Abstract
Andersen's syndrome, an autosomal dominant disorder related to mutations of the potassium channel Kir2.1, is characterized by cardiac arrhythmias, periodic paralysis, and dysmorphic bone structure. The aim of our study was to find out whether heteromerization of Kir2.1 channels with wild-type Kir2.2 and Kir2.3 channels contributes to the phenotype of Andersen's syndrome. The following results show that Kir2.x channels can form functional heteromers: (i) HEK293 cells transfected with Kir2.x–Kir2.y concatemers expressed inwardly rectifying K+ channels with a conductance of 28–30 pS. (ii) Expression of Kir2.x–Kir2.y concatemers in Xenopus oocytes produced inwardly rectifying, Ba2+ sensitive currents. (iii) When Kir2.1 and Kir2.2 channels were coexpressed in Xenopus oocytes the IC50 for Ba2+ block of the inward rectifier current differed substantially from the value expected for independent expression of homomeric channels. (iv) Coexpression of nonfunctional Kir2.x constructs, in which the GYG region of the pore region was replaced by AAA, with wild-type Kir2.x channels produced both homomeric and heteromeric dominant-negative effects. (v) Kir2.1 and Kir2.3 channels could be coimmunoprecipitated in membrane extracts from isolated guinea pig cardiomyocytes. (vi) Yeast two-hybrid analysis showed interaction between the N- and C-terminal intracellular domains of different Kir2.x subunits. Coexpression of Kir2.1 mutants related to Andersen's syndrome with wild-type Kir2.x channels showed a dominant negative effect, the extent of which varied between different mutants. Our results suggest that differential tetramerization of the mutant allele of Kir2.1 with wild-type Kir2.1, Kir2.2, and Kir2.3 channels represents the molecular basis of the extraordinary pleiotropy of Andersen's syndrome.
Recently, Andersen's syndrome (1) has been added to the growing list of inherited disorders related to mutations of ion channels, which are also referred to as channelopathies (2). Andersen's syndrome is a rare autosomal dominant disease characterized by cardiac arrhythmias, periodic paralysis, and dysmorphic bone structure such as scoliosis (curvature of the spine), low-set ears, wide-set eyes, small chin, and broad forehead (3–5), but expression of these traits is highly variable. The disorder has been attributed to mutations in the inward rectifier potassium channel Kir2.1 because examination of 16 affected families showed mutations in the coding region of Kir2.1 in 13 cases (1).
On the basis of sequence homology inward rectifier K+ (Kir) channels have been classified into subfamilies Kir1 to Kir7 (6–9). The potassium channels with the most pronounced inward rectification belong to the Kir2.x subfamily, which consists of four members (Kir2.1–Kir2.4). In many cell types, including neurons and cardiac muscle cells, Kir2 channels play an important role in setting the membrane potential and modulating excitability (7). In the heart, inward rectifier channels are particularly important because they largely determine the shape of the cardiac action potential (10–13). During the plateau of the action potential the inward rectifier channels are mostly closed, thus reducing the inward current required to maintain depolarization. During the terminal phase of repolarization and during diastole the inward rectifier channels provide the dominant membrane conductance. Reduction of inward rectifier current is expected to increase the propensity for arrhythmias (1, 14).
Expression of three different inward rectifier channel subunits, Kir2.1, Kir2.2, and Kir2.3, has been found in human heart (15–17). Recent cell-specific reverse transcriptase–PCR experiments have shown that these three subunits are coexpressed in ventricular cardiomyocytes of the guinea pig, whereas expression of Kir2.4 is restricted to neuronal cells in the heart (18). It is generally accepted that Kir subunits form tetrameric channels, but the question whether members of the Kir2 subfamily can form heteromers has been controversial for many years (19–21), and definite proof for functional heteromerization is still lacking.
The aim of the present study was to find out whether heteromerization of the mutant Kir2.1 channels found in Andersen's syndrome with Kir2.2 and Kir2.3 may contribute to the extraordinary variability of the patients' phenotype. To clarify this question we first sought to establish unequivocally whether heterologously expressed channels of the Kir2 subfamily can form heteromers. Using dominant-negative mutants of various Kir2 subunits, Kir2.x-Kir2.y concatemers, coimmunoprecipitation, and yeast two-hybrid analysis, we obtained evidence for heteromerization of Kir2 channels. Coexpression of the mutants of Kir2.1 responsible for Andersen's syndrome with Kir2.2 and Kir2.3 subunits revealed a strong dominant-negative effect. Our results suggest that in patients with Andersen's syndrome not only the current carried by the inward rectifier channel Kir2.1 but also the current carried by Kir2.2 and Kir2.3 is suppressed in cardiac muscle, skeletal muscle, and possibly other cell types.
Materials and Methods
DNA Constructs.
The coding regions of guinea pig Kir2 (18) and human Kir2.1 and Kir2.3 cDNAs were cloned into the Xenopus oocyte expression vector pSGEM (gift of M. Hollmann, Ruhr University, Bochum, Germany) or the mammalian expression vector pcDNA3.1 (Invitrogen). The QuikChange site-directed mutagenesis kit (Stratagene) was used to mutate GYG motif in the pore region or introduce mutations related to Andersen's syndrome (1). To construct Kir2.x-Kir2.y concatemers we introduced HindIII sites in-frame after the mutated stop codons (TAG → TGG) of the Kir2.x subunit and in front of the coding region of the Kir2.y subunit (containing the last three base pairs of the 5′ noncoding region) and ligated both subunits at this restriction site. Thus, the linker of the two subunits consisted of four amino acids (WKLA). All mutations and DNA constructs were confirmed by sequencing.
Heterologous Expression in Xenopus Oocytes and HEK293 Cells.
Preparation of oocytes, mRNA injection, and electrophysiological measurements have been described (18, 22). Two-microelectrode voltage-clamp experiments were performed 12 h after injection of cRNA. The oocytes were superfused with a solution (KD60) containing 60 mM KCl, 38 mM NaCl, 1.8 mM CaCl2, 2 mM MgCl2, and 5 mM Hepes, titrated to pH 7.4 with NaOH.
HEK293 cells were transfected with 1 μg of Kir2.x–Kir2.y cDNA in the pCiNeo vector (gift of J. Eggermont, Catholic University, Leuven, Belgium) by using Lipofectamine 2000 (Life Technologies, Grand Island, NY), as described (18). Whole-cell measurements were carried out with a pipette solution containing 65 mM K-glutamate, 50 mM KCl, 10 mM KH2PO4, 7.9 mM MgCl2, 5 mM EDTA, 5 mM Hepes, 1.9 mM K2ATP, and 0.2 mM Na3GTP (pH 7.2) and a bath solution containing 85 mM NaCl, 60 mM KCl, 1 mM MgCl2, 5 mM Hepes, 0.33 mM NaH2PO4, and 10 mM glucose (pH 7.4). Cell-attached measurements were carried out with a pipette solution containing 145 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 5 mM Hepes and a bath solution containing 135 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 0.33 mM NaH2PO4, 10 mM Hepes, and 10 mM glucose (pH 7.4). The temperature was 20–22°C.
Immunoprecipitation and Western Blotting.
Polyclonal Kir2.3 antibodies were prepared against the less conserved C terminus of the rat ortholog [amino acids 372–439; GenBank accession no. X87635 (23)], expressed as recombinant protein with pGEX-4T-1 as vector, affinity-purified, and characterized as detailed for Kv subunits (24). Affinity-purified antibodies for Kir2.1 were purchased from Alomone (Jerusalem). HEK293 cells were cultured in 60-mm dishes and transfected with 5 μg plasmid DNA per dish with Lipofectamine. The transfected cells were washed with PBS and collected by centrifugation about 24 h after transfection. Cardiomyocytes were isolated from guinea pig heart by collagenase digestion (18). The membrane preparation was done according to Zeng and Wess (25). The solubilized membrane proteins were either resolved on 8% (wt/vol) acrylamide gels (6 μg protein per lane) and electroblotted to nitrocellulose membrane or used for immunoprecipitation. Immunoprecipitation was performed with Dynabeads-Protein A conjugates (Dynal, Hamburg, Germany) and 15 or 50 μg of membrane proteins from transfected HEK293 cells or cardiomyocytes according to the manufacturer's instructions. Staining of the Western blot was done with 1:500 primary antibody dilution and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate.
Yeast Two-Hybrid Interaction.
Protein–protein interactions were monitored with the yeast Matchmaker two-hybrid system from CLONTECH. EcoRI and BamHI sites were incorporated by PCR into the 5′ and 3′ ends, respectively, of each Kir2 subunit fragment, for cloning in-frame to the yeast shuttle vectors pGBT9 and pGAD424. The following N- and C-terminal regions and deletion constructs (without the two transmembrane domains and the pore region) of the Kir2 subunits were made: N-2.1 (amino acids 1–86 of Kir2.1), N-2.2 (amino acids 1–85 of Kir2.2), N-2.3 (amino acids 1–60 of Kir2.3), C-2.1 (amino acids 180–427 of Kir2.1), C-2.2 (amino acids 181–433 of Kir2.2), C-2.3 (amino acids 171–443 of Kir2.3), NC-2.1 (replacement of amino acids 87–177 of Kir2.1 with four alanines), NC-2.2 (replacement of amino acid 86–178 of Kir2.2 with four alanines), and NC-2.3 (replacement of amino acids 61–169 of Kir2.3 with five alanines). As a positive control we used PDZ domains 1–3 of PSD-95/SAP90 (amino acids 1–401) in pGAD424 vector (26). Protein–protein interactions were tested in the host yeast strain HF7C by cotransformation with pairs of pGBT9 and pGAD424 fusion constructs according to the manufacturer's protocol. Transformations in HF7C were plated on medium lacking tryptophan, leucine, and histidine and incubated for 4–6 days at 30°C. Yeast transformed with interacting fusion proteins grow as a result of activation of the HIS3 gene. The β-galactosidase activity assay on filter paper was performed according to the manufacturer's protocol (CLONTECH).
Results
Expression of Kir2.x–Kir2.y Concatemers.
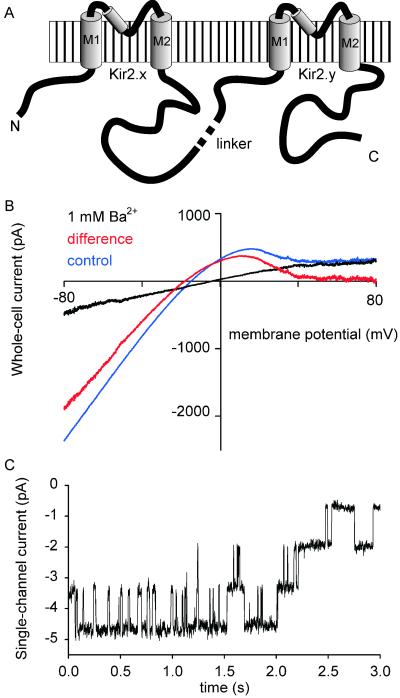
Different members of the Kir2 subfamily of potassium channels are coexpressed in cardiac ventricular muscle (16, 18, 27–30), skeletal muscle (28, 31, 32), neurons of some brain regions (33–35), and several other cell types. To test whether guinea pig Kir2.x subunits are able to form functional heteromeric channels we constructed concatemers with four transmembrane domains, in which two different Kir2 subunits were connected (Fig. 1A). Expression of Kir2.x–Kir2.y concatemers in HEK293 cells produced inwardly rectifying currents that could be blocked by 1 mM Ba2+ (n = 17; Fig. 1B). Cell-attached measurements in symmetrical K+ solution (Fig. 1C) showed inwardly rectifying channels with properties similar to wild-type Kir channels, as illustrated in Fig. 1C and Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org. The mean conductance was 30.0 ± 0.6 pS (n = 4) for Kir2.2–Kir2.1 and 30.1 ± 0.5 pS (n = 5) for Kir2.1–Kir2.2 concatemers; the lumped mean value was 30.0 ± 0.4 pS (n = 9). Kir2.1–Kir2.3 concatemers showed a mean conductance of 28.1 ± 1.5 pS (n = 3).
Figure 1.
Expression of Kir2.2–Kir2.1 concatemers in HEK293 cells. (A) Schematic diagram of Kir2.x–Kir2.y concatemers. (B) Current–voltage relation of Kir2.2–Kir2.1 expressed in HEK293 cells; blue, control; black, with 1 mM Ba2+; red, difference curve. (C) Cell-attached recording (at −60 mV) of single inwardly rectifying channels, carried out 24 h after transfection of HEK293 cells with Kir2.2–Kir2.1 concatemers.
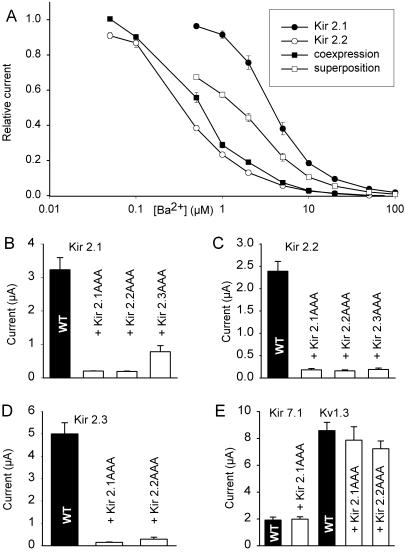
All possible four transmembrane domains–concatemers of Kir2.1, Kir2.2, and Kir2.3 were expressed in Xenopus laevis oocytes. Because homomeric Kir2 channels differ substantially in their sensitivity to Ba2+ block (18) the concentration dependence of Ba2+ block in the steady state was studied in the different concatemers. We found that the Ba2+ sensitivity of Kir2.x–Kir2.y concatemers was not statistically different from that of the alternative concatemers, Kir2.y–Kir2.x, and therefore the data of the two alternative concatemers of each combination were lumped. The data summarized in Table 1 show that the IC50 value for Ba2+ block of the concatemers was near the IC50 value of the subunit with higher Ba2+ sensitivity.
Table 1.
Inhibition of inward rectifier currents in Xenopus oocytes by Ba2+
| Channel | IC50, μM | SEM | n |
|---|---|---|---|
| Homomeric channels | |||
| Kir2.1 | 3.2 | 0.36 | 5 |
| Kir2.2 | 0.5 | 0.04 | 5 |
| Kir2.3 | 10.3 | 0.61 | 5 |
| Concatemers | |||
| Kir2.1–Kir2.2 or Kir2.2–Kir2.1 | 0.68 | 0.04 | 10 |
| Kir2.1–Kir2.3 or Kir2.3–Kir2.1 | 3.39 | 0.48 | 7 |
| Kir2.2–Kir2.3 or Kir2.3–Kir2.2 | 1.73 | 0.29 | 6 |
| Coexpression | |||
| Kir2.1 + Kir2.2 | 0.64 | 0.05 | 5 |
| Kir2.1 + Kir2.3 | 6.32 | 0.29 | 5 |
| Kir2.2 + Kir2.3 | 1.94 | 0.18 | 5 |
The IC50 values were measured after expression of Kir2.x subunits or Kir2.x–Kir2.y concatemers, and after coexpression of different Kir2.x channel subunits. To calculate the IC50 the data were fitted with the function IBa/Icon = 1/(1 + [Ba2+]/IC50), where IBa and Icon are the currents measured in the presence and absence of Ba2+, respectively.
Because inward rectifier channels form tetramers, association of two concatemers is required to form a functional channel. To verify this, we coexpressed normal (GYG) concatemers with nonfunctional (AAA) concatemers, in which one of the pore signature motifs GYG was replaced by AAA. Coinjection of cRNA of Kir2.2–Kir2.1–GYG and Kir2.2–Kir2.1–AAA concatemers at equal amounts (keeping the total amount of cRNA constant) reduced the amplitude of the current from 2.21 ± 0.17 μA (control; n = 17) to 0.61 ± 0.06 μA (n = 21), as illustrated in Fig. 7, which is published as supporting information on the PNAS web site. This reduction to 27.6% of control is close to the result expected for random association of the two concatemers (25%).
Coexpression of Different Kir2 Subunits.
To confirm that Kir2 subunits can form functional heteromeric channels we coinjected cRNA of different Kir2 channel subunits into Xenopus oocytes and studied their sensitivity to block by Ba2+ ions, which differs markedly between different subunits (18). Fig. 2A shows an example in which Kir2.1 and Kir2.2 were coexpressed. The injection of cRNA was titrated to give equal current amplitudes when either of the two subunits was expressed individually. The half-maximal inhibitory concentration (IC50) for Ba2+ block is lower for Kir2.2 (0.5 μM; ref. 18) than for Kir2.1 (3.2 μM; ref. 18). In the case of independent homomeric expression of the two subunits one would expect an intermediate IC50 for Ba2+ block. However, the IC50 of coexpressed Kir2.1/Kir2.2 channels differed substantially from the IC50 expected for independent expression of the two subunits; in fact, it was very close to the IC50 of Kir2.2 (see Table 1). These findings corroborate the idea that different Kir2 subunits can interact to form functional heteromeric channels.
Figure 2.
Coexpression of Kir2 channels in Xenopus oocytes. (A) Concentration dependence of Ba2+ block of the inward rectifier current of different Kir2 subunits expressed in Xenopus oocytes [○, Kir2.2 alone (18); ●, Kir2.1 alone (18)]; □, mean value of Kir2.1 and Kir2.2 currents [(IKir2.1 + IKir2.2)/2)]; ■, Ba2+ block observed after coinjection of Kir2.1 and Kir2.2 cRNA. Whole-cell currents were normalized with respect to the maximum current observed in the absence of Ba2+. (B–D) Coexpression of Kir2.x subunits with dominant-negative mutants (Kir2.xAAA) in Xenopus oocytes. Equal amounts of total mRNA were injected, either 100% Kir2.x (control) or 50% Kir2.x + 50% Kir2.xAAA. Wild-type (WT) channels are shown as black bars, the coexpressed dominant-negative mutants are indicated. (E) Coexpression of wild-type Kir7.1 and Kv1.3 with dominant-negative mutants of Kir2.1. The amplitude of inward rectifier currents was measured at −100 mV with an extracellular K+ concentration of 60 mM.
Coexpression with Dominant-Negative Constructs.
Dominant-negative mutants of Kir2 subunits were produced by replacing the GYG motif in the pore loop by AAA. When these mutants (denoted Kir2.xAAA) were expressed alone in Xenopus oocytes, no inward rectifier current was seen (not shown). Coexpression of the Kir2.xAAA mutants with wild-type Kir2.1, Kir2.2, or Kir2.3 (keeping the total amount of injected cRNA constant) reduced the amplitude of the currents to 2–8% of control (Fig. 2 B–D), except for one combination, Kir2.1 plus Kir2.3AAA (to 24%; Fig. 2B). In contrast, coexpression of Kir2.xAAA subunits with the voltage-activated K+ channel Kv1.3 or with the inward rectifier channel Kir7.1 had no dominant-negative effect (Fig. 2E). These findings confirm that Kir2.1, Kir2.2, and Kir2.3 subunits can form heteromers and indicate that the dominant-negative effect represents a specific interaction between Kir2.x channel subunits. The strong reduction of current observed after coexpression (Fig. 2 B–D) suggests that one dominant-negative subunit is sufficient to completely suppress channel function.
Coimmunoprecipitation of Kir2.1 and Kir2.3.
To examine the heteromerization of Kir2.1/Kir2.3 proteins more directly we carried out coimmunoprecipitation experiments with affinity-purified antibodies against Kir2.1 and Kir2.3. The antibodies were tested in membrane extracts of HEK293 cells transfected with Kir2.1 and/or Kir2.3. Western blot analysis showed that the antibodies were specific (Fig. 3 A and B) and also recognized Kir2.1 and Kir2.3 in membrane extracts from isolated cardiomyocytes. When membrane extracts of HEK293 cells cotransfected with Kir2.1 and Kir2.3 were precipitated with Kir2.3 antibodies the Kir2.1 protein could be detected in the precipitate (Fig. 3C). When the membrane extracts were precipitated with Kir2.1 antibodies the Kir2.3 protein could be detected in the precipitate (Fig. 3D). HEK293 cells transfected with only one of the Kir2 channel subunits served as negative controls. Fig. 3 C and D also shows that Kir2.1 and Kir2.3 subunits could be coimmunoprecipitated from membranes of isolated cardiac muscle cells. These findings confirm the idea that different Kir2.x subunits can coassemble in vitro and in vivo. However, it should be noted that both in HEK293 cells and cardiomyocytes Kir2.1 and Kir2.3 were not coprecipitated completely; up to 50% of the channel proteins remained in the supernatant (data not shown).
Figure 3.
Coimmunoprecipitation of Kir2.1 and Kir2.3 subunits. (A and B) Western blot analysis of membrane extracts from HEK293 cells transiently transfected with Kir2. 1, Kir2.3, or both, and of membrane extracts from isolated cardiomyocytes (CM). Note that the Kir2.1 protein of cardiomyocyte membranes migrated somewhat more slowly than that of transfected HEK293 cells. (C and D) Membrane extracts of HEK cells, transfected with Kir2.1, Kir2.3, or both, were precipitated with antibodies against Kir2.3 (C) or Kir2.1 (D) and subjected to Western blot analysis using antibodies against Kir2.1 (C) or Kir2.3 (D). IP, immunoprecipitation.
Yeast Two-Hybrid Analysis.
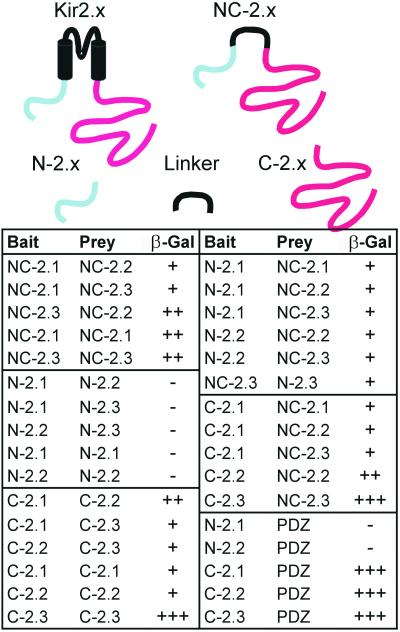
To find out whether the cytosolic domains of the channel proteins play a role in the heteromeric assembly we studied the interaction between the N- and C-terminal domains of Kir2.x subunits by using the yeast two-hybrid system. The transmembrane domains were deleted to facilitate nuclear translocation of the fusion proteins (Fig. 4). We used constructs containing only the amino-terminal cytosolic domains (N-2.x), constructs containing only the carboxyl-terminal cytosolic domains (C-2.x), and constructs in which the amino-terminal and carboxyl-terminal cytosolic domains were connected (NC-2.x). It was found that the amino-terminal domains alone did not interact, whereas the carboxyl-terminal domains interacted in all combinations (Fig. 4 and Fig. 8, which is published as supporting information on the PNAS web site). The NC-2.x constructs interacted with all other constructs. The strongest interaction detected was between bait and prey of C-2.3 constructs. Interaction was also found between the different NC-2.x constructs (NC-2.1/NC-2.2, NC-2.2/NC-2.3, and NC-2.1/NC-2.3). These findings suggest that the cytosolic domains participate in the assembly of heteromers.
Figure 4.
Interaction of Kir2.x channels studied with the yeast two-hybrid system. The schematic diagrams show the constructs used in the yeast two-hybrid screens; in all cases the transmembrane domains were deleted. Protein–protein interactions were tested in the host yeast strain HF7C by cotransformation with bait and prey vectors and subsequent plating on a selective medium (see Fig. 8). Yeast colonies (which appeared as red colored) were transferred to paper filters and tested for β-galactosidase (β-gal) activity. Colonies turning blue within 2 days were scored as positive (+, ++, or +++). Noninteracting fusion proteins did not produce blue color and were scored as negative (−). PDZ denotes domains 1–3 of PSD-95/SAP90 (amino acids 1–401).
Kir2 Mutations Related to Andersen's Syndrome.
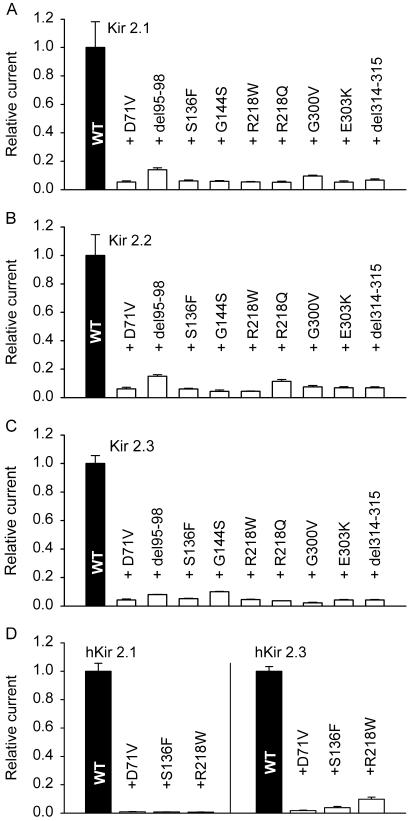
The nine mutations found in Andersen's syndrome (1) were introduced in guinea pig Kir2.1 (which differs from human Kir2.1 only in one amino acid). The mutant channels were coexpressed in Xenopus oocytes with wild-type guinea pig Kir2.1, Kir2.2, or Kir2.3 channels by injecting equal amounts of wild-type and mutant cRNA. The mutants all had a strong dominant-negative effect (Fig. 5 A–C), reducing the inward current measured at −100 mV to 2–15% of control (injection of wild-type Kir2.x alone). In view of the high degree of homology between orthologous Kir2.x channels it is likely that heteromerization in human Kir2.x channels occurs in the same way as in guinea pig. To verify this notion we coexpressed three human (h) Kir2.1 mutants related to Andersen's syndrome with wild-type hKir2.1 and hKir2.3 (Fig. 5D). When coinjected at equal amounts, the mutants reduced the current produced by hKir2.1 to very low values and reduced the current produced by hKir2.3 to values between 2% and 10% of control (illustrated in Fig. 9, which is published as supporting information on the PNAS web site), consistent with the results obtained with the guinea pig orthologs. Taken together, these findings indicate that the mutations related to Andersen's syndrome suppress the functional expression not only of Kir2.1 homotetramers but also of Kir2.x heteromers.
Figure 5.
Coexpression with Kir2.1 mutants related to Andersen's syndrome. (A–C) In each experiment, one of the wild-type (WT) Kir2.x subunits present in guinea pig heart was coexpressed with one of the guinea pig orthologs of the mutants related to Andersen's syndrome (1). The bars represent the mean ± SEM of 11–27 experiments. (D) Three hKir2.1 mutants related to Andersen's syndrome (D71V, S136F, and R218W) were coexpressed with wild-type hKir2.1 and hKir2.3. Equal amounts of wild-type and mutant cRNA were injected in each case (n = 18–25 for each mutant).
Discussion
The following findings indicate that Kir2 channels can form functional heteromers: (i) heterologous expression of Kir2.x–Kir2.y concatemers produced Ba2+-sensitive inward rectifier currents; (ii) cell-attached recordings showed inwardly rectifying channels after transfection of HEK293 cells with Kir2.x–Kir2.y concatemers; (iii) the IC50 for Ba2+ block of inward rectifier currents observed after coexpression of Kir2.1 and Kir2.2 channels differed substantially from the value expected for independent expression of homomeric channels; and (iv) Kir2.xAAA mutants produced both homomeric and heteromeric dominant-negative effects. Coinjection of equal amounts of wild-type and guinea pig Kir2.xAAA cRNA in most combinations reduced the amplitude of the current measured in Xenopus oocytes to 2–10% as compared with control, which suggests that one mutated (AAA) subunit in a tetramer may be sufficient to completely suppress channel function.
Our coprecipitation experiments showed that Kir2.1 and Kir2.3 channels associate not only in cotransfected HEK293 cells but also in isolated cardiomyocytes, indicating that heteromerization of Kir2.x channels also occurs in vivo. Previous work using an in vitro protein–protein interaction assay suggested that interaction between N and C termini of Kir6.x channels may contribute to tetrameric channel assembly (36). Our yeast two-hybrid analysis suggests that interaction of N- and C-terminal intracellular domains also takes place between different Kir2.x subunits. By analogy to the cytosolic tetramerization domain of voltage-activated K+ channels (37, 38), this interaction may play a role in homomeric and heteromeric assembly of Kir2.x channels.
Taken together, our results provide evidence for functional heteromerization of Kir2.x channels both in the expression systems and in vivo. Our findings resolve an issue that has been controversial for many years. Tinker et al. (20) failed to show a dominant-negative effect of Kir2.1AAA on Kir2.2 or Kir2.3 and concluded that functional heteromerization does not occur. However, in the same publication, coprecipitation of Kir2.1 and Kir2.2 subunits was reported, albeit to a limited extent. On the other hand, Fink et al. (19) found that some portions of Kir2.1 and Kir2.3 can interact to form functional channels. Using murine knockout models, Zaritsky et al. (21) have shown that elimination of Kir2.1 completely abolished the inward rectifier current in neonatal cardiomyocytes, whereas elimination of Kir2.2 reduced the inward rectifier current to about 50%, and it has been speculated that this surprising electrophysiological phenotype may also be attributable to functional heteromerization of Kir channel subunits (13, 21).
The identification of the molecular composition of the native channels found in different cell types is an important goal of cellular physiology. However, the correlation between cloned channels studied in heterologous expression systems and native channels studied in freshly isolated cells is notoriously difficult because of missing accessory channel subunits, local interacting proteins, differential glycosylation, or other influences of the cellular milieu. Our finding that the widely distributed channels of the Kir2 subfamily can form heteromers adds an additional complication because this may alter the biophysical characteristics of the channels. In cardiac muscle cells, for example, Kir2.1, Kir2.2, and Kir2.3 channels are coexpressed (18) and will most probably coassemble. Thus, there may be a multitude of different Kir2.x heteromers, and the wide range of conductances of inward rectifier channels observed in guinea pig (18), rat (39), mouse (40), and human (16) cardiomyocytes cells may be attributable to the different stoichiometry of the tetrameric channels.
The mutations of Kir2.1 found in patients with Andersen's syndrome have been reported to have a dominant-negative effect on currents carried by Kir2.1 (1). In the present study we show that Andersen mutations also suppress the currents carried by Kir2.2 and Kir2.3. This finding provides an explanation for the complex phenotype of Andersen's syndrome. Heterozygous Kir2.1 mutations will impair not only the function of Kir2 channels encoded by the wild-type allele of Kir2.1, but also the function of coexpressed Kir2.2 and/or Kir2.3, for example in cardiomyocytes (16, 18, 27–30) (which are responsible for arrhythmias), in skeletal muscle cells (28, 31, 32) (which are responsible for the periodic paralysis occasionally observed in Andersen's syndrome), and in neuronal cells of some regions of the brain (33–35). In osteoclasts [which may be responsible for the impairment of bone structure (14)] Kir2.1 has been identified (41), but expression of Kir2.2 and Kir2.3 has not yet been tested.
In conclusion, our results show that Kir2 channels can form functional heteromers. Our yeast two-hybrid analysis indicates that the N-terminal and the C-terminal domains of different Kir2.x channels can interact, and it is likely that this interaction is involved in the assembly of tetramers. If the process of homomeric and/or heteromeric assembly of Kir2.x channels is affected by the mutations associated with Andersen's syndrome, then different mutations might produce a large variety of phenotypes. At one extreme, the mutant subunits might assemble normally, and one mutated subunit may be sufficient to completely suppress channel function. In this case, the functional expression of all Kir2.x homotetramers and heterotetramers would be greatly reduced, resulting in a severe phenotype. At the other extreme, the mutant allele may not be able to participate in either homomeric or heteromeric assembly of Kir2.x channels. This type of mutation would only moderately reduce the number of functional channels containing Kir2.1 and would be expected to produce a mild phenotype. In the (probably more common) intermediate cases, the mutations may suppress channel function if coassembled, but may at the same time modulate the extent of homomeric and/or heteromeric assembly, which would be expected to produce a spectrum moderate to severe phenotypes. These examples illustrate that differential multimerization represents a mechanism by which different mutations of one gene can give rise to a variety of phenotypes. Indeed, the localization of the Andersen mutations on the channel protein is quite heterogeneous: one is located on the N terminus, one is a deletion in M1, two are in the pore loop, and five are on the C terminus (1), and the extent of the dominant-negative effect varied by at least a factor of 3 between different mutants and between different subunits. The data presented here suggest that differential tetramerization of different mutants of Kir2.1 represents the molecular basis of the extraordinary pleiotropy of Andersen's syndrome.
Supplementary Material
Acknowledgments
We thank Anette Hennighausen for technical support and Niels Decher for useful discussions. This work was supported by Deutsche Forschungsgemeinschaft (Da 177/7-3 and Ve 187/1-2), “Ernst und Berta Grimmke Stiftung,” “Karl und Lore Klein Stiftung,” and “P. E. Kempkes Stiftung.”
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Plaster N M, Tawil R, Tristani-Firouzi M, Canun S, Bendahhou S, Tsunoda A, Donaldson M R, Innaccone S T, Brunt E, Barohn R, et al. Cell. 2001;105:511–519. doi: 10.1016/s0092-8674(01)00342-7. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft F M, editor. Ion Channels and Disease. San Diego: Academic; 2000. [Google Scholar]
- 3.Canun S, Perez N, Beirana L G. Am J Med Genet. 1999;85:147–156. doi: 10.1002/(sici)1096-8628(19990716)85:2<147::aid-ajmg9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 4.Sansone V, Griggs R C, Meola G, Ptacek L J, Barohn R, Innaccone S, Bryan W, Baker N, Janas S J, Scott W, et al. Ann Neurol. 1997;43:305–312. doi: 10.1002/ana.410420306. [DOI] [PubMed] [Google Scholar]
- 5.Tawil R, Ptacek L J, Pavlakis S G, DeVivo D C, Penn A S, Ozdemir C, Griggs R C. Ann Neurol. 1994;35:326–330. doi: 10.1002/ana.410350313. [DOI] [PubMed] [Google Scholar]
- 6.Doupnik C A, Davidson N, Lester H A. Curr Opin Neurobiol. 1995;5:268–277. doi: 10.1016/0959-4388(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 7.Nichols C G, Lopatin A N. Annu Rev Physiol. 1997;59:171–191. doi: 10.1146/annurev.physiol.59.1.171. [DOI] [PubMed] [Google Scholar]
- 8.Töpert C, Döring F, Wischmeyer E, Karschin C, Brockhaus J, Ballanyi K, Derst C, Karschin A. J Neurosci. 1998;18:4096–4105. doi: 10.1523/JNEUROSCI.18-11-04096.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Döring F, Derst C, Wischmeyer E, Karschin C, Schneggenburger R, Daut J, Karschin A. J Neurosci. 1998;18:8625–8636. doi: 10.1523/JNEUROSCI.18-21-08625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimoni Y, Clark R B, Giles W R. J Physiol (London) 1992;448:709–727. doi: 10.1113/jphysiol.1992.sp019066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols C G, Makhina E N, Pearson W L, Sha Q, Lopatin A N. Circ Res. 1996;78:1–7. doi: 10.1161/01.res.78.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Ishihara K, Ehara T. J Physiol (London) 1998;510:755–771. doi: 10.1111/j.1469-7793.1998.755bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopatin A N, Nichols C G. J Mol Cell Cardiol. 2001;33:625–638. doi: 10.1006/jmcc.2001.1344. [DOI] [PubMed] [Google Scholar]
- 14.Jongsma H J, Wilders R. Curr Biol. 2001;11:R747–R750. doi: 10.1016/s0960-9822(01)00437-7. [DOI] [PubMed] [Google Scholar]
- 15.Raab-Graham K F, Radeke C M, Vandenberg C A. NeuroReport. 1994;5:2501–2505. doi: 10.1097/00001756-199412000-00024. [DOI] [PubMed] [Google Scholar]
- 16.Wible B A, De Biasi M, Majumder K, Taglialatela M, Brown A M. Circ Res. 1995;76:343–350. doi: 10.1161/01.res.76.3.343. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Yue L, White M, Pelletier G, Nattel S. Circulation. 1998;98:2422–2428. doi: 10.1161/01.cir.98.22.2422. [DOI] [PubMed] [Google Scholar]
- 18.Liu G X, Derst C, Schlichthörl G, Heinen S, Seebohm G, Brüggemann A, Kummer W, Veh R W, Daut J, Preisig-Müller R. J Physiol (London) 2001;532:115–126. doi: 10.1111/j.1469-7793.2001.0115g.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fink M, Duprat F, Heurteaux C, Lesage F, Romey G, Barhanin J, Lazdunski M. FEBS Lett. 1996;378:64–68. doi: 10.1016/0014-5793(95)01388-1. [DOI] [PubMed] [Google Scholar]
- 20.Tinker A, Jan Y N, Jan L Y. Cell. 1996;87:857–868. doi: 10.1016/s0092-8674(00)81993-5. [DOI] [PubMed] [Google Scholar]
- 21.Zaritsky J J, Redell J B, Tempel B L, Schwarz T L. J Physiol (London) 2001;533:697–710. doi: 10.1111/j.1469-7793.2001.t01-1-00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerche C, Scherer C R, Seebohm G, Derst C, Wei A D, Busch A E, Steinmeyer K. J Biol Chem. 2000;275:22395–22400. doi: 10.1074/jbc.M002378200. [DOI] [PubMed] [Google Scholar]
- 23.Falk T, Meyerhof W, Corrette B J, Schäfer J, Bauer C K, Schwarz J R, Richter D. FEBS Lett. 1995;367:127–131. doi: 10.1016/0014-5793(95)00527-g. [DOI] [PubMed] [Google Scholar]
- 24.Veh R W, Lichtinghagen R, Sewing S, Wunder F, Grumbach I M, Pongs O. Eur J Neurosci. 1995;7:2189–2205. doi: 10.1111/j.1460-9568.1995.tb00641.x. [DOI] [PubMed] [Google Scholar]
- 25.Zeng F Y, Wess J. J Biol Chem. 1999;274:19487–19497. doi: 10.1074/jbc.274.27.19487. [DOI] [PubMed] [Google Scholar]
- 26.Nehring R B, Wischmeyer E, Döring F, Veh R W, Sheng M, Karschin A. J Neurosci. 2000;20:156–162. doi: 10.1523/JNEUROSCI.20-01-00156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morishige K, Takahashi N, Jahangir A, Yamada M, Koyama H, Zanelli J S, Kurachi Y. FEBS Lett. 1994;346:251–256. doi: 10.1016/0014-5793(94)00483-8. [DOI] [PubMed] [Google Scholar]
- 28.Perier F, Radeke C M, Vandenberg C A. Proc Natl Acad Sci USA. 1994;91:6240–6244. doi: 10.1073/pnas.91.13.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang W, Yang X C. FEBS Lett. 1994;348:239–243. doi: 10.1016/0014-5793(94)00612-1. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Yue L, White M, Pelletier G, Nattel S. Circulation. 1998;98:2422–2428. doi: 10.1161/01.cir.98.22.2422. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi N, Morishige K, Jahangir A, Yamada M, Findlay I, Koyama H, Kurachi Y. J Biol Chem. 1994;269:23274–23279. [PubMed] [Google Scholar]
- 32.Tang W, Qin C L, Yang X C. Receptors Channels. 1995;3:175–183. [PubMed] [Google Scholar]
- 33.Horio Y, Morishige K, Takahashi N, Kurachi Y. FEBS Lett. 1996;379:239–243. doi: 10.1016/0014-5793(95)01519-1. [DOI] [PubMed] [Google Scholar]
- 34.Karschin C, Dissmann E, Stuhmer W, Karschin A. J Neurosci. 1996;16:3559–3570. doi: 10.1523/JNEUROSCI.16-11-03559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stonehouse A H, Pringle J H, Norman R I, Stanfield P R, Conley E C, Brammar W J. Histochem Cell Biol. 1999;112:457–465. doi: 10.1007/s004180050429. [DOI] [PubMed] [Google Scholar]
- 36.Tucker S J, Ashcroft F M. J Biol Chem. 1999;274:33393–33397. doi: 10.1074/jbc.274.47.33393. [DOI] [PubMed] [Google Scholar]
- 37.Bixby K A, Nanao M H, Shen N V, Kreusch A, Bellamy H, Pfaffinger P J, Choe S. Nat Struct Biol. 1999;6:38–43. doi: 10.1038/4911. [DOI] [PubMed] [Google Scholar]
- 38.Yi B A, Minor D L, Jr, Lin Y F, Jan Y N, Jan L Y. Proc Natl Acad Sci USA. 2001;98:11016–11023. doi: 10.1073/pnas.191351798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura T Y, Artman M, Rudy B, Coetzee W A. Am J Physiol. 1998;274:H892–H900. doi: 10.1152/ajpheart.1998.274.3.H892. [DOI] [PubMed] [Google Scholar]
- 40.Picones A, Keung E, Timpe L C. Biophys J. 2001;81:2035–2049. doi: 10.1016/S0006-3495(01)75853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komarova S V, Dixon S J, Sims S M. Curr Pharma Des. 2001;7:637–654. doi: 10.2174/1381612013397799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.