Abstract
Membrane surface electrostatic interactions impose structural constraints on imported proteins. An unprecedented sensitive dependence on these constraints was seen in the voltage-gated import and channel formation by the C-terminal pore-forming domain of the bacteriocin, colicin E1. At physiological ionic strengths, significant channel current was observed only in a narrow interval of anionic lipid content ([L−]), with the maximum current (Imax) at 25–30 mol% (dioleoyl)-phosphatidylglycerol ([L−]max) corresponding to a surface potential of the lipid bilayer in the absence of protein, ψ = −60 ± 5 mV. Higher ionic strength shifted [L−]max to larger values, but ψ
= −60 ± 5 mV. Higher ionic strength shifted [L−]max to larger values, but ψ remained approximately constant. It is proposed that the channel current (i) increases and (ii) decreases at |ψo| values <55 mV and >65 mV, because of (i) electrostatic interactions needed for effective insertion of the channel polypeptide and (ii) constraints due to electrostatic forces on the flexibility needed for cooperative insertion into the membrane. The loss of flexibility for |ψo| ≫ 65 mV was demonstrated by the absence of thermally induced intraprotein distance changes of the bound polypeptide. The anionic lipid content, 25–30 mol%, corresponding to the channel current maxima, is similar to that of the target Escherichia coli cytoplasmic membrane and membranes of mesophilic microorganisms. This suggests that one reason the membrane surface potential is tuned in vivo is to facilitate protein import.
remained approximately constant. It is proposed that the channel current (i) increases and (ii) decreases at |ψo| values <55 mV and >65 mV, because of (i) electrostatic interactions needed for effective insertion of the channel polypeptide and (ii) constraints due to electrostatic forces on the flexibility needed for cooperative insertion into the membrane. The loss of flexibility for |ψo| ≫ 65 mV was demonstrated by the absence of thermally induced intraprotein distance changes of the bound polypeptide. The anionic lipid content, 25–30 mol%, corresponding to the channel current maxima, is similar to that of the target Escherichia coli cytoplasmic membrane and membranes of mesophilic microorganisms. This suggests that one reason the membrane surface potential is tuned in vivo is to facilitate protein import.
It is well known that electrostatic forces are involved in the binding of membrane-active proteins or peptides to anionic lipid membrane surfaces (1). However, the nature of protein–lipid interactions that allow protein insertion into the lipid bilayer is not understood. Although the specificity of assembly of integral membrane proteins is organized and directed by specific proteinaceous translocation complexes (2), several general properties of membrane proteins and protein translocation are specified by the lipid environment: (i) the length of the transmembrane segments of integral membrane proteins, ∼20 residues in an α-helical conformation, dictated by the width, ∼35 Å, of the hydrophobic lipid bilayer (3); (ii) the position of aromatic Trp and Tyr residues of the transmembrane helical domains in the lipid head group interfacial region (3–5); (iii) the cis-positive rule for the transmembrane orientation of integral membrane proteins and signal peptides (6), explained by the existence of the anionic membrane surface and resulting negative surface potential, ψo, of biological membranes (7, 8). (iv) The rate of protein unfolding at the membrane surface, in the case of the C-terminal channel polypeptide of colicin E1, is increased by larger values of the negative membrane surface potential and decreased by higher viscosity of the membrane (9).
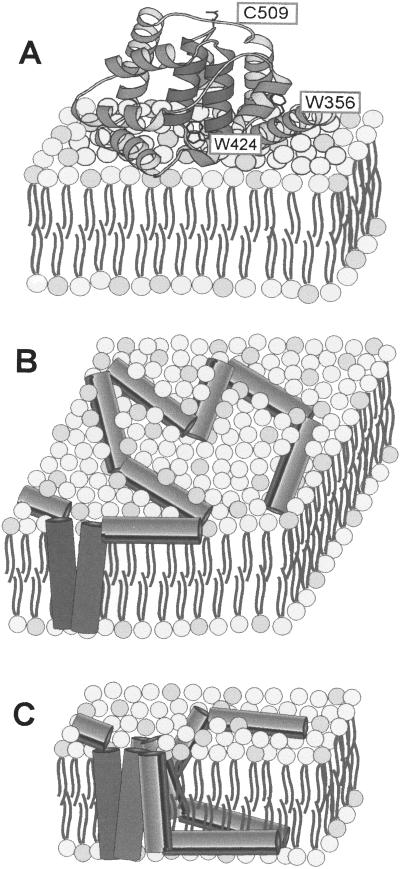
Voltage-gated colicin channels (10–12) provide a useful system to study the effects of protein–lipid interactions on protein insertion into membranes. The basis of the bactericidal action of the pore-forming voltage-gated colicins is the highly conductive nature of the colicin channel (13), which depolarizes and de-energizes target E. coli cells (14). After binding to the outer membrane receptor(s), and translocating across the periplasmic space, using the Tol/Ton intermembrane translocation systems (10, 12), colicin molecules are bound to the cytoplasmic membrane surface, inserted into the interfacial layer, and imported from the surface into the membrane bilayer to form a voltage-gated channel (Fig. 1 A–C). In vitro studies with liposomes and planar bilayers imply that voltage-gated opening of the colicin channel in the cytoplasmic membrane occurs through voltage-driven import of the channel polypeptide (16–18).
Figure 1.
Sequence of events in membrane import of the colicin channel domain. (A) Surface-bound globular non-inserted P178 (15). The approximate positions of Trp-356, Trp-424, and Cys-509 are shown. (B) An extended two-dimensional helical array in the membrane interfacial layer, including hydrophobic helical hairpin (darker shading), as a precursor to the open channel state. (C) Depiction of imported open channel pore-forming state (16, 17).
High-resolution structures have been determined of the pore-forming colicin Ia (19), a large part of colicin N (20), and of the ten helix channel-forming domain of colicins A (21) and E1 (ref. 15; Fig. 1A), all in the soluble state. The binding of the 178-residue C-terminal channel domain of colicin E1, P178 (C-terminal 178-residue colicin E1 channel-forming polypeptide), to the membrane surface depends on attractive electrostatic and hydrophobic interactions (22). The conversion of the P178 to an integral channel protein involves a large conformational change from the soluble structure to an intermediate state in which the channel domain binds and unfolds on the membrane surface, with a concomitant large increase of helical content and insertion into the membrane interfacial layer (Fig. 1B; ref. 23). This surface-bound state in the membrane interfacial layer was characterized by spectroscopic, fluorescence resonance energy transfer (FRET), calorimetric, and surface plasmon resonance analysis as a flexible two-dimensional helical array in which the individual ten helices are extended by an average of 25% from their length in the soluble state (Fig. 1B; refs. 9, 23, and 24). Flexibility of the surface-bound helical array was implied by reversible thermally induced changes of intraprotein distances of the colicin channel domain in the membrane interfacial layer (23). It was proposed that helix extension and P178 flexibility are necessary prerequisites for its insertion into the membrane bilayer in the transition to open channel state (23).
The present study shows a unique sensitive dependence of colicin channel current on membrane anionic lipid content ([L−]). Compensatory changes of ionic strength and anionic lipid content associated with large amplitude channel current demonstrate that these currents are observed over a narrow range of the membrane surface potential. The anionic lipid content corresponding to maximum channel currents in vitro at physiological ionic strengths are similar to that in many biological membranes. Together with the well documented response of microorganisms to increase of salinity by higher level of anionic lipid content, this implies that the membrane surface potential in vivo may be regulated by requirements for protein import.
Materials and Methods
Preparation and Mutagenesis of the C-Terminal Channel Polypeptide of Colicin E1.
The 178-residue C-terminal channel domain of colicin E1, P178, was prepared by thermolysin proteolysis of intact colicin E1 (25). P178 from wild-type colicin, containing three tryptophans at positions 424, 460, and 495, and a single C505, was used in the planar bilayer experiments. Mutants W356, W424, and W− refer to P178 with point mutations Y356W/W460F/W495F/C505A/D509C, W460F/W495F/C505A/D509C, and W424F/W460F/W495F/C505A/D509C, respectively (9).
Planar Lipid Bilayer Measurements.
Membranes were formed with varying molar ratios of (dioleoyl)-phosphatidylglycerol (DOPG) and (dioleoyl)-phosphatidylcholine (DOPC) in n-decane (total concentration, 2%; Avanti Polar Lipids) in a 0.5-mm-diameter membrane aperture in a bilayer chamber (Warner Instruments, Hamden, CT) by a brush technique (26). Bilayer formation was monitored by measuring capacitance that was typically 500–600 pF, and macroscopic current was measured at 23°C in a voltage-clamp mode. The bathing solution in both compartments was 0.1–1.0 M KCl/20 mM HOAc/NaOAc (pH 4.0) in most experiments. P178 (final concentration, 100–400 ng/ml) was added to the cis-compartment that was held at virtual ground potential. Solutions in both compartments were stirred magnetically for 20 min before measurements were started. The sign of the membrane potential refers to that in the trans-compartment.
Calculation of Membrane Surface Potential, ψo.
ψo was calculated as a function of anionic lipid content and ionic strength, using the Gouy-Chapman formalism for monovalent solutes, Fψo/2RT = Aσ; A = (8ɛoɛRTc)−1/2 (1), where c is the bulk electrolyte concentration (mol/l), F and R are the Faraday and gas constants, T is temperature, σ is the surface charge density (charges per Å2), ɛ and ɛo are the medium dielectric constant and permittivity of free space, respectively, and the constant A = 136.2 (c)−1/2.
Trans-Monolayer Potential Measurements in Lipid Monolayers.
The term “trans-monolayer potential” is used to define the potential across the air to aqueous interface of the monolayer. This potential consists of surface and dipole components. Monolayers were made of 100% DOPG and were formed by spreading 0.5 mM lipid solutions in chloroform on the aqueous solution surface in a Langmuir trough equipped with a Wilhelmy balance (both homemade). Trans-monolayer potentials were measured with a homemade Kelvin probe sensor with the three-electrode arrangement consisting of a gold-coated vibrating electrode, Ag/AgCl reference electrode, and a stainless steel grounding electrode (27).
FRET.
The sulfhydryl-reactive dye, 5-[[[(2-iodoacetyl) amino]ethyl]amino]naphthalene-1-sulfonic acid (AEDANS), whose excitation spectrum (λex, 337 nm) overlaps the Trp emission spectrum of P178 (λem, 325 and 330 nm for W424 and W356, respectively), was attached to Cys-509 as described (9, 23). AEDANS-labeled P178 (1 μM) was mixed with large unilamellar vesicles (LUV; equivalent to 0.4 mM total lipid) prepared from the synthetic lipids, DOPG and DOPC, by multiple extrusion through a 0.1 μm pore size filter (Nucleopore, Costar; ref. 28). Temperature-dependent changes in the ratio of AEDANS fluorescence at 490 nm excited at 290 and 337 nm, were measured using a photon counting spectrofluorimeter (Fluorolog3, ISA, Edison, NJ) equipped with a programmed temperature-control unit, and were analyzed as described (9, 23). To correct for the FRET contribution from Tyr (nine Tyr in wild-type P178), AEDANS fluorescence at 490 nm in the W− mutant excited at 290 nm was subtracted from the signal from W356 or W424 mutants for every temperature data point. The “intramolecular ruler” for this system was calibrated from the distance between W424 and D509 in the 2.5-Å x-ray structure of the colicin E1 channel polypeptide (15). The Förster distance for 50% energy transfer, R0 = 22 Å (29), was used in the FRET analysis.
Results
Dependence of Colicin E1 Channel Activity on Anionic Lipid Content.
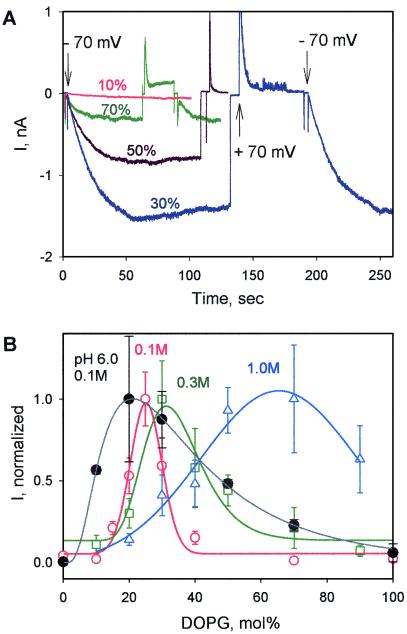
Channels of the 178-residue C-terminal channel-forming domain of colicin E1, P178, incubated with the bilayer membrane for 20 min to ensure equilibration of binding, were opened upon imposition of a trans-negative potential, and closed upon switching to trans-positive (Fig. 2A). Channel formation was greatly enhanced by the presence of anionic lipid (22), for example DOPG, which allows electrostatic interactions. This is shown in the comparison of the channel current at 30 vs. 10 mol% DOPG (Fig. 2A). The channel current reached a pronounced maximum, Imax, at a lipid content, [L−]max = 30 mol% DOPG, and decreased at higher DOPG contents, 50 and 70 mol% (Fig. 2A).
Figure 2.
Dependence of colicin E1 channel activity on anionic lipid content. (A) Time course of channel formation in planar lipid bilayers with different anionic lipid content. Channels opened and closed at trans-negative and trans-positive potentials, respectively, after incubation with P178 (200 ng/ml). [L−] = 10 (red), 30 (blue), 50 (brown), or 70 (green) mol% DOPG. Chamber solution: 20 mM HOAc/NaOAc, pH 4.0, 0.3 M KCl. (B) Macroscopic steady-state current in planar bilayers as a function of anionic lipid (DOPG) content and pH. Data were normalized relative to Imax at 25, 30, and 70 mol% DOPG for 0.1 (○, red), 0.3 (□, green), and 1.0 M (▵, blue) KCl, respectively, at pH 4.0, and relative to Imax at 20 mol% for 0.1 M KCl at pH 6.0 (●, black). The protein concentration was 400, 200, and 100 ng/ml at 0.1, 0.3, and 1.0 M KCl, so that Imax = 1–5 nA. The standard deviations were based on n trials with different planar membranes, with n = 10–15 near the peaks, and n = 5–7 elsewhere. In the pH-6 experiments, planar bilayers were preincubated with P178 at pH 4.0 before pH was shifted to 6.0 by addition of KOH. Buffer: 10 mM MES/10 mM β-alanine/0.1 M KCl.
The dependence of the channel current on (i) lipid concentration and (ii) on ambient pH was similar (i) with the anionic lipid, DOPA, and (ii) at pH 6 after initial binding at pH 4 (Fig. 2B, black), implying (i) the lipid effect is not specifically related to the nature of the lipid headgroup, but rather to its anionic character and the value of negative membrane surface potential, ψo. (ii) It is also not a unique effect of the use of pH 4 to initiate the binding interaction between cationic P178 and the membrane surface.
The biphasic dependence of the current on [L−] at ionic strengths of 0.1 M (red), 0.3M (green), and 1.0 M (blue), with phases of increasing and decreasing current that are particularly steep at ionic strengths of 0.1 and 0.3 M, is summarized in Fig. 2B. The lipid “bandwidth,” Δ[L−]1/2, of the P178 channel current response at pH 4.0 and 0.1 M ionic strength is very narrow, Δ[L−]1/2 = 10–15 mol% with [L−]max = 25 mol% (Fig. 2B, red). The channel current declined precipitously for [L−] > 25 mol%, although higher [L−] is linked to a faster rate of binding and unfolding at the membrane surface (9, 23). At 0.3 and 1.0 M KCl, respectively, the critical value of [L−] shifted to 30 mol% with Δ[L−]1/2 = 25 mol% (Fig. 2B, green), and to 60–70 mol% with Δ[L−]1/2 ≈ 50 mol% (Fig. 2B, blue). This implies that control of voltage-gated colicin channel formation depends on the value of the membrane surface potential, ψo, whose magnitude is diminished by counter ion screening at high ionic strength. The dependence of the channel current on [L−] at pH 6.0 has a similar [L−]max, although the bandwidth was broader (Fig. 2B, black).
Determination of Surface Potential, ψo.
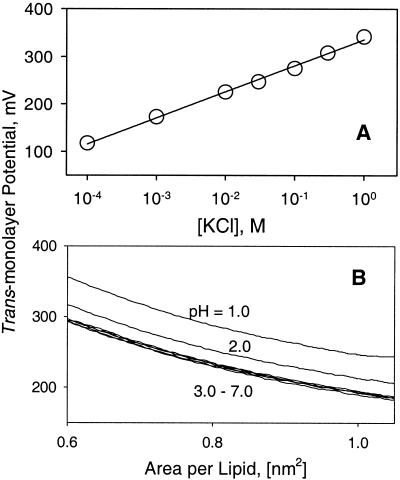
The applicability of the Gouy-Chapman formalism (1) to the present conditions (pH, lipid and ion composition) was tested by direct measurement of the electrical potential in Langmuir monolayers. The trans-monolayer potential was measured as a function of lipid head group area in monolayers made of 100 mol% DOPG, where deviation from Guoy-Chapman theory is most likely. The trans-monolayer electrical potential consists of two components, the surface potential, ψo, and the lipid dipole potential. It was assumed that the dipole potential, being an inherent feature of lipid molecules organized in a bilayer, does not depend on the solution variables of ionic composition, ionic strength, and pH. Therefore, changes in trans-monolayer electrical potential were attributed to variation in ψo. The linear dependence of the trans-monolayer potential on the logarithm of the salt concentration had a slope of 55 ± 1 mV per decade change of concentration (Fig. 3A), in reasonable agreement with the theoretical value of 58 mV at 20°C. Moreover, trans-monolayer potential isotherms in these experiments did not depend on pH in the range 3–7 (Fig. 3B), showing that corrections for specific ion binding, including protonation of phosphate groups at the pH 4.0 used in bilayer experiments, were not required.
Figure 3.
Applicability of Gouy-Chapman formalism to calculations of surface potential and near-membrane Cl− concentration by using lipid monolayers made of 100 mol% DOPG. (A) Linearity of trans-monolayer potential dependence on log [KCl]. Data taken at a lipid monolayer compression of 0.65 nm2 per lipid molecule after measurement of trans-monolayer potential isotherms as a function of area/lipid and [KCl] at pH 6.0. (B) Trans-monolayer potential isotherms as a function of pH. Na-Mes, NaOAc, HCl or HClO4, and KCl were used to adjust the ionic strength to 0.1 M and the pH to 1.0–7.0.
The Phase of Decreasing Channel Current for [L−] > [L−]max.
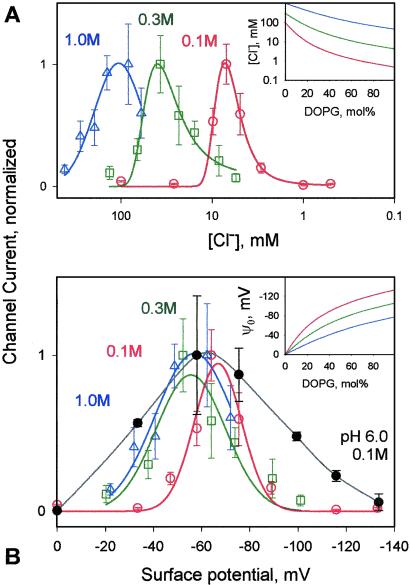
The sharp peak of the P178 channel current as a function of anionic lipid content and the steep decrease in colicin channel current for [L−] > [L−]max (Fig. 2B) are unprecedented in the literature on membrane–protein interactions. Because the colicin E1 ion channel is known to be selective for monovalent anions at acidic pH (30), the phase of decreasing channel current for |ψo| > |ψ | at each ionic strength might be explained by the decrease in the Cl− concentration in the near-membrane space that accompanies larger negative surface potentials (Fig. 4A Inset, [Cl−] vs. [L−] calculated using Guoy-Chapman theory). This possibility could be excluded because colicin channel currents measured at the three ionic strengths were not correlated with a unique or even similar Cl− concentration (Fig. 4A).
| at each ionic strength might be explained by the decrease in the Cl− concentration in the near-membrane space that accompanies larger negative surface potentials (Fig. 4A Inset, [Cl−] vs. [L−] calculated using Guoy-Chapman theory). This possibility could be excluded because colicin channel currents measured at the three ionic strengths were not correlated with a unique or even similar Cl− concentration (Fig. 4A).
Figure 4.
Dependence of colicin E1 channel activity on surface potential and Cl− concentration in near-membrane space as a function of ionic strength. (A) Macroscopic steady-state current in planar bilayers as a function of Cl− concentration in the near-membrane space calculated using Gouy-Chapman theory (Inset). (B) Steady-state current in planar bilayers as a function of surface potential calculated using Gouy-Chapman theory (Inset), using data from Fig. 2. Ionic strengths, 0.1 M (○, red), 0.3 M (□, green), or 1.0 M (▵, blue) at pH 4.0, and 0.1 M (●, black) at pH 6.0.
However, there was a correlation between the Imax generated by P178 and a narrow range of values of ψ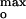 obtained over the wide range of ionic strengths, 0.1–1.0 M (Fig. 4B). Imax at 25, 30–35, and 60–70 mol% DOPG in 0.1, 0.3, and 1.0 M KCl, respectively (Fig. 2B), corresponded to surface potentials, ψ
obtained over the wide range of ionic strengths, 0.1–1.0 M (Fig. 4B). Imax at 25, 30–35, and 60–70 mol% DOPG in 0.1, 0.3, and 1.0 M KCl, respectively (Fig. 2B), corresponded to surface potentials, ψ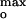 = −65 ± 2, −56 ± 3, and −59 ± 3 mV (Fig. 4B Inset). At pH 6.0 and 0.1 M KCl, Imax at 20 mol% corresponded to ψ
= −65 ± 2, −56 ± 3, and −59 ± 3 mV (Fig. 4B Inset). At pH 6.0 and 0.1 M KCl, Imax at 20 mol% corresponded to ψ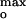 = −58 ± 3 mV (Fig. 4B, black). The average value of the surface potential at pH 4.0 corresponding to the current maxima at the three ionic strengths is ψ
= −58 ± 3 mV (Fig. 4B, black). The average value of the surface potential at pH 4.0 corresponding to the current maxima at the three ionic strengths is ψ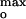 = −60 ± 5 mV (Fig. 4B). The channel current was much smaller when the surface potential ψo ≠ ψ
= −60 ± 5 mV (Fig. 4B). The channel current was much smaller when the surface potential ψo ≠ ψ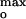 ; for example, at 0.1 M ionic strength, the channel current was effectively zero at |ψo| < 40 mV and > 90 mV. Thus, the surface potential appears to be “tuned” to allow a significant amplitude of the colicin channel current that results from effective import of the channel polypeptide. The rate of increase of the colicin channel current was found to have a similar dependence (data not shown). At optimum tuning, ψ
; for example, at 0.1 M ionic strength, the channel current was effectively zero at |ψo| < 40 mV and > 90 mV. Thus, the surface potential appears to be “tuned” to allow a significant amplitude of the colicin channel current that results from effective import of the channel polypeptide. The rate of increase of the colicin channel current was found to have a similar dependence (data not shown). At optimum tuning, ψ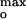 = −60 mV corresponds to a favorable free energy of electrostatic interaction of the channel domain with the membrane surface, ΔGel = −9.7 ± 0.8 kcal/mol, with P178 having an effective electrical charge of +7 at pH 4.0 (22). The interaction energy of the bound polypeptide with the membrane surface is, in first approximation, proportional to the membrane surface potential, ψo. As a bound polyvalent ion, the channel polypeptide has a larger affinity with lipid headgroups than do monovalent ions. This is seen by comparison of its Kd for monovalent cations (>0.1 M), and the high affinity (Kd < 10 nM) for the membrane surface under these conditions (22).
= −60 mV corresponds to a favorable free energy of electrostatic interaction of the channel domain with the membrane surface, ΔGel = −9.7 ± 0.8 kcal/mol, with P178 having an effective electrical charge of +7 at pH 4.0 (22). The interaction energy of the bound polypeptide with the membrane surface is, in first approximation, proportional to the membrane surface potential, ψo. As a bound polyvalent ion, the channel polypeptide has a larger affinity with lipid headgroups than do monovalent ions. This is seen by comparison of its Kd for monovalent cations (>0.1 M), and the high affinity (Kd < 10 nM) for the membrane surface under these conditions (22).
Determination of Conformational Flexibility of Membrane-Bound P178 as a Function of ψo by Using FRET.
The conserved free energy of the P178-membrane surface electrostatic interaction at the current maximum, Imax, at different ionic strengths implies a specific structural requirement for this interaction and for membrane insertion of P178. The dependence of the channel current on ψo for |ψo| < 55 mV (Figs. 2B and 3B) is attributed to electrostatic interactions required for unfolding of soluble P178 on the membrane surface (9, 22, 23) to form the precursor (Fig. 1B) of the inserted open channel (Fig. 1C). For |ψo| > 65 mV, the increased strength of the protein–membrane electrostatic interactions may constrain degrees of freedom of the protein that are necessary for insertion.
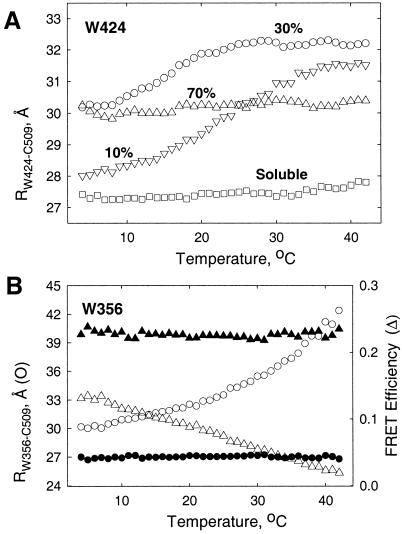
The distances between Trp- and Cys-AEDANS in single-Trp/single-Cys mutants, RWi-C509—distance from single tryptophan residue to Cys509; Wi, single Trp mutants of P178 (e.g., W356, W424)—has previously been shown to undergo reversible “thermal melting” over the temperature range 10–40°C (23). Distances were measured by FRET, using the single-Trp W424 (helix V) and W356 (helix I) mutants of the channel polypeptide (Fig. 5) as energy donors, with dye (AEDANS)-labeled single Cys-509 (helix IX) as the energy transfer acceptor (9, 23). The 2.5-Å resolution x-ray structure of the channel domain (15) provided a reference to normalize the intraprotein distances, and a W− mutant was used to determine and correct for the background contribution of nine Tyr to the FRET (9). This resulted in a FRET-determined RW424-C509 of 27 Å at 20°C, similar to the distance from the Cζ of W424 to Cγ of D509 determined in the wild-type protein by x-ray diffraction (9, 15). The FRET efficiency from tyrosine residues to AEDANS in the W− mutant was found to be independent of temperature (data not shown). Thus, the temperature-dependent distance changes seen in the presence of 10% and 30%, but not 70%, anionic lipid content (Fig. 5) arise from changes in distances between Trp residues and C509.
Figure 5.
Intra-residue distances, R, of P178 on the membrane surface as a function of anionic lipid content, R(L), and temperature, R(T). FRET efficiency, E, and intraprotein distances, RWi-C509 (9, 24), were measured between Trp-424 in helices V (A) or Trp-356 in helix I (B) and AEDANS-labeled Cys-509 in helix IX (15). Reversible temperature-dependent distance changes obtained from FRET efficiencies were corrected for contribution of nine Tyr residues by using a W− mutant. Buffer: 20 mM NaOAc/0.1 M KCl, pH 4.0. Heating rate: 0.5°/min. (A) R(T) function for soluble W424 mutant (□), or bound to LUV with 10 mol% (ψo = −32 mV; ▿), 30 mol% (ψo = −75 mV; ○), or 70 mol% (ψo = −115 mV; ▵) DOPG. (B) R(T) (circles) and E(T) (triangles) functions for W356 mutant bound to LUV with 30 mol% (ψo = −75 mV; open symbols) or 70 mol% (ψo = −115 mV; filled symbols) DOPG.
The extent of the thermal melting was found in the present study to depend on the anionic lipid content (Fig. 5). The increase in R from 4 to 42°C was ΔRW424-C509 = 3.3 ± 0.6, 1.9 ± 0.5, and 0.4 ± 0.3 Å (Fig. 5A; averages calculated from three trials) for [L−] = 10 (ψo = −32 mV), 30 (ψo = −75 mV), and 70 mol% (ψo = −115 mV), respectively. ΔRW356-C509 = 13 Å and 0 Å (Fig. 5B, blue and green) for L− = 30 and 70 mol%, respectively. The distance changes were completely reversible. Moreover, as manifested by the absence of temperature-dependent distance changes, P178 bound to membranes containing 70 mol% anionic lipid (ψo = −115 mV) shows no indication of flexibility. The absence of thermal melting implies that large values of ψo impose major conformational constraints on the membrane-bound protein. This presumably limits the mechanical freedom that is necessary for its insertion into the membrane bilayer (Fig. 1 B and C). The absence of thermal melting of soluble P178 and of P178 bound to 70 mol% anionic lipid membranes serves as a control to show that the thermal effects are not caused by possible temperature-dependent changes in the quantum yield of tryptophan or AEDANS.
Discussion
Formation of the Colicin Channel; Role of Protein Import.
Voltage-gated opening of the ion channel of the pore-forming colicins is known to be mechanistically different from that of the classical integral membrane channels. Whereas gating of the latter involves small conformational changes of the transmembrane segments (31), gating of colicin channels involves import of a large part of the protein from the membrane surface into and across the hydrophobic core of the bilayer (refs. 9, 16–18, and 23; see Fig. 1). It is concluded in the present study that the complex dependence of colicin E1 channel current on anionic lipid content and membrane surface potential is a consequence of the effects on protein import of electrostatic interactions between the colicin channel domain and the membrane surface.
Narrow Bandwidth Dependence of Channel Current on Anionic Lipid Content.
The increase of channel current, I, with increasing [L−] at low [L−] (Fig. 2), can be understood in terms of a requirement for a minimum electrostatic interaction with the membrane surface to achieve proper unfolding. For example, when |ψo| < |ψ | at 10 mol% DOPG, where interhelix interactions may dominate those between the protein helices and the membrane surface, the unfolding of P178 is incomplete. This is indicated by the smaller value of RW424-C509, which at 4°C = 28 Å for 10 mol% DOPG vs. 30 Å at 30 mol% (Fig. 5A).
| at 10 mol% DOPG, where interhelix interactions may dominate those between the protein helices and the membrane surface, the unfolding of P178 is incomplete. This is indicated by the smaller value of RW424-C509, which at 4°C = 28 Å for 10 mol% DOPG vs. 30 Å at 30 mol% (Fig. 5A).
The surface-bound precursor of the inserted open channel is an extended two-dimensional helical array that is stabilized by electrostatic interactions between elongated helices and the membrane surface, and by interhelix interactions (23). The pronounced inhibitory effect of membrane surface charge on protein import for |ψo| > |ψ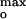 | correlates with an increase in electrostatic interactions of the basic residues of hydrophilic or amphipathic helices with the anionic membrane surface. The attractive electrostatic interactions between the many basic residues results in local kinetic barriers to the surface flexibility that is necessary for protein insertion. The interaction of each Lys residue contributes ∼1 kcal/mol at [L−] = 33 mol% (32), which creates a barrier to the movement of the Lys away from the anionic surface. However, because insertion is a cooperative event involving movement of all protein segments on the surface, the total barrier energy is the sum of the individual interaction energies. For [L−] above that required to initiate unfolding, this results in a large dependence of the activation energy for insertion on [L−] and ψο, and thus in small steady-state currents. It is important to note that the dependence of colicin channel current on anionic lipid content is qualitatively the same as previously obtained for the dependence of colicin-induced solute efflux from liposomes (22).
| correlates with an increase in electrostatic interactions of the basic residues of hydrophilic or amphipathic helices with the anionic membrane surface. The attractive electrostatic interactions between the many basic residues results in local kinetic barriers to the surface flexibility that is necessary for protein insertion. The interaction of each Lys residue contributes ∼1 kcal/mol at [L−] = 33 mol% (32), which creates a barrier to the movement of the Lys away from the anionic surface. However, because insertion is a cooperative event involving movement of all protein segments on the surface, the total barrier energy is the sum of the individual interaction energies. For [L−] above that required to initiate unfolding, this results in a large dependence of the activation energy for insertion on [L−] and ψο, and thus in small steady-state currents. It is important to note that the dependence of colicin channel current on anionic lipid content is qualitatively the same as previously obtained for the dependence of colicin-induced solute efflux from liposomes (22).
The absence of thermally induced flexibility of P178 bound to membranes with [L−] = 70 mol% (ψo = −115 mV; Fig. 5) implies that the decrease of current for [L−] > [L−]max and |ψo| > −65 mV relates to a loss of conformational freedom of the surface-bound channel domain that is necessary for translocation of the protein from the membrane surface into the bilayer (Fig. 1). It appears that the channel activity is associated with flexibility of the surface-bound precursor state that is obligatory for protein insertion into the membrane. The similar dependence of the colicin channel current on membrane surface potential as a function of ionic strength implies that a proper balance of interhelix and helix–membrane interactions is attained at a surface potential that corresponds to ψo ≈ −60 mV in the absence of protein.
Content of Anionic Lipid in Biological Membranes.
The channel domain of colicin E1 targets the cytoplasmic membrane of E. coli, whose anionic lipid content, PG and cardiolipin, is 20–30 mol% (33–37). This content is similar to the [L−]max in the present study. The polar lipid component of the cytoplasmic membrane of E. coli is typical for phospholipid membranes of mesophilic microorganisms (15–40 mol% of the total polar lipid content; refs. 33, 37, and 38), although the magnitude of the surface potential can be altered by bound protein and nonpolar lipids. For example, the plasma membrane of the unicellular eukaryote Saccharomyces cerevisiae has a higher [L−] than its subcellular membranes, ∼50 and 15–35 mol%, respectively. However, it is highly enriched in ergosterol (ergosterol:phospholipid, 3.3 and 0–0.7 mol/mol; ref. 38).
Physiological Response of Cells to Increase of Salinity in Media.
The magnitude of ψo is also influenced by salt concentration. Microorganisms respond to increased salinity by increasing the [L−] of the cytoplasmic membrane (39–41). An increase in PG and/or cardiolipin is compensated by a concomitant decrease in PE content. The same response to increased salt concentration in the media is observed in halophilic archaebacteria that grow at extremely high salinity. Halophiles respond to increased salinity by increasing the content of PGP−3 at the cost of PG−1 (42, 43).
Cellular Tuning of Surface Potential for Regulation of Protein Import and Other Membrane Protein Functions.
These regulatory reactions to changes in salinity achieve an approximately constant ψο. Anionic membrane lipids are involved in the regulation of many intracellular processes: initiation of DNA replication (44), cotranslation targeting of proteins (45), signal transduction (32). Thus, in addition to ensuring proper function of membrane-bound biosynthetic, transport, and signaling proteins, the cellular rationale to tune ψo may be the facilitation of efficient protein import.
Acknowledgments
We thank M. Hasson, M. Lindeberg, M. Sherman, and H. Weiner for helpful discussions, and Ms. Carol Greski for assistance with the manuscript. This work was supported by National Institutes of Health Grant GM-18457, The Henry Koffler Professorship (W.A.C.), and Fogarty Grant TW001235.
Abbreviations
- FRET
fluorescence resonance energy transfer
- AEDANS
5-[[[(2-iodoacetyl) amino]ethyl]amino]naphthalene-1-sulfonic acid
- P178
C-terminal 178-residue colicin E1 channel-forming polypeptide
- DOPG
(dioleoyl)-phosphatidylglycerol
- [L−]
anionic lipid content
- PGP
phosphatidylglycerol-phosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McLaughlin S. Annu Rev Biophys Biophys Chem. 1989;18:113–136. doi: 10.1146/annurev.bb.18.060189.000553. [DOI] [PubMed] [Google Scholar]
- 2.Driessen A J M, Manting E H, van der Does C. Nat Struct Biol. 2001;8:492–498. doi: 10.1038/88549. [DOI] [PubMed] [Google Scholar]
- 3.Deisenhofer J, Michel H. EMBO J. 1989;8:2149–2169. doi: 10.1002/j.1460-2075.1989.tb08338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Killian J A, von Heijne G. Trends Biochem Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 5.Wimley W C, White S H. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 6.von Heijne G. Annu Rev Biophys Biomol Struct. 1994;23:167–192. doi: 10.1146/annurev.bb.23.060194.001123. [DOI] [PubMed] [Google Scholar]
- 7.Krishtalik L I, Cramer W A. FEBS Lett. 1995;369:140–143. doi: 10.1016/0014-5793(95)00756-y. [DOI] [PubMed] [Google Scholar]
- 8.van Klompenburg W, de Kruijff B. J Membr Biol. 1998;162:1–7. doi: 10.1007/s002329900336. [DOI] [PubMed] [Google Scholar]
- 9.Lindeberg M, Zakharov S D, Cramer W A. J Mol Biol. 2000;295:679–692. doi: 10.1006/jmbi.1999.3396. [DOI] [PubMed] [Google Scholar]
- 10.Lazdunski C J, Bouveret E, Rigal A, Journet L, Lloubes R, Benedetti H. J Bacteriol. 1998;180:4993–5002. doi: 10.1128/jb.180.19.4993-5002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakes K J, Kienker P J, Finkelstein A. Q Rev Biophys. 1999;32:189–205. doi: 10.1017/s0033583599003492. [DOI] [PubMed] [Google Scholar]
- 12.Lakey J H, Slatin S L. Curr Top Microbiol Immunol. 2001;257:131–161. doi: 10.1007/978-3-642-56508-3_7. [DOI] [PubMed] [Google Scholar]
- 13.Bullock J O, Cohen F S, Dankert J R, Cramer W A. J Biol Chem. 1983;258:9908–9912. [PubMed] [Google Scholar]
- 14.Gould J M, Cramer W A. J Biol Chem. 1977;252:5491–5497. [PubMed] [Google Scholar]
- 15.Elkins P, Bunker A, Cramer W A, Stauffacher C V. Structure (London) 1997;5:443–458. doi: 10.1016/s0969-2126(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 16.Qiu X-Q, Jakes K S, Kienker P, Finkelstein A, Slatin S L. J Gen Physiol. 1994;107:313–328. doi: 10.1085/jgp.107.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kienker P K, Jakes K S, Finkelstein A. J Gen Physiol. 2000;116:587–597. doi: 10.1085/jgp.116.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrill A R, Cramer W A. Biochemistry. 1990;29:8529–8534. doi: 10.1021/bi00489a004. [DOI] [PubMed] [Google Scholar]
- 19.Wiener M, Freymann D, Ghosh P, Stroud R M. Nature (London) 1997;385:461–464. doi: 10.1038/385461a0. [DOI] [PubMed] [Google Scholar]
- 20.Vetter I R, Parker M W, Tucker A D, Lackey J H, Pattus F, Tsernoglou D. Structure. 1998;6:863–874. doi: 10.1016/s0969-2126(98)00088-4. [DOI] [PubMed] [Google Scholar]
- 21.Parker M W, Postma J P M, Pattus F, Tucker A D, Tsernoglou D. J Mol Biol. 1992;224:639–657. doi: 10.1016/0022-2836(92)90550-4. [DOI] [PubMed] [Google Scholar]
- 22.Zakharov S D, Heymann J B, Zhang Y-L, Cramer W A. Biophys J. 1996;70:2774–2783. doi: 10.1016/S0006-3495(96)79847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zakharov S D, Lindeberg M, Griko Y, Salamon Z, Tollin G, Prendergast F G, Cramer W A. Proc Natl Acad Sci USA. 1998;95:4282–4287. doi: 10.1073/pnas.95.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zakharov S D, Lindeberg M, Cramer W A. Biochemistry. 1999;38:11325–11332. doi: 10.1021/bi9903087. [DOI] [PubMed] [Google Scholar]
- 25.Bishop L J, Bjes E S, Davidson V L, Cramer W A. J Bacteriol. 1985;164:237–244. doi: 10.1128/jb.164.1.237-244.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller P, Rudin D O, Tien H, Wescott W C. J Phys Chem. 1963;67:534–535. [Google Scholar]
- 27.Shapovalov V, Tronin A. Langmuir. 1998;13:4870–4875. [Google Scholar]
- 28.Hope M J, Bally M B, Webb G, Cullis P R. Biochim Biophys Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 29.Steer B A, Merrill R A. Biochemistry. 1994;33:1108–1115. doi: 10.1021/bi00171a009. [DOI] [PubMed] [Google Scholar]
- 30.Shirabe K, Cohen F S, Xu S, Peterson A A, Shiver J W, Nakazawa A, Cramer W A. J Biol Chem. 1989;264:1951–1957. [PubMed] [Google Scholar]
- 31.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer; 2001. pp. 603–662. [Google Scholar]
- 32.Ben-Tal N, Honig B, Peitzsch R M, Denisov G, McLaughlin S. Biophys J. 1996;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gennis R B. Biomembranes: Molecular Structure and Function. New York: Springer; 1989. pp. 20–35. [Google Scholar]
- 34.Shibuya I. Prog Lipid Res. 1992;31:245–299. doi: 10.1016/0163-7827(92)90010-g. [DOI] [PubMed] [Google Scholar]
- 35.Morein S, Andersson A-S, Lindblom G. J Biol Chem. 1996;271:6801–6809. doi: 10.1074/jbc.271.12.6801. [DOI] [PubMed] [Google Scholar]
- 36.de Kruijff B, Killian A, Rietveld A G, Kusters R. Curr Top Membr. 1997;44:477–515. [Google Scholar]
- 37.Datta D B. A Comprehensive Introduction to Membrane Biochemistry. Madison, WI: Floral Publishing; 1987. p. 72. [Google Scholar]
- 38.Zinser E, Sperka-Gottlieb C D M, Fasch E-V, Kohlwein S D, Paltauf F, Daum G. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanna K, Bengis-Garber C, Kushner D J, Kogut M, Kates M. Can J Microbiol. 1984;30:669–675. [Google Scholar]
- 40.Miller K. J Bacteriol. 1985;162:263–270. doi: 10.1128/jb.162.1.263-270.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vreeland R H, Anderson R, Murray R G E. J Bacteriol. 1984;160:879–883. doi: 10.1128/jb.160.3.879-883.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langworthy T A. In: The Bacteria: Archaebacteria. Woese C R, Wolfe R S, editors. Vol. 8. New York: Academic; 1985. pp. 459–497. [Google Scholar]
- 43.Nicolaus B, Lanzotti V, Trincone A, De Rosa M, Grant W D, Gambacorta A. FEMS Microbiol Lett. 1989;59:157–160. [Google Scholar]
- 44.Xia W, Dowhan W. Proc Natl Acad Sci USA. 1995;92:783–787. doi: 10.1073/pnas.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Leeuw E, te Kaat K, Moser C, Menestrina G, Demel R, de Kruijff B, Oudega B, Luirink J, Sinning I. EMBO J. 2000;19:531–541. doi: 10.1093/emboj/19.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]