Abstract
A previously undescribed gene, Saitohin (STH), has been discovered in the intron between exons 9 and 10 of the human tau gene. STH is an intronless gene that encodes a 128-aa protein with no clear homologs. The tissue expression of STH is similar to tau, a gene that is implicated in many neurodegenerative disorders. In humans, a single nucleotide polymorphism that results in an amino acid change (Q7R) has been identified in STH and was used in a case control study. The Q7R polymorphism appears to be over-represented in the homozygous state in late onset Alzheimer's disease subjects.
Alzheimer's disease (AD) is the most common late onset dementia, affecting several million people in the developed countries of the world. AD has two major neuropathological hallmarks, extracellular aggregates called amyloid plaques and intracellular aggregates called neurofibrillary tangles. Although the processes of AD could be triggered by many environmental insults, genetic studies have shown that mutations and polymorphisms of particular genes can confer susceptibility to this degenerative process. Genetic studies of AD have identified several early onset disease (<65 years old) risk factors, mutations in the amyloid precursor protein, Presenilin 1, and Presenilin 2, and a late-onset disease (>65 years old) risk factor, the E4 allele of apolipoprotein E (APOE4). In the late onset AD (LOAD) population, only 50% of the subjects have been shown to carry the APOE E4 allele, which is compelling evidence for the existence of additional genetic risk factors. Recently, linkage studies of LOAD-affected sibling pairs have identified loci on chromosome 9 and 10 as potentially harboring risk factor genes (1). So far, the risk factors for AD have shown to modulate amyloid production or deposition, but none have demonstrated a role in the formation of neurofibrillary tangles (NFT).
There has been extensive research on NFT through the study of the conversion of tau from a normally soluble protein into a hyperphosphorylated insoluble protein found in NFT. Tau, a microtubule associate protein is important in establishing and maintaining neuronal morphology. In addition to its role in normal cells, tau protein is also involved in many neurodegenerative diseases in addition to AD, as the main component of intraneuronal aggregates. Some of the more common diseases with tau pathology include frontotemporal dementia (FTD), Pick's disease, and progressive supranuclear palsy (PSP) (2). The involvement of tau in these disorders has prompted much investigation into its function and role in their progression. Recently, several tau-coding mutations have been shown to segregate with FTD, and there have been some intriguing results with transgenic mice expressing the FTD–tau mutation (2), P301L that develop tau aggregates (3). However, in AD, there have been no mutations found in the tau gene, suggesting that other factors are likely involved in the formation of tau aggregates, which could have implications for other neurodegenerative disorders with tau pathology (4).
In FTD and PSP subjects, recent genetic studies of the tau locus have shown that several mutations and a polymorphism segregate with these disorders, respectively (2). The identification and investigation of neighboring genes in the tau locus could be valuable in the study of molecular genetic risk factors in PSP, because there have been no reported tau coding or intronic mutations (5, 6). This information could also be useful not only for PSP but possibly other neurodegenerative disorders, such as AD.
During the examination of expressed sequence tags (ESTs) in the human tau locus, our study has identified a previously uncharacterized gene, Saitohin (STH) (named in honor of late Dr. Tsunao Saitoh and his lab). Nucleotide and amino acid sequence analyses of the STH gene show it encodes for a 128-aa protein that appears to have no clear homology to any protein in humans or other organisms. In the genetic investigation of STH in human subjects, our study identified a nucleotide polymorphism (A → G) that changes a glutamine to arginine at amino acid position 7 (Q7R) of STH. Initial genetic studies of the Q7R polymorphism show an association with LOAD, suggesting that STH could be a contributing factor in AD and could have implications for other neurodegenerative disorders.
Materials and Methods
Cloning of STH.
The cerebellar EST28266 (GenBank accession no. AA325304) was found to be homologous to a segment of the intron between exons 9 and 10 of the human tau gene by using the standard nucleotide–nucleotide blast program (7). The EST was confirmed to be expressed and was cloned from the Marathon cerebellar cDNA library by nested PCR using primers to the EST (F Cel I 5′-CCC TGT AAA CTC TGA CCA CAC-3′ and R Cel I 5′-ACA GGG AAG CTA CTT CCC ATG-3′) and 5′ or 3′ AP linkers according to the manufacturer's protocols (CLONTECH). The PCR products were cloned into TA cloning vector (Invitrogen) and several clones of each product were sequenced (GeneWiz, New York).
Expression of Saitohin and Tau.
The tissue expression of Saitohin and tau transcripts was examined through the use of the Human Rapid Scan Panels (Origene). The cDNA was amplified using the “Touchdown” PCR method (8). The primers used were F-Cel I and R-Cel I, the same as for the cloning. PCR was in a 50-μl reaction mixture containing 20 mM Tris⋅HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.2 μM each primer, 0.2 mM dNTPs, and 1 unit of Taq polymerase (Invitrogen). The touchdown PCR method for Saitohin was as follows: after an incubation at 94°C for 3 min, steps of 94°C for 30 s, 65°C for 30 s for the initial annealing cycle (in each subsequent cycle the annealing temperature was decreased by 0.2°C), and then 72°C for 30 s for polymerization. This was repeated for 25 cycles. Finally, an additional set of 10 cycles were performed, consisting of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; the last cycle was followed by an incubation at 72°C for 30 s.
For the amplification of tau transcripts, the PCR mixture and touchdown program were the same as above except the Tau sense primer was a 21-mer 5′-GCC ACC AGG ATT CCA GCA AAA-3′ and the antisense primer was a 21-mer 5′-TTT ACT TCC ACC TGG CCA CCT-3′ and longer PCR polymerization time of 55 s were used. Both Saitohin and tau PCR products were run on a 2.0% agarose gel with ethidium bromide.
Identification of the Q7R STH Polymorphism and Genotyping Assay.
A total of 81 subjects were used for the case-control study. The AD (n = 51) and the normal control groups of subjects (n = 30) have all been autopsy-confirmed and age-matched. The genomic DNA of the subjects was extracted from frozen brain tissue from AD and normal control subjects by using a genomic DNA extraction kit according to the manufacturer's protocols (Qiagen). For the amplification of coding and noncoding DNA sequence of the STH gene, the PCR mixture and touchdown program were the same as above, except the STH sense primer was a 21-mer FF-Cel 2 (5′-CCA AGT TCA GTT GCC ATC TCC-3′), the antisense primer was a 21-mer OR Cel 2 (5′-CTC TTG TGC ATG GAC CCT GTA-3′), and a longer PCR polymerization time of 55 s was used. Sequencing of the PCR products identified a nucleotide polymorphism A → G changing the amino acid at position 7 from a glutamine (Q) to arginine (R) thereby creating a new HinFI restriction enzyme site.
For the genotyping assay, the PCR products from the primers: F-Cel I and R-CelI (using protocol described in the expression of STH) were digested with 5 units of HinFI (5′-GANTC-3′) restriction enzyme (New England Biolabs) for 3 h at 37°C, then run on a 4.0% agarose gel with ethidium bromide. The Fisher's Exact test was used for the comparisons of the allele (R vs. Q) and genotype (RR vs. non-RR) frequencies in the AD and normal control groups.
The STH genotyping and DNA extraction of the non-AD cases (n = 32) was carried out as above and included clinically and/or pathologically diagnosed subjects with dementia lacking distinctive histopathology (n = 7), FTP (n = 4), Pick's disease (n = 9), PSP (n = 6), corticobasal degeneration (n = 4), and amyotrophic lateral sclerosis (n = 1).
APOE genotyping was carried out as described (9). The Fisher's Exact test was used for the comparisons of the allele and genotype frequencies for the groups included.
MAb Generation and Immunoblot Analyses.
Saitohin monoclonal antibodies: 11F11 (IgG2B), TS6 (IgM), and 10B3 (IgM) were generated as described by immunizing mice with both recombinant and synthetic peptides of the amino acids 8–24 and 112–128 of the Saitohin protein (10). Brain protein samples of normal (NC) and AD subjects with the QQ, QR, and RR genotypes were homogenized in 1XTBS (2 mM PMSF), and STH was partially purified by size fractionation. Subsequently, the prepared samples were incubated in a urea sample buffer for 30 min at 37°C and were run on a SDS/15% PAGE gel. Enhanced chemiluminescence immunoblot analysis was performed by using described protocols (10).
Immunohistochemistry was carried out on paraffin-embedded brain sections as described (10), except that microwave antigen retrieval was performed; sections were stained with the antibodies TS6 and 10B3, then counterstained with Toluidine blue, washed, and were subsequently dehydrated and mounted.
Results
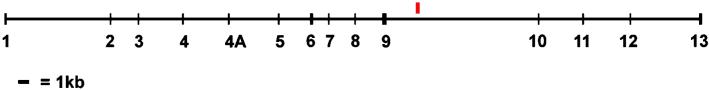
In February, the Human Genome Project completed the sequencing for the genomic clones overlapping the tau locus on Chromosome 17q21, and an EST (GenBank accession no. AA325304) was identified in a tau intron ≈2.5 kb downstream of exon 9 of the tau gene that is shown in Fig. 1. This location is of particular interest because most of the mutations in FTD are in or around this intron and adjacent exons of tau (2). The EST was found to be expressed in the Marathon cerebellar cDNA library (CLONTECH) and in a human oligodendroglioma cell line (that expresses tau), but mRNA isolated from COS7 cells failed to yield an reverse transcription (RT)-PCR product, suggesting that the mRNA was not expressed in COS7 cells (data not shown). We used RNA from a human oligodendroglioma cell line, to clone the 5′ and 3′ ends of the putative gene with the Gene Racer kit (Invitrogen). The sequence of the full-length clones revealed a new gene, Saitohin (STH), which was intron-less and in the sense direction relative to tau. In Fig. 2, nucleotide and amino acid sequence analyses of the STH gene show it encodes for a 128-aa protein that appears to have no clear homology to genes, proteins, signal sequences, or motifs (7, 11). Because the location of STH within the tau locus could provide insight into its function, if they share a similar expression pattern, tau and STH could function together as in the example of the choline acetyltransferase/acetylcholine transporter gene locus (12).
Figure 1.
Tau locus, Saitohin location. The vertical red bar shows the physical location of the Saitohin gene in the intron downstream of exon 9 of the tau gene.
Figure 2.
The ORF for Saitohin protein. The predicted amino acids (three letter abbreviation) for the nucleotide sequences are shown. The red-boxed glutamine (gln) codon [CAA] at amino acid 7 is where the nucleotide polymorphism (A → G) changes the codon [CGA] to an arginine in Saitohin.
To further characterize STH, we analyzed the gene expression in 24 human tissues through the use of the Human Tissue Rapid-Scan panel (Origene). The expression of STH was highest in placenta, muscle, fetal brain, and adult brain, and was lower in heart, kidney, stomach, testis, and adrenal gland, as shown in Fig. 3A. Because STH and tau share a common genetic locus, they could be coordinately expressed; therefore the expression of tau was also examined. To determine whether the location of STH upstream of the alternatively spliced exon 10 would affect its expression, the tau isoforms with exon 10 (Fig. 3A Upper) and without exon 10 (Fig. 3A Lower) were also examined. In Fig. 3B, the expression of tau was found to be highest in heart, kidney, muscle, testis, salivary glands, adrenal glands, adult, and fetal brain, and was lower in placenta, thyroid, prostate, and skin. This finding is in agreement with other reports of tau expression (13).
Figure 3.
Human expression of Saitohin and Tau in multiple tissues and the CNS. Reverse transcription (RT)-PCR was performed on the Human Tissue Rapid-Scan panel (A and B) and the Human Brain Rapid-Scan panel (C and D), Saitohin expression is shown in the panels as a single band (A and C). (B and D) The expression of the tau isoforms as two bands, the Upper band consists of isoforms with exon 10, and the Lower band contain the isoforms without exon 10. The lanes of multiple tissue panels in A and B are: 1, brain; 2, heart; 3, kidney; 4, spleen; 5, liver; 6, colon; 7, lung; 8, small intestine; 9, muscle; 10, stomach; 11, testis; 12, placenta; 13, salivary; 14, thyroid; 15, adrenal; 16, pancreas; 17, ovary; 18, uterus; 19, prostate; 20, skin; 21, pbl; 22, bone marrow; 23, fetal brain; 24, fetal liver. The lanes for the Human Brain panel are: 1, frontal lobe; 2, temporal lobe; 3, cerebellum; 4, hippocampus; 5, substantia nigra; 6, caudate nucleus; 7, amygdala; 8, thalamus; 9, hypothalamus; 10, pons; 11, medulla; 12, spinal cord.
As a result of tau and STH having a significant overlap in general tissue expression, an expanded study was performed by using the Human Brain Rapid-Scan panel to determine the central nervous system (CNS) expression patterns and whether STH and tau are coordinately expressed in the brain. Fig. 3C shows that STH was expressed in the CNS with higher expression in the temporal lobe, hypothalamus, medulla, and spinal cord, and with lower expression in the other brain regions. Fig. 3D shows that the expression of tau was high in most of the CNS samples, except in the pons, where the expression was lower. From the general tissue and CNS tissue expression, these observations suggest that tau shares some tissue expression with STH; however, there are some differences. Taken together, these results indicate that STH is not under the regulation of the tau promoter but could share some regional regulatory elements with the tau gene, which could have implications for the function of STH. The location and expression of STH upstream of the exon 10 does not appear to correlate with the splicing of exon 10 of tau in the CNS or the other tissues.
The location of STH warranted investigation into its possible role in neurodegenerative disorders, because there is genetic evidence implicating the tau locus involvement in many of these diseases. During the sequencing of the STH gene from human subjects, a nucleotide polymorphism (a → g) was identified that changes a glutamine (Q) to arginine (R) at amino acid position 7 (Q7R) of STH, shown in Fig. 2. The “g” polymorphism (R allele) creates a novel HinfI restriction enzyme site generating a distinctive fragment pattern as compared with the “a” nucleotide (Q allele); a representative gel is shown in Fig. 4. The distribution of AD and normal control subjects with different STH alleles and frequencies were tabulated in Table 1. The two groups of subjects were age-matched and were limited to Caucasian subjects. The RR genotype was found at a significantly higher frequency in the AD group (16%) as compared with the normal control subjects (0%) (P = 0.0232, odds ratio 11.920 by Fisher's Exact test). The R allele does occur in the normal subjects at frequency of 13%, but at a significant lower percentage compared with the AD subjects in which the R allele is 32% (P = 0.0085, odds ratio 3.109 by Fisher's Exact test). The average age of onset for the RR subjects is 83.1 with a range of 77–93, which provides evidence that RR genotype is a risk factor for LOAD. In addition to the STH genotype, the APOE genotype of the subjects was also determined because the allele 4 is an important risk factor for AD, and also to investigate whether the APOE allele 4 could synergize with the STH genotype (14). The APOE genotypes were in close agreement with the frequencies in the general population, thereby providing evidence against sampling bias (15). In agreement with previous reports, the APOE 4 allele was found to be over-represented in our AD group as compared with normal control subjects (data not shown); however, the APOE alleles were evenly distributed among the RR, QR, and QQ subjects of the AD group in table 2, suggesting that there is no association between the APOE and STH genotypes (15).
Figure 4.
A representative alleleotyping gel of HinFI-digested PCR products of the genotypes: QQ, QR, and RR. The polymorphism creates a HinF1restriction enzyme site. HinF1-digested PCR product yields two bands (171 bp and 55 bp) in subjects with a Q allele, and three bands (55, 74, and 97 bp) in individuals with an R allele. Two QQ homozygotes, two QR heterozygotes, and two RR homozygotes are shown.
Table 1.
STH polymorphism in AD and normal
| Saitohin | AD | Normal |
|---|---|---|
| Genotype | n = 51 | n = 30 |
| 26 (51%) | 22 (73%) | |
| QR | 17 (33%) | 8 (27%) |
| RR | 8 (16%) | 0 (0%) |
| Alleles | n = 102 | n = 60 |
| Q | 69 (67.6%) | 52 (86.7%) |
| R | 33 (32.4%) | 8 (13.3%) |
Normal (average age = 78.83, range 55–97), AD (average age = 80.51, range 56–98) patients. A total of 81 subjects were used for the case-control study. The AD (n = 51) and normal control groups (n = 30) of subjects have all been autopsy confirmed and age-matched. Fisher's Exact test was used for the comparisons of the allele and genotype frequencies for the groups included.
In addition to the AD group, a small number of subjects with other neurodegenerative disorders, most of which have tau pathology, were also examined for the polymorphism. The results in Table 3 illustrate that the polymorphism is not only AD specific, and show data on groups that might be of interest for further study. For example, one subject with dementia lacking distinctive histopathology had the RR genotype, suggesting this genotype is not AD specific. Also, PSP subjects seem to have an over-representation of the QQ genotype, where FTD and Pick's disease have a higher percentage of QR genotype, as compared with normal controls. Further investigation of these trends with larger groups of subjects is required to determine their significance.
Table 3.
STH polymorphism in several non-AD neurodegenerative disorders
| Saitohin genotype | DLDH n = 7 | PSPn = 6 | FTDn = 4 | CBD n = 4 | PICKS n = 9 | ALS n = 1 |
|---|---|---|---|---|---|---|
| 4 (57%) | 5 (83%) | 1 (25%) | 4 (100%) | 3 (33%) | 1 (100%) | |
| QR | 2 (29%) | 1 (17%) | 3 (75%) | 0 (0%) | 6 (67%) | 0 (0%) |
| RR | 1 (14%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
The numbers of subjects were too small for statistical analysis. Subjects with dementia lacking distinctive histopathology (DLDH), frontotemporal dementia (FTD), Pick's disease (PICKS), progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), and amyotrophic lateral sclerosis (ALS).
The experiments demonstrating STH expression so far rely on the use of PCR, which is so sensitive that it is difficult to be sure that bands result from legitimate mRNA transcripts or come from a trace contamination of DNA. When working with an intron-less gene, where the sequence of the mRNA matches up exactly with the genomic DNA encoding the gene, DNA contamination remains a concern. The consistent failure to find evidence of STH mRNA in COS7 cells and in several tissues of the multiple tissue cDNA panel argues against contamination. We generated monoclonal antibodies to the predicted protein, produced as either a 6-His tagged recombinant protein, or as a glutatione S-transferase fusion protein in bacteria, to demonstrate the existence of a STH protein product. By using a combination of synthetic peptides from the STH sequence and a deletion mutant protein, the epitopes recognized by these monoclonal antibodies have been mapped to three different regions of STH: N terminus, a central region, and C terminus. In Fig. 5, three representative immunoblots with antibodies to GST (DT12), 6XHis, and STH (TS6) demonstrate the specificity of the monoclonal antibodies.
Figure 5.
Western blot analysis of normal (NC) and AD subjects with the QQ, QR, and RR genotypes. (A–C) Immunoblots of whole lysates of IPTG-induced bacteria of expressing recombinant 6XHis tagged saitohin (6H-SA) or 6XHis tagged tau or glutatione S-transferase (GST) or GST-saitohin fusion protein (GST-SA) with antibodies to GST (DT12), 6XHIS, and Saitohin (TS6). (D) Immunoblots of partial purified brain homogenates from QQ, QR, or RR genotypes of AD (QQ-AD, AD-QR, and AD-RR) and normal (QQ-NC, QR-NC) subjects with Saitohin monoclonal antibodies 11F11, TS6, and 10B3 (Left, Center, and Right, respectively).
In Fig. 5D, STH protein was detected by immunoblots of brain homogenates from AD and normal subjects with antibodies to N terminus (11F11, an IgG2B), C terminus (10B3, an IgM), and intervening sequence (TS6, an IgM) of STH protein. The immunoblots show a protein with an apparent molecular mass of about 16 kDa, 2 kDa more than the calculated size of 13.6 kDa. A similar protein is recognized by all three antibodies, which are reactive with three different epitopes of STH, in addition, other N-terminal antibodies to STH show a similar blotting result (data not shown). This size difference and the presence of the higher molecular mass bands common to all of the blots suggest that STH could be posttranslationally modified and/or aggregated with itself or other proteins. Further investigation is needed to determine which hypothesis or hypotheses are correct.
Expression of STH protein was examined by immunocytochemistry on paraffin sections of human brain tissues. Two antibodies, 10B3 and TS6, recognizing different epitopes of the protein, both show staining of neurons in hippocampus and temporal cortex (Fig. 6). Staining of what appeared to be reactive astrocytes was also noted in some sections. Further studies are needed to examine STH expression in both normal and AD brain tissue, in relation to Saitohin genotype and AD pathology.
Figure 6.
Immunohistochemistry of STH. Immunolabelling of the cytoplasm of neurons and some astrocytes in human brain sections with TS6 and 10B3.
Discussion
The Human Genome Project facilitated the cloning of STH gene and the further investigation of the Tau/STH locus. Despite the sequence analysis of STH gene yielding no clues to the function of STH, several things are known about STH. First, STH is a nested gene in the tau locus and has a very similar expression pattern to tau, which suggests that these two proteins are not only expressed jointly but might function together in a pathway. A well-studied example of this situation is the choline acetyltransferase (CHAT)/vesicular acetylcholine transporter (VCHAT) gene locus. The VCHAT gene resides within the intron between the exons 1 and 2 of the CHAT gene, and these coordinately expressed genes function in packaging of the neurotransmitter acetylcholine (ACH) into vesicles and in the synthesis of ACH, respectively (12). An attractive hypothesis starts to emerge that STH and tau might not only function together normally but also in disease states.
Second, the expression studies have also provided preliminary evidence that STH expression is not linked to the splicing of exon 10 of the tau gene. However, further study is required to confirm this finding and determine whether this is true in the disease state. In FTD, the splicing of exon 10 is altered by some of the mutations in the tau gene (2), the expression of the STH gene could also be affected by these tau mutations or by the R allele of STH.
Third, the discovery of STH expression in a human oligodendroglioma cell line and human brain suggests that the STH gene could be investigated in vitro and in vivo.
Fourth, a Q7R polymorphism in the STH gene was identified in human subjects and was found to be over-represented in the homozygous state in the LOAD population. There have been however, several genetic studies of the tau locus in AD that have provided evidence for an association (16–18) where others have not (4, 19–21). Incorporating the STH polymorphism screening in these studies could elucidate the apparent inconsistencies in these reports and help determine whether there is an association between the tau/STH locus and AD.
The RR genotype in the AD subjects is strongly suggestive of a loss of function mutation because of its homozygous state; however, the possibility that the Q7R polymorphism is a gain of function mutation remains. STH presumably has a normal cellular function and, although sequence analysis has yielded no clues, there may be hints to the function of STH and its possible role in AD and other dementias. Because STH lacks a consensus targeting signal sequence and the immunostaining of neuronal and perhaps astrocytic cell bodies, it is a putative cytosolic protein, placing it in the same compartment as tau. Unlike tau, STH has no proline directed phosphorylation sites. There are however, putative phosphorylation sites on STH for protein kinase C and casein kinase II, kinases that normally have significant roles in the CNS and are implicated in tau phosphorylation and AD (22). The higher molecular weight species of the STH protein on the Western blots provide evidence for postranslational modifications (e.g., phosphorylation, glycation, etc.) and/or multimerization/aggregation; these observations are suggestive and further work is necessary.
Because the R allele of STH may be a risk factor for AD, STH could be modulating neurofibrillary tangle and/or amyloid plaque formation, the major hallmarks of this disease. Immunostaining studies of AD cases are needed to determine whether there is colocalization of STH with amyloid plaques and/or neurofibrillary tangles. These immunohistochemical experiments and follow up genetic studies of STH in neurodegenerative diseases with a variety of tau or amyloid pathologies could help to determine whether STH plays a role in the formation of neurofibrillary tangles and/or amyloid plaques.
Table 2.
STH genotype vs. APOE genotype
| STH genotype | AD (n = 51)
|
Normal (n = 30)
|
||
|---|---|---|---|---|
| APOE ɛ4 negative (n = 25) | APOE ɛ4 positive (n = 26) | APOE ɛ4 negative (n = 26) | APOE ɛ4 positive (n = 4) | |
| 13 | 13 | 20 | 3 | |
| QR | 8 | 9 | 6 | 1 |
| RR | 4 | 4 | 0 | 0 |
A logistic regression analysis was performed with the statistical program spss for the determination of associations of APOE ɛ4 with RR genotype in the AD and normal populations.
Acknowledgments
This work was supported by grants from National Institutes of Mental Health (38623) and by Molecular Geriatrics Corporation.
Abbreviations
- AD
Alzheimer's disease
- FTD
frontotemporal dementia
- PSP
progressive supranuclear palsy
- STH
Saitohin
- Q
glutamine
- R
arginine
- APOE
apolipoprotein E
- LOAD
late onset AD
- NFT
neurofibrillary tangles
- EST
expressed sequence tag
- CNS
central nervous system
References
- 1.Myers A J, Goate A M. Curr Opin Neurol. 2001;14:433–440. doi: 10.1097/00019052-200108000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof P R. Brain Res Brain Res Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 3.Lewis J, Dickson D W, Lin W L, Chisholm L, Corral A, Jones G, Yen S H, Sahara N, Skipper L, Yager D, et al. Science. 2001;293:1487–1491. doi: 10.1126/science.1058189. [DOI] [PubMed] [Google Scholar]
- 4.Russ C, Powell J F, Zhao J, Baker M, Hutton M, Crawford F, Mullan M, Roks G, Cruts M, Lovestone S. Neurosci Lett. 2001;314:92–96. doi: 10.1016/s0304-3940(01)02289-3. [DOI] [PubMed] [Google Scholar]
- 5.Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J, Hardy J, Lynch T, Bigio E, Hutton M. Hum Mol Genet. 1999;8:711–715. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- 6.Poorkaj P, Kas A, D'Souza I, Zhou Y, Pham Q, Stone M, Olson M V, Schellenberg G D. Mamm Genome. 2001;12:700–712. doi: 10.1007/s00335-001-2044-8. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecker K H, Roux K H. BioTechniques. 1996;20:478–485. doi: 10.2144/19962003478. [DOI] [PubMed] [Google Scholar]
- 9.Wenham P R, Price W H, Blandell G. Lancet. 1991;337:1158–1159. doi: 10.1016/0140-6736(91)92823-k. [DOI] [PubMed] [Google Scholar]
- 10.Jicha G A, Weaver C, Lane E, Vianna C, Kress Y, Rockwood J, Davies P. J Neurosci. 1999;19:7486–7494. doi: 10.1523/JNEUROSCI.19-17-07486.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thornton J M. Science. 2001;292:2095–2097. doi: 10.1126/science.292.5524.2095. [DOI] [PubMed] [Google Scholar]
- 12.Oda Y. Pathol Int. 1999;49:921–937. doi: 10.1046/j.1440-1827.1999.00977.x. [DOI] [PubMed] [Google Scholar]
- 13.Gu Y, Oyama F, Ihara Y. J Neurochem. 1996;67:1235–1244. doi: 10.1046/j.1471-4159.1996.67031235.x. [DOI] [PubMed] [Google Scholar]
- 14.Saunders A M, Strittmatter W J, Schmechel D, George-Hyslop P H, Pericak-Vance M A, Joo S H, Rosi B L, Gusella J F, Crapper-MacLachlan D R, Alberts M J. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 15.Corder E H, Lannfelt L, Bogdanovic N, Fratiglioni L, Mori H. Cell Mol Life Sci. 1998;54:928–934. doi: 10.1007/s000180050223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford F, Freeman M, Town T, Fallin D, Gold M, Duara R, Mullan M. Neurosci Lett. 1999;266:193–196. doi: 10.1016/s0304-3940(99)00303-1. [DOI] [PubMed] [Google Scholar]
- 17.Lilius L, Froelich F S, Basun H, Forsell C, Axelman K, Mattila K, Andreadis A, Viitanen M, Winblad B, Fratiglioni L, et al. Neurosci Lett. 1999;277:29–32. doi: 10.1016/s0304-3940(99)00833-2. [DOI] [PubMed] [Google Scholar]
- 18.Bullido M J, Aldudo J, Frank A, Coria F, Avila J, Valdivieso F. Neurosci Lett. 2000;278:49–52. doi: 10.1016/s0304-3940(99)00893-9. [DOI] [PubMed] [Google Scholar]
- 19.Roks G, Dermaut B, Heutink P, Julliams A, Backhovens H, Van de B M, Serneels S, Hofman A, Van Broeckhoven C, van Duijn C M, et al. Neurosci Lett. 1999;277:137–139. doi: 10.1016/s0304-3940(99)00861-7. [DOI] [PubMed] [Google Scholar]
- 20.Baker M, Graff-Radford D, Wavrant D F, Graff-Radford N, Petersen R C, Kokmen E, Boeve B, Myllykangas L, Polvikoski T, Sulkava R, et al. Neurosci Lett. 2000;285:147–149. doi: 10.1016/s0304-3940(00)01057-0. [DOI] [PubMed] [Google Scholar]
- 21.Kwon J M, Nowotny P, Shah P K, Chakraverty S, Norton J, Morris J C, Goate A M. Neurosci Lett. 2000;284:77–80. doi: 10.1016/s0304-3940(00)00972-1. [DOI] [PubMed] [Google Scholar]
- 22.Jin L W, Saitoh T. Drugs Aging. 1995;6:136–149. doi: 10.2165/00002512-199506020-00006. [DOI] [PubMed] [Google Scholar]