miltenyi Biotec) in the magnetic field of a MACS separator (Miltenyi Biotec) (1 column per biological donor) and place a 15 mL falcon tube underneath to collect flow-through.
Note: A maximum of 2 x 109 cells can be run in 1 column.
-
40.
Rinse the LS columns with 3 mL of FACS buffer and allow it to pass through and collect in the 15 mL falcon tube.
Note: Always wait until all flow-through is collected and the column reservoir is empty before proceeding to the next step.
-
41.
Apply cell suspension onto the columns.
-
42.
Collect unlabeled cells and wash columns with 3 mL of FACS buffer.
-
43.
Wash columns with 3 mL of FACS buffer two more times.
-
44.
Remove the column from the separator and place it on a new 15 mL falcon tube.
-
45.
Pipette 5 mL of FACS buffer onto the column and immediately flush by firmly pushing the plunger into the column.
-
46.
Manually count the cells with a hemocytometer and spin at 350 x g for 4 min.
-
47.
Resuspend pellets in complete IMDM media (See materials) at 5 x 105 cells/mL.
-
48.
Add 50 ng/mL of recombinant human M-CSF (PeproTech).
-
49.
Proceed to differentiation and polarization protocol for human.
Macrophage differentiation and polarization (mouse)
Timing: 1 week before ADCP assay
-
50.
Transfer 20 mL of the cell suspension to a 150 × 15 mm Petri dish (See key resources table).
Note: Seeding density should be about 10 million cells per plate.
Note: If different polarization states are planned (M1 vs M2 vs Unpolarized), distribute macrophages in separate dishes per polarization condition.
-
51.
Every two days, aspirate the supernatant and replace it with fresh complete media supplemented with 50 ng/mL of murine M-CSF, starting two days after the initial bone marrow isolation (Day 0).
-
52.On Day 6 after, aspirate supernatant and replace with new complete DMEM media supplemented with polarization reagents (See key resources table):
-
a.M1 polarization: murine IFN-γ (90 ng/mL) and LPS (10 ng/mL).
-
b.M2 polarization: murine IL-4 (20 ng/mL) and murine IL-10 (50 ng/mL).Note: Further M2 subtype (M2a, M2c, etc.) differentiation requires optimization and is beyond the scope of this protocol.
-
c.For undifferentiated macrophages, add 50 ng/mL of murine M-CSF.Note: If no macrophage polarization is required, do not replace the media on Day 6. The macrophages should be ready to use on Day 7.Note: Polarization can go for 18–24 h at minimum, and 48 h at maximum.
-
a.
-
53.
Incubate with polarization media for 24 h.
-
54.
Proceed to cancer cell preparation and co-culture studies.
Macrophage differentiation and polarization (human)
Timing: 1 week before ADCP assay
-
55.
Transfer 20 mL of the cell suspension to a 150 × 15 mm Petri dish (See key resources table).
CRITICAL: Use complete IMDM media for human macrophages (See materials and equipment).
-
56.
Every two days, aspirate supernatant and replace with fresh complete media supplemented with 50 ng/mL of human M-CSF, starting two days after the initial bone marrow isolation (Day 0).
-
57.On Day 6 after, aspirate supernatant and replace with new complete IMDM media supplemented with polarization reagents (See key resources table):
-
a.M1 polarization: human IFN-γ (90 ng/mL) and LPS (10 ng/mL).
-
b.M2 polarization: human IL-4 (20 ng/mL), human IL-10 (50 ng/mL), and TGF-β1 (50 ng/mL).
-
c.For undifferentiated macrophages, add 50 ng/mL of human M-CSF.
-
a.
-
58.
Incubate with polarization media for 24 h.
Note: Throughout differentiation, macrophage morphology will change as indicated in Figure 4.
Figure 4.
Phenotypic characterization of macrophages
(A) Representative pictures of human MDMs polarized into M0, M1, and M2 macrophages on Day 8. Scale bar: 100 um.
(B) Representative flow cytometry histogram plots indicating the levels of expression of cell surface markers used to differentiate M1 from M2 macrophages. M1 macrophages express high levels of CD80 and CD86 while M2 macrophages express high levels of CD204 and CD163.
-
59.
Proceed to cancer cell preparation and co-culture studies.
Cancer cell culture and expansion
Timing: 3 days before ADCP assay
Note: The steps described are applicable to both mouse and human experiments.
-
60.
Thaw vials with indicated tumor cells per laboratory protocols (See key resources table for tumor cell lines used).
-
61.
Plate cancer cells in a T150 flask (Corning) with 25 mL of indicated complete media.
-
62.
Place in a 37°C and 5% CO2 incubator and expand cells to yield a tumor-to-macrophage ratio of 3:1.
-
63.
Once the desired number of tumor cells has grown to 80% confluency, harvest the cells to use in phagocytosis assay.
Note: Target cancer cells should be able to grow in complete RPMI media to perform co-culture studies (See materials and equipment). This protocol is compatible with adherent and non-adherent tumor cells.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brilliant Violet 421(TM) anti-mouse F4/80 (1:100) | BioLegend | Cat# 123137; RRID: AB_2563102 |
| Brilliant Violet 711(TM) anti-mouse/human CD11b (1:100) | BioLegend | Cat# 101242; RRID: AB_2563310 |
| Alexa Fluor(R) 700 anti-human CD14 (1:100) | BioLegend | Cat# 367114; RRID: AB_2566716 |
| Brilliant Violet 785(TM) anti-mouse/human CD11b (1:100) | BioLegend | Cat# 101243; RRID: AB_2561373 |
| TruStain FcX (anti-mouse CD16/32) Antibody (1:10) | BioLegend | 101320 |
| Human TruStain FcX (Fc receptor blocking solution) (1:10) | BioLegend | 422302 |
| MIAP301-mIgG2a (Fc activating anti-mCD47) | Juan Osorio, Memorial Sloan Kettering, USA | NA |
| MIAP301-mIgG1 D265A (Fc null anti-mCD47) | Juan Osorio, Memorial Sloan Kettering, USA | NA |
| 5F9-IgG1 GAALIE (Fc activating anti-hCD47) | Juan Osorio, Memorial Sloan Kettering, USA | NA |
| 5F9-IgG1-GRLR (Fc null anti-hCD47) | Juan Osorio, Memorial Sloan Kettering, USA | NA |
| Biological samples | ||
| Human lymphocytes (buffy coat) | New York Blood Center | NA |
| Chemicals, peptides, and recombinant proteins | ||
| CellTrace CFSE Cell Proliferation Kit, for flow cytometry | Invitrogen | C34554 |
| CellTrace Violet Cell Proliferation Kit, for flow cytometry | Invitrogen | C34557 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation | Invitrogen | L34966 |
| DAPI and Hoechst nucleic acid stain | Thermo Scientific | D1306 |
| CD14 MicroBeads, human | Miltenyi Biotec | 130-050-201 |
| Lymphoprep | STEMCELL Technologies | 07851 |
| Ficoll 400 | MilliporeSigma | F2637 |
| RBC lysis buffer (1X) | MilliporeSigma | R7757-100ML |
| Fetal bovine serum | MilliporeSigma | F2442 |
| DMEM (1X) | Gibco | 11995–065 |
| IMDM (1X) | Gibco | 12440–053 |
| RPMI medium 1640 (1X) | Gibco | 11875–093 |
| Penicillin Streptomycin (P/S) | Gibco | 15140–122 |
| PBS (1X) | Gibco | 14190–144 |
| 0.05% Trypsin-EDTA (1X) | Gibco | 25300–062 |
| 0.25% Trypsin-EDTA (1X) | Gibco | 25200–058 |
| Recombinant murine M-CSF | PeproTech | 315–02 |
| Recombinant murine IFN-γ | PeproTech | 315–05 |
| Lipopolysaccharides | MilliporeSigma | L61-43 |
| Recombinant murine IL-4 | PeproTech | 214–14 |
| Recombinant murine IL-10 | PeproTech | 210–10 |
| Recombinant human M-CSF | PeproTech | 300–25 |
| Recombinant human IFN-γ | PeproTech | 300–02 |
| Recombinant human IL-4 | PeproTech | 200–04 |
| Recombinant human IL-10 | PeproTech | 200–10 |
| Recombinant human TGF-β1 (HEK293 derived) | PeproTech | 100–21 |
| Experimental models: Cell lines | ||
| Mouse: MC-38 (mouse colon adenocarcinoma tumor model) | Kerafast | Cat# ENH204-FP; RRID: CVCL_B288 |
| Human: Raji (Burkitt lymphoma tumor model) | ATCC | Cat# CCL-86; RRID: CVCL_0511 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J mice, age 4–6 weeks, either gender | JAX | IMSR_JAX: 000664 |
| Software and algorithms | ||
| FlowJo software | Tree Star | https://www.flowjo.com/ |
| Other | ||
| Protein LoBind tubes 1.5 mL | Eppendorf | 022431081 |
| Posi-Click 1.7 mL microcentrifuge tubes | Thomas Scientific | 1159M35 |
| Nunclon Sphera 96-well, Nunclon Sphera-treated, U-shaped-bottom microplate | Thermo Scientific | 174925 |
| Falcon® 150 mm × 15 mm not TC-treated bacteriological Petri dish | Corning | 351058 |
| Falcon 150 cm2 Rectangular canted neck cell culture flask with vented cap | Corning | 355001 |
| Corning Small cell scraper | Corning | 3010 |
| MACS MultiStand | Miltenyi Biotec | 130-042-303 |
| MidiMACS Separator | Miltenyi Biotec | 130-042-302 |
| LS columns | Miltenyi Biotec | 130-042-401 |
Materials and equipment
Complete DMEM media
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | N/A | 445 mL |
| FBS | 10% | 50 mL |
| P/S | 1% | 5 mL |
| Total | N/A | 500 mL |
Note: Store at 4°C for up to 1 month.
Complete IMDM media
| Reagent | Final concentration | Amount |
|---|---|---|
| IMDM | N/A | 445 mL |
| FBS | 10% | 50 mL |
| P/S | 1% | 5 mL |
| Total | N/A | 500 mL |
Note: Store at 4°C for up to 1 month.
Complete RPMI media
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI | N/A | 445 mL |
| FBS | 10% | 50 mL |
| P/S | 1% | 5 mL |
| Total | N/A | 500 mL |
Note: Store at 4°C for up to 1 month.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1x | 98 mL |
| FBS | 2% | 2 mL |
| Total | N/A | 100 mL |
Note: Store at 4°C for up to 1 month.
Step-by-step method details
Cancer cell collection, staining, and antibody opsonization
Timing: 1.5 h
On the day of the ADCP assessment, tumor cells are washed, dissociated, and stained before co-incubating with macrophages. Therapeutic antibodies and other experimental conditions are also added before co-incubating with macrophages. Here, we will use antibodies targeting CD47, which are known to induce phagocytosis of cancer cells.2 To evaluate their capacity to induce ADCP through the antibody Fc domain, we will use antibodies that are capable of engaging Fc receptors on macrophages to induce ADCP,1 and compare them to Fc variants that lack binding to any Fc receptor and therefore incapable of inducing ADCP.
Note: The steps described are applicable to both mouse and human experiments.
-
1.
Aspirate medium from culture flask where cancer cells are grown and wash the cells with 10 mL PBS.
-
2.
Add 5 mL 0.05% trypsin-EDTA (Gibco) and incubate at 37°C for 5 min.
-
3.
Add 12 mL of warm complete media against the bottom surface of the flask until the cells have detached.
Note: Gently tapping the flask before adding media will help dislodge cells from the bottom of the flask.
-
4.
Collect and count the cells and spin at 300 x g for 5 min.
-
5.
Resuspend cancer cells in PBS at 1 x 106 cells/mL.
-
6.
Aliquot one million cells into a 15 mL falcon tube and keep on ice.
Note: This will be used for an unstained control and a live/dead control.
-
7.
Centrifuge the remaining cells at 300 x g for 5 min.
-
8.
Wash cells twice with PBS.
-
9.
Prepare the cancer cell staining according to the manufacturer’s protocol.
Note: Here, we typically use the CellTrace CFSE (Invitrogen) or CellTrace Violet stain (Invitrogen). For this experiment, we used 1:5000 Dilution of CFSE in PBS, for every 5 million cells.
Note: If fluorescent cancer cells (i.e., GFP+ or mCherry+ cancer cells) are used, skip this step and proceed to step 14.
-
10.
Resuspend cells with CellTrace staining and incubate for 10 min in the dark at 20°C–25°C.
Note: CellTrace staining varies across tumor cell lines and could be very bright. Consider performing a titration experiment to evaluate the ideal concentration for the specific cell type.
CRITICAL: Do not overstain cells with CFSE. Maximum incubation time is 15 min.
-
11.
Wash cells with FACS twice.
-
12.
Aliquot one million cells out to use as a CFSE-only control.
-
13.
Resuspend the cells in RPMI FBS-free media to a concentration of 1 x 106 cells/mL.
-
14.
Add 150 μL of resuspended cells to each well in a low-adherent 96-well microplate (150,000 cells per well) (Thermo Scientific).
-
15.
Add the antibodies, isotype control, and other experimental conditions to the specified well for opsonization as per the pre-planned outline sheet (Figure 1).
Note: For antibody dosing concentration, we usually recommend a starting concentration of 10 μg/mL. However, for experiments including dose-dependent conditions using serial dilutions is strongly recommended (See key resources table for monoclonal antibodies used).
Note: Plate treatment conditions in duplicates or triplicates for technical replicates.
CRITICAL: Make sure to designate wells for unstained controls, live/dead controls, and CFSE-only stained controls (Figure 1).
-
16.
Incubate opsonized cells for 30 min in the dark at 37°C and 5% CO2.
-
17.
While incubating, proceed to macrophage collection and staining.
Macrophage collection and co-incubation with cancer cells
Timing: 2.5 h
For ADCP assays, we recommend the ratio of cancer cells to macrophages per well be 3:1. Here, we will prepare each well with 150.000 tumor cells to be co-incubated with 50.000 macrophages.
Note: The steps described are applicable to both mouse and human experiments.
-
18.
While cancer cells are incubating, carefully aspirate media from the Petri dishes containing macrophages.
Note: Live macrophages are very adherent to the bottom of each dish.
-
19.
Add enough warm FBS-free RPMI media to cover the bottom of each dish (about 10 mL).
-
20.
Using a cell scraper (Corning), gently and unidirectionally scrape the bottom of the dish until all cells are detached.
Note: Be sure to submerge the scraper in media while passing over cells. Tilt the dish to check if cells have detached from the bottom surface.
-
21.
Using a pipette, transfer the cell suspension to a 50 mL falcon tube.
-
22.
Repeat Steps 18-20 if many cells are still attached to the bottom of the well.
Note: To detach more cells, add 5 mL 0.25% Trypsin-EDTA (Gibco) to each dish and incubate for 5 min at 37°C. Check under a microscope to see if the cells detached. Collect macrophages in separate 50 mL falcon tubes for each polarization state.
-
23.
Spin cells down at 300 x g for 5 min.
-
24.
Resuspend macrophages in RPMI FBS-free media.
-
25.
Manually count the cells and centrifuge at 300 x g for 5 min.
-
26.
Resuspend in RPMI FBS-free media at a concentration of 1 x 106 macrophages/mL.
-
27.
Add 50 μL of resuspended macrophages (50,000 macrophages) to specified wells of plated cancer cells.
Note: The ratio of cancer cells to macrophages should be 3:1 for optimal results. However, higher or lower ratios may be tested. In a 96-well plate, do not exceed 200,000 cells total per well.
-
28.
Incubate the plate in the dark at 37°C for 2 h.
Note: The 2-h incubation is based on a time-course analysis of ADCP, indicating that at 2 h we find the highest rate of phagocytosis following treatment and co-incubation (Figure 7). However, this time point may vary depending on the target cancer cell line and treatment conditions.
-
29.
Proceed to immunostaining and flow cytometry analysis.
Figure 7.
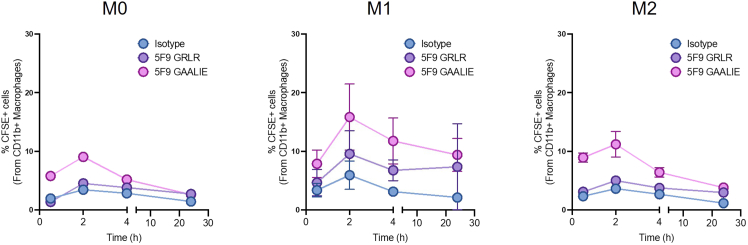
Assessment of antibody-dependent cellular phagocytosis of tumor cells by primary human macrophages with different polarization states across time
Quantification of ADCP (%) at 0.5, 2, 4, and 24 h. Results demonstrate that the Fc-competent antibody (5F9 IgG1-GAALIE) significantly enhances phagocytosis compared to Isotype control or Fc-null antibody (5F9 IgG1-GRLR), particularly at 2 h. These results are seen across all polarization states (M0, M1 and M2 macrophages). Average % ± SEM.
Immunostaining and flow cytometry analysis
Timing: 2–3 h
To measure ADCP, macrophages from the co-culture experiment are stained with antibodies targeting myeloid receptors (CD14 and/or CD11b). Phagocytosis is determined by the percentage of live CD11b+CD14+ macrophages that are positive for CFSE/GFP or another CellTrace stain.
Note: The steps described are applicable to both mouse and human experiments.
-
30.
While the cancer cells and macrophages co-incubate perform compensation in the flow cytometer using unstained, live/dead, and fluorophore-stained controls.
-
31.
After 2 h of co-incubation, spin down the 96-well plate containing both macrophages and cancer cells at 300 x g for 5 min.
Note: Two h of incubation is optimal for the tumor cell lines used in this protocol (Figure 7). Further titration experiments for timing may be required depending on target cells and the treatment used.
-
32.
Aspirate media and wash the cells twice with PBS.
-
33.
Prepare the live/dead stain per the recommended manufacturer’s protocol.
Note: We typically use Live/Dead Aqua (Invitrogen) at a 1:1000 dilution.
Alternatives: DAPI (Invitrogen) staining can be performed at this step or with antibody cocktail staining.
-
34.
Resuspend each well in 200 μL of live/dead stain.
-
35.
Incubate at 20°C–25°C in the dark for 15 min.
-
36.
Wash cells twice with FACS buffer.
-
37.
Prepare Fc block (mouse or human; BioLegend) at a concentration of 1:10 in FACS buffer.
-
38.
Resuspend each well in 50 μL of Fc block.
-
39.
Incubate at 20°C–25°C in the dark for 10 min.
-
40.
In FACS buffer, prepare the anti-macrophage antibody cocktail so that the final concentration is 1:100 after adding it to the Fc block volume (total volume determined by number of samples times 50 μl cocktail per sample).
Note: For the mouse macrophage cocktail, we performed staining with BV421-conjugated anti-F4/80 antibody and BV711-conjugated anti-CD11b antibody. For the human macrophage cocktail, we performed staining with AF700-conjugated anti-CD14 antibody and BV785-conjugated anti-CD11b antibody. (See key resources table)
Note: Some tumor cells may express myeloid markers (CD14, CD11b, etc.), make sure the cancer cell line used does not share these macrophage markers.
-
41.
Add 50 μL of the macrophage antibody cocktail to each sample and pipette up and down to mix.
-
42.
Stain the cells for 30 min in the dark at 4°C.
-
43.
Add 200 μL of FACS and spin down at 300 x g for 5 min.
-
44.
Repeat the wash from step 43 twice and discard the supernatant.
-
45.
After the second wash, resuspend cells in 200 μL of FACS buffer and assess the phagocytosis efficiency by flow cytometry.
-
46.
Collect at least 20,000 cells per tube condition.
-
47.
Use the following gating strategy to determine phagocytosis efficiency (Figure 5).
Note: It is important to refine the gating strategy in single populations (macrophage only and tumor cell only) to ensure accurate discrimination of macrophages, tumor cells, and phagocytosis.
Note: Phagocytosis is determined by the percentage of CFSE+/GPF+ cells within the macrophage cell gate (CD11b+F4/80+ for mouse BMDMs, and CD11b+CD14+ for MDMs). Results can be presented as the number of macrophages positive for CFSE/GFP divided by the total number of macrophages, or as a fold change in which the percentage of phagocytosis in the treatment condition is divided by the percentage of phagocytosis in the isotype/control condition.1
Note: Alternatively, the median fluorescence intensity (MFI) of CFSE in BMDMs or MDMs can be used to quantify antibody-dependent cellular phagocytosis.
Figure 5.
Gating strategy for evaluating phagocytosis of cancer cells by macrophages
Representative flow cytometry plots indicating the gating strategy used to assess phagocytosis of human cancer cells by monocyte-derived macrophages (MDMs).
(A) Histogram plot of number of cells over time confirming that no peaks are generated during sample acquisition.
(B) Gating of MDMs only; or (C) tumor cells only and their respective gates for single cells. Use the macrophage singlets gate for the co-culture analysis to prevent doublets of macrophage-to-cancer cells.
(D) In co-incubation studies, live cells are identified by gating on LD Aqua-negative cells. Macrophages were further gated based on CD11b+ expression. Finally, CFSE positivity was used to determine phagocytosis of CFSE-labeled tumor cells by macrophages.
Expected outcomes
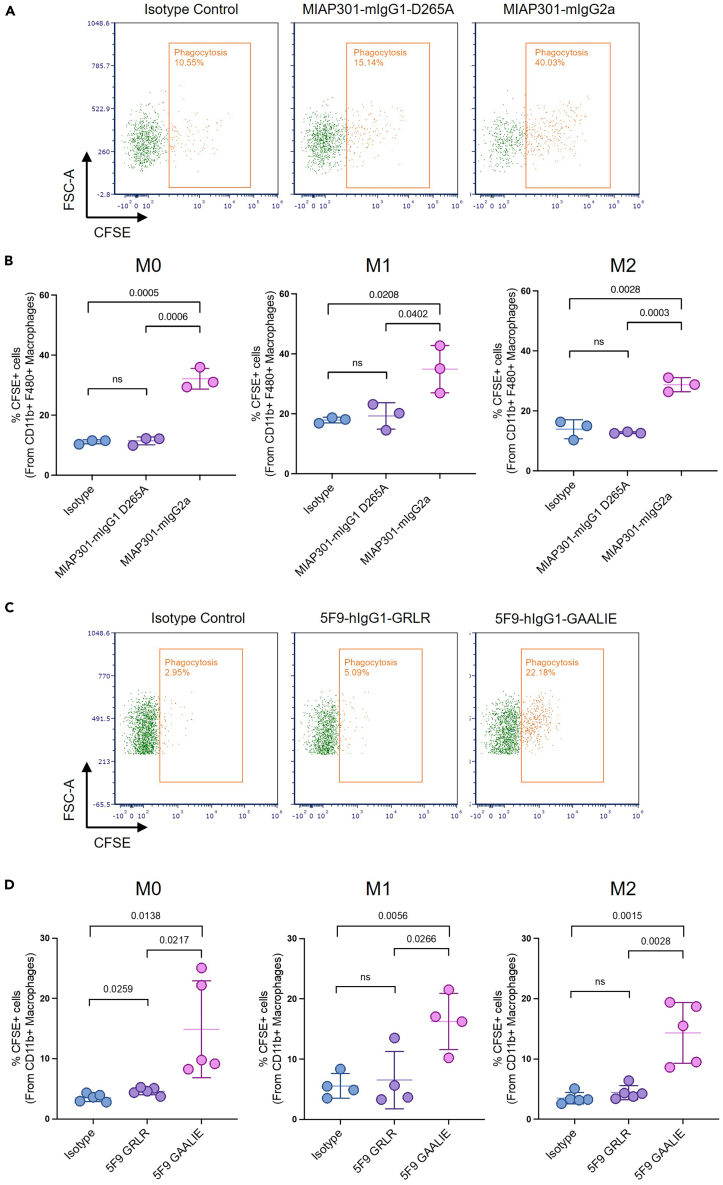
This ADCP assay determines the in vitro capacity of macrophages (M0, M1, and M2) to engulf and phagocytose cancer cells following treatment with antibody therapies. In our results presented in Figure 6, mouse BMDMs treated with antibodies targeting mouse CD47 antibodies that engage Fc receptors (MIAP301-mIgG2a) display significantly higher ADCP when compared to antibodies that lack binding to Fc receptors (MIAP301-mIgG1 D265A) (Figure 6A).1 Of note, M1 BMDMs show the highest rates of phagocytosis across all experimental conditions when compared to M0 and M2 (Figure 6B).1 Similar results are seen with human MDMs, in which treatment with Fc-competent antibodies targeting human CD47 (5F9-IgG1 GAALIE) leads to a significant increase in ADCP when compared to Fc-null antibodies targeting human CD47 (5F9-IgG1GRLR) (Figures 6C and 6D). While 5F9-GAALIE induced higher phagocytosis across unpolarized and polarized macrophages, phagocytosis efficiency varied across different polarization states (Figure 6D). Furthermore, we observed more variability in the phagocytosis capacity of human biological replicates (MDMs) compared to mice (BMDMs). To determine the 2-h time point to assess ADCP, a time course was performed across all polarization states of MDMs co-incubated with Raji cells (Figure 7).
Figure 6.
Assessment of antibody-dependent cellular phagocytosis of tumor cells by primary murine and human macrophages with different polarization states
(A) Representative flow cytometry plots indicating the percentage of murine cancer cell phagocytosis by mouse bone marrow-derived macrophages (BMDM). Treatment groups with mouse antibodies are indicated above the plot.
(B) Quantification of ADCP (%) demonstrating that the Fc-competent antibody (MIAP01-mIgG2a) significantly enhances phagocytosis compared to Isotype control or Fc-null antibody (MIAP301-mIgG1-D265A) by unpolarized (M0), M1 and M2 macrophages. Comparison by unpaired t-test. P-value indicated; ns, not significant.
(C) Representative flow cytometry plots illustrating the percentage of human cancer cell phagocytosis by human monocyte-derived macrophages (MDM). Treatment groups with human antibodies are indicated above the plots.
(D) Quantification of ADCP (%) demonstrating that the Fc-competent antibody (5F9 IgG1-GAALIE) significantly enhances phagocytosis compared to Isotype control or Fc-null antibody (5F9 IgG1-GRLR) by unpolarized (M0), M1 and M2 macrophages. Each dot represents a biological replicate. Comparison by unpaired t-test. P-value indicated; ns, not significant.
Limitations
Stimulating with macrophage polarization factors for less than 24 h or more than 48 h could affect macrophage phenotype and subsequent phagocytic activity.
Detaching macrophages from plates mechanically using cell scrapers causes a significant amount of cell death. For bone marrow-derived macrophages, the yield is high enough to offset death from scraping but the yield from human monocytes may not be great enough. Another option (utilized in the protocol) is to trypsin the cells. Trypsin may consequently destroy some epitopes, so consider using a low concentration (0.05%) and minimizing the amount of time needed to detach cells. A mix of mechanical detachment and trypsin may be utilized; gently pipetting up and down or tapping the plate may speed up the detachment process.
While flow cytometry analysis using the gating strategy outlined in this protocol provides an accurate assessment of ADCP, complementary quantification methods, such as live imaging, should be considered to further validate experimental findings and capture additional aspects of phagocytosis dynamics.
Troubleshooting
Problem 1
Cell contamination of BMDMs is possible.
Potential solution
To minimize the risk, remove the legs from the mouse outside the flow hood, and clean and trim the bones inside the hood (Step 5, macrophage isolation (mouse)).
Problem 2
It is easy to disrupt the Lymphoprep layer when layering buffy coats on top of it. This can affect the density gradient after centrifuging and decrease the total cells acquired.
Potential solution
Hold the falcon tube with the Lymphoprep at 45 degrees or lower. Change the pipette to the slowest setting. Place the pipette tip against the side of the falcon tube near the top, and pipette as slowly as possible. Once the first part of the layer is created, the falcon can be tilted back to 45 degrees and the leukocytes pipetted slightly faster. Minimize layer disruption by minimizing the time the leukocyte layer sits on top of the Lymphoprep layer by centrifuging as soon as possible after layering (Step 25, macrophage isolation (human)).
Problem 3
Sometimes the macrophages grow too confluent while differentiating.
Potential solution
Check the cells every time the media is replaced and split the cells if necessary to maintain 70–80% confluency (Steps 51 and 56, macrophage differentiation and polarization).
Problem 4
The number of monocytes acquired from buffy coats after CD14 MACS isolation can vary between donors.
Potential solution
To ensure there are enough cells to complete the phagocytosis assay, it is possible to reduce the number of macrophages to less than 50.000 per well but retaining the 3:1 cancer cell to macrophages ratio is maintained (e.g., using 20,000 macrophages and 60,000 cancer cells per condition instead of 50,000 and 150,000 respectively) (Step 27, macrophage collection and co-incubation with cancer cells).
Problem 5
Different fluorophores can be used to stain for the same markers, but certain combinations have higher channel bleed-through than others.
Potential solution
Always optimize flow panels beforehand using stained cells and/or compensation beads to test panels and minimize bleed-through. To ensure proper compensation, it is suggested to have unstained cells (cancer cells and macrophages) and live/dead-only stained cells (instead of compensation beads) to gate on positive/negative cells (Step 30, immunostaining and flow cytometry analysis).
It is not necessary to polarize the macrophages for ADCP unless looking for the effects of polarization on phagocytosis activation in the context of specific antibody treatments (Steps 52 and 57, macrophage differentiation and polarization).
While the ADCP assay can be used to visualize the effects of other antibodies or markers on phagocytosis, the authors suggest to always stain the cancer cells with one fluorophore and the macrophages with a verified macrophage marker (F480 for murine macrophages, and Cd11b for human macrophages) (Step 40, immunostaining and flow cytometry analysis).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Juan C. Osorio (osorioj@mskcc.org).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contacts, Dr. Juan C. Osorio (osorioj@mskcc.org) and Carlo Miguel Sevilla (sevillac@mskcc.org).
Materials availability
All requests for unique reagents generated in this study should be directed to the lead contact, Juan C. Osorio (osorioj@mskcc.org).
Data and code availability
This study does not generate unique datasets or code.
Acknowledgments
We thank all the members of the J.V.R. Laboratory of Molecular Genetics and Immunology for helpful discussions and for sharing experiment materials. Research reported in this publication was supported by the National Cancer Institute (NCI) under award number K08CA266740 to J.C.O., the Memorial Sloan Kettering Cancer Center (MSK) Gerstner Physician Scholars Program, and the NCI Cancer Center Support Grant P30 CA008748.
Author contributions
J.C.O. conceptualized and designed the experiments, analyzed the data, and wrote and edited the manuscript. C.M.S., A.M., and B.S. performed the experiments, assembled the data, and wrote and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Osorio J.C., Smith P., Knorr D.A., Ravetch J.V. The antitumor activities of anti-CD47 antibodies require Fc-FcgammaR interactions. Cancer Cell. 2023;41:2051–2065.e2056. doi: 10.1016/j.ccell.2023.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logtenberg M.E.W., Scheeren F.A., Schumacher T.N. The CD47-SIRPalpha Immune Checkpoint. Immunity. 2020;52:742–752. doi: 10.1016/j.immuni.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study does not generate unique datasets or code.