Abstract
The events of the cell cycle, the stages at which the cell proliferates and divides, are facilitated and controlled by multiple signaling pathways. Among the many regulatory enzymes that contribute to these processes is the polo-like kinase (Plk). Plks have been reported to mediate multiple mitotic processes, including bipolar spindle formation, activation of Cdc25C, actin ring formation, centrosome maturation, and activation of the anaphase-promoting complex. To investigate its functions in mammalian cells further, we used the recently developed small interfering RNA technique specifically to deplete Plk1 in cultured cells. We find that Plk1 depletion results in elevated Cdc2 protein kinase activity and thus attenuates cell-cycle progression. About 45% of cells treated with Plk1 small interfering RNA show the formation of a dumbbell-like DNA organization, suggesting that sister chromatids are not completely separated. About 15% of these cells do complete anaphase but do not complete cytokinesis. Finally, Plk1 depletion significantly reduces centrosome amplification in hydroxyurea-treated U2OS cells. These data provide direct evidence that Plk is required for multiple mitotic processes in mammalian cells and their significance is discussed.
Reversible protein phosphorylation is an important regulatory mechanism in the control of cell cycle progression. Central to this regulation are the cyclin-dependent kinases and their activating proteins-cyclins (1). The mitotic cyclin-dependent kinase complex consists of a Cdc2 catalytic subunit and a cyclin B regulatory subunit. A critical event in the regulation of Cdc2/cyclin B involves its dephosphorylation by Cdc25C (1, 2). Kumagai and Dunphy (3) reported that the Xenopus polo-like kinase Plx1 phosphorylates and activates Cdc25C leading to Cdc2/cyclin B activation.
Polo-like kinases (Plk) have been shown to be important contributors to several other cell-cycle events (4, 5). Genetic and biochemical experiments in various organisms indicate that polo-like kinases regulate diverse cellular events at multiple mitotic stages. The report of Kumagai and Dunphy (3) suggests that Plx1 is a critical trigger that initiates the onset of the G2/M transition. Further studies showed that Plx1 is a member of the Cdc2/cyclin B amplification loop in meiotic maturation in Xenopus oocytes (6–8). During the metaphase–anaphase transition, Plx1 is required for the destruction of targets of the anaphase-promoting complex (APC) that drives mitosis exit (9). Genetic studies in Drosophila and yeast indicate plks function in centrosome assembly and separation during the formation of the bipolar spindle. Drosophila polo mutants reveal phenotypes of hypercondensed chromosomes, monopolar spindles, disorganized spindle poles, and abnormal chromosome segregation (10). Schizosaccharomyces pombe plo1− displays similar phenotypes, such as the formation of monopolar spindles or a failure in septum formation after nuclear division (11). The budding yeast polo-like kinase homolog, Cdc5, seems to play an important role in actin ring formation and cytokinesis (12). Other studies show the APC-dependent destruction of Clb2 depends on Cdc5 activity (13). In mammalian cells, antibody microinjection suggests a role for Plk1 in centrosome maturation (14). Mammalian Plk1 was further shown to phosphorylate specifically at least three components of APC, and to activate APC to ubiquitinate cyclin B in an in vitro-reconstituted system (15). More recent studies demonstrated that polo kinase activity plays a pivotal role in the separation of sister chromatids during mitosis. Cdc5 is required to phosphorylate the cohesin subunit Scc1, to facilitate its cleavage by separase at the onset of anaphase (16). Plx1 is also the key regulator for the cleavage-independent dissociation of cohesin from chromosome in Xenopus (17).
To investigate the functions of Plk1 in mammalian cells further, we took advantage of a recently developed small interfering RNA (siRNA) technique to deplete specifically Plk1 in human cells (18). We find that cells treated with Plk1 siRNA had greatly reduced levels of Plk, but nevertheless maintained an elevated level of Cdc2/cyclin B activity relative to control M-phase cells treated with nocodazole. About 45% of treated cells did not seem to complete anaphase, and about 15% did not complete cytokinesis. In addition, Plk1 is required for the centrosome duplication normally induced in U2OS cells treated with hydroxyurea.
Materials and Methods
RNA Preparation.
Twenty-one-nucleotide double-stranded RNAs were synthesized by Dharmacon Research (Lafayette, CO). The targeting sequence of human Plk1 (accession no. NM 005030) was AAGGGCGGCTTTGCCAAGTGCTT, corresponds to the coding region 181–203 relative to the first nucleotide of the start codon. siRNA, targeting a sequence in the firefly (Photinus pyralis) luciferase (accession no. X65324) mRNA, served as a control.
Cell Culture, Synchronization, and Transfections.
HeLa and U2OS cells were maintained in DMEM supplemented with 10% (vol/vol) FBS, 100 units/ml penicillin, and 100 units/ml streptomycin at 37°C in 5% CO2. To obtain mitotically synchronized HeLa cells, cells were treated with 100 ng/ml nocodazole for 16 h. To synchronize U2OS cells, cells were either blocked with 0.3 mM mimosine for 16 h to arrest at G1 phase or 100 ng/ml nocodazole for 16 h to arrest at M-phase. Incubation with 4 mM hydroxyurea for 40 h results in U2OS cells arrested at S-phase.
Cells were transfected with Oligofectamine reagent (Invitrogen). Cells were seeded in 6-well plates on the day before transfection at the concentration of 1 × 105 cells per well. Opti-MEM I (200 μl) was mixed with 7 μl of 20 μM siRNA duplex. In a second tube, 54 μl of Opti-MEM I was incubated with 7 μl of Oligofectamine for 8 min at room temperature. The two mixtures above were combined, gently mixed, and incubated for another 22 min at room temperature. After addition of 32 μl of Opti-MEM I medium to the mixture, the entire mixture was added to the cells in 1.2 ml of 10% FBS containing DMEM without antibiotics. Cells were assayed after 48 h transfection.
Immunoprecipitation and Immunoblotting.
Transfected HeLa cells were lysed in TBSN buffer (20 mM Tris, pH 8.0/150 mM NaCl/1.5 mM EDTA/5 mM EGTA/0.5% Nonidet P-40/0.5 mM Na3VO4) supplemented with proteinase inhibitors (20 mM p-nitrophenyl phosphate/1 mM Pefabloc/10 μg/ml pepstatin A/10 μg/ml leupeptin/5 μg/ml aprotinin), and the lysates were clarified by centrifugation at 15,000 × g for 30 min. Cell lysates were incubated with anti-Cdc2 antibody (Santa Cruz Biotechnology) for 1.5 h at 4°C, followed by an additional 1 h incubation with Protein A-Sepharose beads. Immunocomplexes were washed three times in TBSN buffer, resolved by SDS/PAGE and transferred to Immobilon-P membranes (Millipore). Proteins transferred to the filter were detected by various antibodies, horseradish peroxidase-linked secondary antibody, and visualized by enhanced chemiluminescence reagents.
Kinase Assay.
Anti-Cdc2 immunoprecipitates were incubated with histone H1 in TBMD buffer (50 mM Tris, pH 7.5/10 mM MgCl2/5 mM DTT/2 mM EGTA/0.5 mM Na3VO4/20 mM p-nitrophenyl phosphate) supplemented with 25 μM ATP and 50 μCi of [γ-32P]ATP. The reaction mixtures were incubated at 30°C for 30 min and resolved on SDS-polyacrylamide gels. The gels were transferred to the membranes and subjected to autoradiography.
Immunofluorescence Staining.
HeLa cells were grown on coverslips, transfected, and cultured for 48 h before fixation. For U2OS cells, 4 mM hydroxyurea was added 24 h after transfection, and the cells were incubated for additional 40 h. Cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with methanol for 2 min. After washing with 0.1% Triton X-100/PBS three times, coverslips were blocked with 10% goat serum/PBS for 30 min, stained with either 10 μg/ml anti-α-tubulin or γ-tubulin primary antibody for 2 h at room temperature, followed by incubation with FITC- or Cy3-conjugated anti-mouse Ig secondary antibody for 30 min. Finally, DNA was stained with either propidium iodide or 4′,6-diamidino-2-phenylindole.
Results
Plk1 Depletion Prevents Cyclin B Degradation and Causes Elevated Cdc2 Activity.
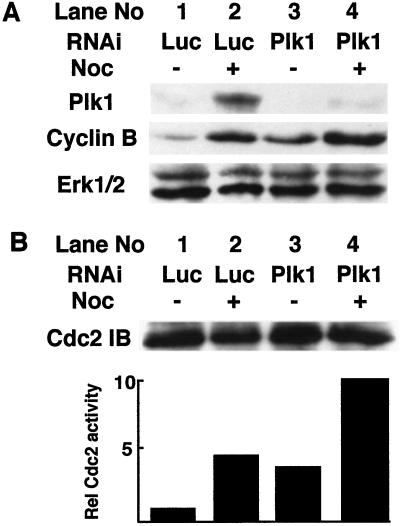
To investigate the function of Plk1 in mammalian cells, we synthesized a RNA duplex directed against the coding region 181–203 relative to the first nucleotide of the start codon. A sequence against the firefly luciferase gene was used as a nonspecific control. The RNA duplexes were transfected into HeLa cells, and the cells were cultured for 36 h, and incubated for additional 12 h in the absence or presence of nocodazole. Cell lysates were prepared and standard Western blotting was performed. Plk1 was efficiently depleted by siRNA (Fig. 1A Top, compare lane 2 with lane 4), whereas the level of Erk1/2 was unchanged (Fig. 1A Bottom). The level of Plk1 protein was reduced by at least 90% after 48 h of siRNA treatment, corresponding to the 90% transfection efficiency in these experiments, which suggests that Plk1 was almost completely depleted in most cells by the siRNA approach.
Figure 1.
Plk1 depletion prevents cyclin B degradation and causes elevated Cdc2 activity. (A) HeLa cells were transfected with RNA oligos against luciferase or Plk1. Thirty-six hours after transfection, cells were treated with 100 ng/ml nocodazole for 12 h and harvested. About 150 μg of cell lysates were directly resolved by SDS/PAGE, and subjected to Western blot by using the antibodies as indicated on the left. (B) Approximately 1 mg of cell lysates were immunoprecipitated with anti-Cdc2 antibody, and the immunoprecipitates were assessed for histone H1 kinase activity. The reaction products were resolved by SDS/PAGE, transferred to membrane, and exposed to the film. The same filter was also probed with anti-Cdc2 antibody to show that equal amounts of proteins were immunoprecipitated.
At the onset of anaphase, polo-like kinases activate the APC by phosphorylating several of its subunits (13, 15). Activated APC catalyzes the polyubiquitination of cyclin B and causes its degradation (15). Thus, we measured the cyclin B level in Plk1 siRNA-treated cells and found an increased level of cyclin B relative to controls (Fig. 1A Middle, compare lane 1 with lane 3, lane 2 with lane 4). The same cell lysates were then subjected to anti-Cdc2 immunoprecipitation, and the immunocomplexes were assayed for histone H1 protein kinase activity. Plk1-depleted cells expressed about 4-fold greater Cdc2 activity than that in control cells (Fig. 1B Bottom), nearly equal to that of nocodazole-treated control cells (5-fold). Nocodazole treatment of Plk1-depleted cells results in 10-fold greater Cdc2 activity than that in untreated control cells (Fig. 1B Bottom). A Cdc2 Western blot indicates equal quantities of protein were assayed for protein kinase activity (Fig. 1B Top).
Plk1 Depletion Inhibits the Separation of Sister Chromatids and Leads to Failure of Cytokinesis.
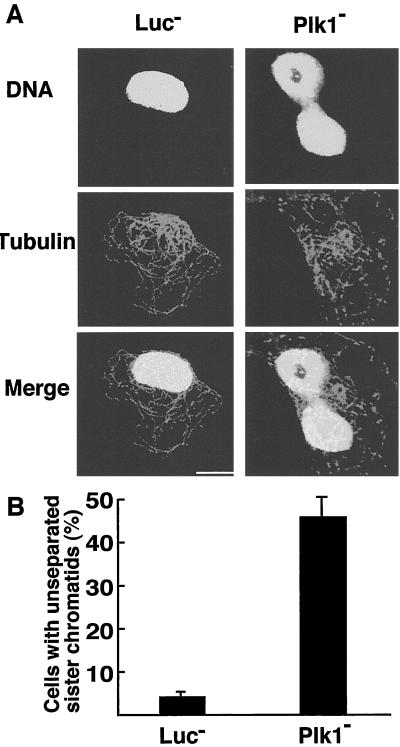
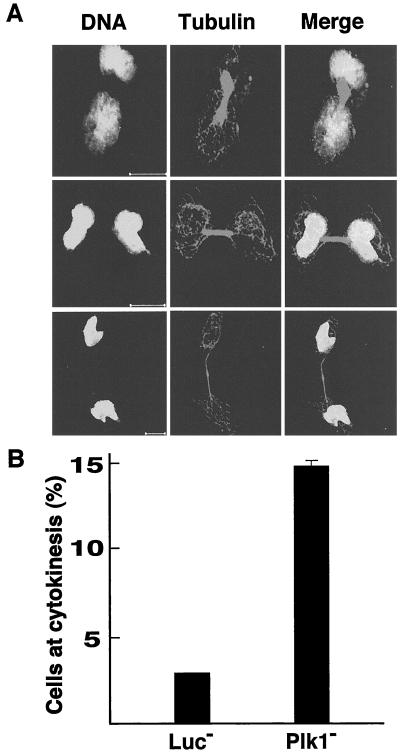
The separation of sister chromatids in anaphase require the proteolysis of cohesin by APC. We found that approximately 45% of Plk1-depleted cells display a dumbbell-like DNA structure, indicating that sister chromatids have not completely separated 48 h after siRNA addition (Fig. 2). Staining with α-tubulin confirms these connected chromosomes are within one cell, and the cage-like microtubule pattern also suggests these cells are in interphase. Less than 3% of untreated cells or those treated with nonspecific RNA displayed the phenotype (Fig. 2B). A role for polo-like kinase in cytokinesis has been demonstrated in several organisms, but has not yet been fully examined in mammalian cells. Examination of HeLa cells 48 h after siRNA addition reveals that 15% of the cells were arrested in cytokinesis, whereas less than 3% of control cells showed this phenotype (Fig. 3 A and B). The arrested cells were connected by a cytoplasmic bridge as revealed by α-tubulin immunofluorescence.
Figure 2.
Plk1 depletion inhibits the separation of sister chromatids. (A) HeLa cells were transfected with RNA oligos against luciferase or Plk1. Forty-eight hours after transfection, cells were fixed and stained with anti-α-tubulin primary antibody, followed with FITC-conjugated anti-mouse secondary antibody. DNA was stained with propidium iodide. (Scale bar: 10 μm.) (B) Histogram shows results from five independent experiments (more than 300 cells each) and bars indicate standard deviations.
Figure 3.
Failure of cytokinesis in Plk1-depleted cells. (A) HeLa cells were transfected with a RNA oligo against Plk1. Forty-eight hours after transfection, cells were fixed and stained with anti-α-tubulin primary antibody, followed with FITC-conjugated anti-mouse secondary antibody. DNA was stained with propidium iodide. Three representative images from different phases of cytokinesis are shown. (Scale bar: 10 μm.) (B) Histogram shows results from three independent experiments (more than 300 cells each) and bars indicate standard deviations. An RNA oligo against firefly luciferase gene was used as a control.
Plk1 Is Required for Centrosome Duplication.
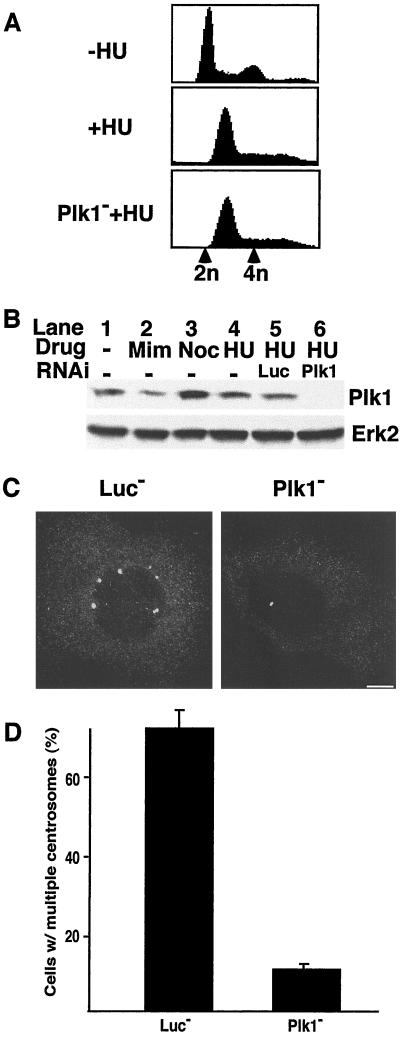
Polo-like kinases are localized to the centrosomes and spindle poles in a variety of organisms and may play a role in centrosome maturation and function (5). The U2OS cells undergo multiple rounds of centrosome duplication on treatment with hydroxyurea. To determine more directly whether Plk1 has a role in centrosome duplication, U2OS cells were treated with Plk1 siRNA and 24 h later hydroxyurea was added. After 40 h of hydroxyurea treatment, U2OS cells almost completely arrested at S-phase (Fig. 4A). We also compared the Plk1 level in U2OS cell at different cell cycle phases. As expected, cells at prometaphase arrested with nocodazole show the highest Plk1 level, whereas G1 phase arrested cells after mimosine treatment have the lowest Plk1 level. However, a significant level of Plk1 (about 50% of nocodazole-treated cells) in hydroxyurea-treated cells was detected, and it was almost completely abolished after siRNA treatment (Fig. 4B). Hydroxyurea-treated cells were fixed, centrosomes were detected by staining with γ-tubulin, and counted. As shown in Fig. 4 C and D, about 70% of hydroxyurea-treated U2OS cells had more than two centrosomes, whereas less than 8% of Plk1 siRNA-treated cells had multiple centrosomes.
Figure 4.
Plk1 is involved in centrosome duplication. (A) fluorescence-activated cell sorting profile of U2OS cells with or without hydroxyurea treatment. Positions of cells with 2n or 4n DNA contents are labeled with arrowheads on the bottom. (B) U2OS cells were transfected with RNA oligos against luciferase or Plk1. Twenty-four hours after transfection, cells were treated with 4 mM hydroxyurea (HU) for an additional 40 h (lanes 4–6). For comparison, cells were either untreated (lane 1), treated with mimosine (lane 2), or treated with nocodazole (lane 3). About 300 μg of total cellular protein was used for the direct Western blotting by using the antibodies indicated on the right. Anti-Plk1 Western blot indicates that Plk1 was easily detected in S-phase arrested cells, and treatment with Plk1 siRNA efficiently reduced the Plk1 level. Anti-erk2 Western blot was served as a loading control. (C) Hydroxyurea-treated cells were fixed and stained with anti-γ-tubulin primary antibody, followed with Cy3-conjugated anti-mouse secondary antibody. DNA was stained with 4′,6-diamidino-2-phenylindole. (Scale bar: 10 μm.) (D) Histogram shows results from five independent experiments (more than 300 cells each) and bars indicate standard deviations.
Discussion
In this communication, we describe the phenotype of human cells that have been depleted of Plk1 by siRNA treatment. At 48 h after transfection, the cells were examined by confocal microscopy after immunostaining, and by fluorescence-activated cell sorting. At this time, approximately 40% of siRNA-treated cells had a G2 DNA content compared with 12% in untreated cells. Further examination of these cells on coverslips revealed that approximately 45% of them were well attached and displayed a dumbbell-shaped chromatin organization as judged by DNA staining. Less than 3% of untreated cells had this phenotype. The images suggest that sister chromatids were unable to be separated. These data are consistent with previous studies that indicate Plk1 plays a role in activating the APC (15). Other reports indicate a role for Polo-like kinases in sister chromatid separation. Cohesin serves to hold sister chromatids together until it is cleaved by separase after activation of the APC (19). The bulk of cohesin is removed from chromosomes in prophase and prometaphase during mitosis in vertebrates. In Xenopus, Plx1 seems to be required for this process, because Plx1 phosphorylation of cohesin reduces its capacity to bind to chromatin in a cleavage-independent event (17). However, in Saccharomyces cerevisiae, the polo-like kinase, Cdc5, has been shown to phosphorylate serine residues adjacent to cleavage sites in the cohesin subunit Scc1 and enhance its cleavage. Thus, polo-like kinases, may have distinct functions in different organisms, whether the cells observed in Fig. 2 have unseparated chromatin structure because of failure of cleavage-independent cohesin dissociation or failure to cleave the remaining cohesin requires additional experimentation.
An additional 15% of the siRNA-treated population consisted of two cells attached by a cytoplasmic bridge indicating that cytokinesis had not been completed. In an untreated population, less than 3% of cells with this phenotype were observed. Failure to complete cytokinesis has also been observed with certain Drosophilia polo mutants (20). Cdc5 also functions in cytokinesis. Overexpression of the C terminus of Cdc5 results in connected cells with incomplete septa and shared cytoplasm (12). Thus, Cdc5, and perhaps other polo-like kinases, may participate in both APC activation and cytokinesis.
The morphology of the cell populations described above that do not express the two phenotypes resembled the untreated controls in Fig. 2. The siRNA transfection is performed on unsynchronized cells; thus, the heterogeneity of the population at 48 h is likely to reflect the point in the cell cycle that Plk1 depletion becomes rate-limiting. We have investigated two other functions that were previously attributed to polo-like kinases in other studies: (i) participation in Cdc2 activation and (ii) contributions to centrosome maturation and duplication. The extent of Cdc2/cyclin B activation was examined in Plk1-depleted cells with or without nocodazole addition and compared with the appropriate controls. These results show that in the siRNA-treated population, in which about 40% cells have a G2 DNA content, the Cdc2 activity was nearly equal to that found in control cells treated with nocodazole. Moreover, Plk1-depleted populations treated with nocodazole had greatly enhanced Cdc2 activity. These data indicate that Cdc2 activation is Plk1-independent in HeLa cells. These data are in contrast to reports that describe experiments with maturing oocytes in which Plx1 seems to phosphorylate and activate Cdc25, which then dephosphorylates and activates Cdc2/cyclin B (3, 6–8). These results may reflect the fact that polo-like kinases play distinct roles in different organisms or that the somatic cells and embryonic cells are unique with regard to these activation events. Further studies are required to determine the activity of Cdc25A or C in Plk1-depleted mammalian cells.
Finally, we have examined the potential of Plk1 to influence centrosome duplication. Several protein kinases have been reported to contribute to centrosome duplication. In Xenopus egg extracts, Cdk2/cyclin E is reported to be required for centrosome duplication during S-phase (21). In somatic cells, however, Cdk2/cyclin A is required for centrosome duplication (22). Subsequently, others reported that Cdc2/cyclin E phosphorylates a centrosomal protein, nucleophosmin. Expression of a nonphosphorylatable form of nucleophosmin in cells blocks centrosome duplication (23, 24). Another mammalian protein, mMps1, has been reported to be required for centrosome duplication (25). More recently, however, others have found that human Mps1 is not required for centrosome duplication, on the basis of antibody microinjection data and siRNA-mediated depletion of the enzyme (26). Mps1, like Plk1, shows maximum activity during M phase. Based on our results, we find that Plk1 is required for centrosome duplication in hydroxyurea-treated U2OS cells, which is in contrast to human Mps1. The data presented in Fig. 4 do not, however, show whether or not Plk1 has a direct or indirect effect on centrosome duplication. It is well established that Plk1 is localized at the centrosomes and spindle poles in prometaphase; thus, a Plk1 substrate may reside in the centrosome. Identification of such a substrate may clarify this issue.
Previous reports describe the inhibition of Plk1 activity as the result of ATM (ataxia-telangiectasia mutated and ATR (ATM and Rad 3-related) activation in response to DNA damage (27, 28). Because HU treatment may activate ATR as a consequence of stalled replication forks (29), Plk1 activity is likely to be attenuated in U2OS cells. The protein kinase activity in immune complex assays of Plk1 from HU-treated cells is about 3- to 5-fold less than that of Plk1 obtained from nocodazole-treated cells shown in Fig. 4 (unpublished data). The significance of this level of activity in supporting centrosome duplication requires additional experimentation; however, the depletion of Plk1 clearly attenuates multiple rounds of centrosome duplication.
Acknowledgments
We thank E. Erikson and J. Blenis for critical reading of this manuscript. This work was supported by National Institutes of Health Grant GM59172. R.L.E. is the John F. Drum American Cancer Society Research Professor.
Abbreviations
- Plk1
polo-like kinase 1
- siRNA
small interfering RNA
- APC
anaphase-promoting complex
References
- 1.Nigg E A. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 2.Kumagai A, Dunphy W G. Cell. 1991;64:903–914. doi: 10.1016/0092-8674(91)90315-p. [DOI] [PubMed] [Google Scholar]
- 3.Kumagai A, Dunphy W G. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 4.Lane H A, Nigg E A. Trends Cell Biol. 1997;7:63–68. doi: 10.1016/S0962-8924(96)10051-9. [DOI] [PubMed] [Google Scholar]
- 5.Glover D M, Hagan I M, Tavares A A M. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- 6.Abrieu A, Brassac T, Galas S, Fisher D, Labbe J, Doree M. J Cell Sci. 1998;111:1751–1757. doi: 10.1242/jcs.111.12.1751. [DOI] [PubMed] [Google Scholar]
- 7.Karaiskou A, Cayla X, Haccard O, Jessus C, Ozon R. Exp Cell Res. 1998;244:491–500. doi: 10.1006/excr.1998.4220. [DOI] [PubMed] [Google Scholar]
- 8.Qian Y, Erikson E, Li C, Maller J. Mol Cell Biol. 1998;18:4262–4271. doi: 10.1128/mcb.18.7.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Descombes P, Nigg E A. EMBO J. 1998;17:1328–1335. doi: 10.1093/emboj/17.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunkel C E, Glover D M. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 11.Ohkura H, Hagan I, Glover D M. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- 12.Song S, Lee K S. J Cell Biol. 2001;152:451–469. doi: 10.1083/jcb.152.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane H A, Nigg E A. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotani S, Tugendreich S, Fujii M, Jorgensen P, Watanabe N, Hoog C, Hieter P, Todokoro K. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- 16.Alexandru G, Uhlmann F, Mechtler K, Poupart M, Nasmyth K. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 17.Sumara I, Vorlaufer E, Stukenberg P T, Kelm O, Redemann N, Nigg E A, Peters J-M. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- 18.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 19.Peters J M. Exp Cell Res. 1999;248:339–349. doi: 10.1006/excr.1999.4443. [DOI] [PubMed] [Google Scholar]
- 20.Carmena M, Riparbelli M G, Minestrini G, Tavares A M, Adams R, Callaini G, Glover D M. J Cell Biol. 1998;143:659–671. doi: 10.1083/jcb.143.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinchcliffe E H, Li C, Thompson E A, Maller J L, Sluder G. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- 22.Meraldi P, Lukas J, Fry A M, Bartek J, Nigg E A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 23.Okuda M, Horn H F, Tarapore P, Tokuyama Y, Smulian A G, Chan P, Knudsen E S, Hofmann I A, Snyder J D, Bove K E, Fukasawa K. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 24.Tokuyama Y, Horn H F, Kawamura K, Tarapore P, Fukasawa K. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- 25.Fisk H A, Winey M. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 26.Stucke V M, Sillje H H, Arnaud L, Nigg E A. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smits V A J, Klompmaker R, Arnaud L, Rijksen G, Nigg E A, Medema R H. Nat Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- 28.van Vugt M A T M, Smits V A J, Klompmaker R, Medema R H. J Biol Chem. 2001;276:41656–41660. doi: 10.1074/jbc.M101831200. [DOI] [PubMed] [Google Scholar]
- 29.Abraham R T. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]