Abstract
Microsomal membranes catalyze the formation of xyloglucan from UDP-Glc and UDP-Xyl by cooperative action of α-xylosyltransferase and β-glucan synthase activities. Here we report that etiolated pea microsomes contain an α-xylosyltransferase that catalyzes the transfer of xylose from UDP-[14C]xylose onto β(1,4)-linked glucan chains. The solubilized enzyme had the capacity to transfer xylosyl residues onto cello-oligosaccharides having 5 or more glucose residues. Analysis of the data from these biochemical assays led to the identification of a group of Arabidopsis genes and the hypothesis that one or more members of this group may encode α-xylosyltransferases involved in xyloglucan biosynthesis. To evaluate this hypothesis, the candidate genes were expressed in Pichia pastoris and their activities measured with the biochemical assay described above. One of the candidate genes showed cello-oligosaccharide-dependent xylosyltransferase activity. Characterization of the radiolabeled products obtained with cellopentaose as acceptor indicated that the pea and the Arabidopsis enzymes transfer the 14C-labeled xylose mainly to the second glucose residue from the nonreducing end. Enzymatic digestion of these radiolabeled products produced results that would be expected if the xylose was attached in an α(1,6)-linkage to the glucan chain. We conclude that this Arabidopsis gene encodes an α-xylosyltransferase activity involved in xyloglucan biosynthesis.
Although considerable progress has been made in elucidating the structure of plant cell wall polysaccharides, the biosynthesis and deposition of these components are still poorly understood. Because the primary cell wall contains a number of complex polysaccharides (for review see refs. 1 and 2), many glycosyltransferases are needed to produce the various sugar linkages contained in them. To date, only a few genes encoding glycosyltransferases have been cloned and characterized at the molecular level (3–5). On the other hand, the recent explosion in bioinformatic and functional genomic methods opens opportunities to identify, by sequence similarity, putative glycosyltransferase genes (see for example http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). The challenge will be to determine which of these genes are involved in cell wall polysaccharide biosynthesis and to determine their function. Identification of gene function can be achieved either by genetic or reverse genetic methods, or with biochemical approaches; the latter is the most direct way.
Cell wall xyloglucan (XyG) is composed of a β(1,4)glucan backbone that is substituted on position 6 by Xyl residues in a regular pattern. Usually, 75% of the backbone Glc units are substituted, but lower substitution patterns have been observed in solanaceous plants and rice. Some Xyl residues are substituted with Gal residues and some of them contain Fuc residues (for reviews see refs. 6 and 7). The biosynthesis of this major hemicellulosic polysaccharide in dicots and nongraminaceous monocot cell walls (≈25% of total primary cell wall) involves at least four biosynthetic enzymes, namely α-fucosyltransferase, β-galactosyltransferase, α-xylosyltransferase, and β(1,4)glucan synthase. Isolated microsomal membranes can synthesize the core of XyG with UDP-Glc and UDP-Xyl as sugar donors (8–11). However, most of the ability to synthesize XyG is lost when microsomal membranes are treated with detergent. One explanation for the loss of activity after solubilization is that XyG synthesis requires cooperative action between α(1,6)xylosyltransferase and XyG β(1,4)glucan synthase, which are thought to form a complex in Golgi membranes. Thus far, the number of the transferases required to form the XyG core and the mechanisms for their cooperation are still unknown. Thus, to understand the XyG biosynthetic process the genes for α(1,6)xylosyltransferase and/or XyG β(1,4)glucan synthase enzymes must be identified. In this report we describe a biochemical assay that allows detection of α-xylosyltransferase activity after its solubilization with detergent. This assay was used to screen candidate genes from Arabidopsis, for possible identification of genes encoding α-xylosyltransferase activity. We identified one gene, the product of which possessed α-xylosyltransferase activity and provided preliminary evidence that it is involved in XyG biosynthesis.
Materials and Methods
Chemical and Plant Material.
UDP-[14C]Xyl (0.925 GBq/mmol), UDP-[14C]Glc (11.21 GBq/mmol), and UDP-[14C]Gal (10.3 GBq/mmol) were purchased from NEN. UDP-Xyl, GDP-Glc, UDP-Glc, β-glucosidase (EC 3.2.1.21), all detergents, malto- and cello-oligosaccharides, Dowex 1-X80 resin, and Driselase were purchased from Sigma. More recently UDP-Xyl was made available by CarboSource (see WallBioNet at http://xyloglucan.prl.msu.edu). Bio-gel P2 was from Bio-Rad. Trichoderma sp. cellulase, tamarind XyG, carboxymethyl-cellulose 4M (CMC), and manno-oligosaccharides were from Megazyme International (Bray, Ireland). Driselase was solubilized by stirring 1 g of the dry powder in 2 ml of sodium acetate buffer (pH 5) containing 0.7 M NaCl. After centrifugation (15,000 × g, 10 min, 4°C), the supernatant was filter-sterilized before use. The α- xylosidase from Sulfolobus solfataricus (XylS; at 0.05 units/mg protein) was a gift from M. Moracci (Institute of Protein Biochemistry and Enzymology/Consiglio Nazionale delle Ricerche, Naples) and was prepared by expressing the cDNA of an XylS in Escherichia coli and purified by heating steps at 55, 75, and 80°C for 30 min and centrifugation as described in ref. 12. β-Glucosidase (≈1.4 units/mg) is from the thermophilic bacterium Caldocellum saccharolyticum and was overproduced in E. coli and purified (13). Garden pea seeds (Pisum sativum L. var. Alaska) were purchased from Olds Seed Company (Madison, WI). They were cleaned, sterilized, and grown in moist vermiculate in the dark as described (5).
Pea Microsome Preparation and Activity Solubilization.
All steps were carried out at 4°C. Microsomal membranes were prepared from the growing region of 7-day-old pea as described (10). The segments were homogenized in extraction buffer (EB) [0.1 M Hepes-KOH (pH 7), containing 0.4 M sucrose, 1 mM EDTA, 1 mM DTT, 5 mM MgCl2, 5 mM MnCl2, 0.1% BSA, 1 mM PMSF, and 10 μg/ml of the following protease inhibitors: aprotinin, pepstatin, and leupeptin]. After filtration, the homogenate was first centrifuged at 1,500 × g for 15 min, and the supernatant layered over 6 ml of 1.8 M sucrose cushion; it was then subjected to centrifugation (Beckman L8–70M) at 100,000 × g for 90 min. The membranes at the interface were collected and pelleted at 100,000 × g (Sorvall RC M120) before resuspension in a desired volume of EB at a protein concentration of 2–5 mg/ml. This membrane preparation was used directly or stored at −80°C.
Solutions of 10% detergents were usually prepared in EB. Microsomes were incubated with detergent at various concentrations on ice for 10 min and then either used directly for the assays or after separation of the detergent-soluble fraction from the detergent-insoluble pellets (residual membranes) by ultracentrifugation at 100,000 × g (Sorvall RC M120) for 15 min at 4°C. The pellets were resuspended in the same volume of EB, and both the supernatants and the pellets were assayed for the xylosyltransferase activity or XyG synthesis as described below. The 13 detergents screened were digitonin, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO), Lyso-α-phosphatidylcholine (Lys-α-PC), Triton X-100, Triton X-114, Brij 58, sodium lauryl sulfate, glycocholate, glycodeoxycholate, Tween 20, Zwittergent 3-14, and taurocholate. Although all these detergents affect XyG synthesis, most of them were effective in solubilizing xylosyltransferase. For instance, Triton X-100 disturbs XyG biosynthesis (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org) but effectively solubilizes the xylosyltransferase without destroying its activity (data not shown).
Enzyme Assays.
The standard XyG polymer synthesis reaction with microsomal fractions was performed essentially as described earlier (10), except that the total reaction volume was 60 μl. The crude membranes (40 μl) were incubated for 1 h at 25°C with 20 μl of the assay medium containing UDP-[14C]Xyl (60,000 cpm), 0.6 mM UDP-Xyl, and 3 mM UDP-Glc. Reactions were stopped by addition of 100 μg of tamarind XyG and then precipitated with cold 67% ethanol. The excess of UDP-[14C]Xyl was removed by 3 washes of the insoluble-ethanol products with 67% ethanol, and the radiolabeled incorporation was measured with a Beckman LS 5000 TD counter after dissolution of the products in 0.2 ml water and 2 ml of liquid scintillation mixture (ICN). In some experiments the membranes were preincubated in the presence of 1 mM UDP-Glc for 30 min and then ultracentrifuged at 100,000 × g for 15 min at 4°C to remove unused UDP-Glc. The pellets were rinsed twice with cold EB before resuspension in the same volume of EB and incubation (1 h, 25°C) in the presence of UDP-[14C]Xyl (60,000 cpm) and 0.1–0.2 mM UDP-Xyl. Control reactions were conducted in the absence of UDP-Glc or with boiled membranes. The same assays were used to test the effect of detergents on XyG biosynthesis in the permeabilized membranes.
When cello-oligosaccharides (usually 100–300 μg per reaction) were used as acceptors for xylosyltransferase activity, the reactions were conducted as above and were stopped by adding 0.2 ml of water, followed by centrifugation. Sufficient Dowex 1-X80 resin (Cl− form) was added to complex the unused UDP-[14C]Xyl before centrifugation at 12,000 × g to pellet the resin. The supernatants that contained radiolabeled products were saved and counted as above. The control did not contain the oligosaccharide acceptors.
When CMC polymer was used as acceptor the reactions were conducted as described above and stopped by ethanol (67%) precipitation and washed twice before resuspension in 0.2 ml of water and counting as above.
Characterization of the Radiolabeled Products.
For product analysis, the reactions were scaled up 10–30 times (15,000–45,000 cpm). The 67% ethanol-insoluble 14C-labeled products were washed as above and resuspended in 50 mM sodium acetate buffer (pH 5) and then treated with 0.5% (vol/vol) Trichoderma cellulase for 16 h at 37°C. The reactions were stopped by boiling for 5 min, and the soluble products fractionated on a Bio-gel P2 column (1 × 100 cm) that was eluted with water containing 0.02% sodium azide and previously calibrated with purified malto-oligosaccharides [degree of polymerization (DP) 2-7]. The radiolabel that coeluted with authentic XyG oligosaccharides was pooled and further separated by high pH anion exchange chromatography (HPAEC) on a CarboPac PA-100 column (Dionex), as described (14). A mixture of oligosaccharides from tamarind and pea XyG were used as standards. Samples from Bio-gel P2 peaks were analyzed further by treatment with 0.2 ml of Driselase in 50 mM sodium acetate buffer (pH 5) for 9–16 h at 37°C and then refractionated again on a Bio-gel P2 column.
When cellopentaose (G5) was used as acceptor, the products formed were first fractionated on Bio-gel P2. The radiolabeled products eluted around DP6 were pooled and treated for 16 h with XylS (0.02 units), β-glucosidase (3 units), or Driselase (0.2 ml) in 50 mM sodium acetate buffer (pH 5) at 37°C, except for XylS, for which the incubation was at 65°C. After each treatment the reactions were stopped by heating for 5 min, centrifuged (15,000 × g, 5 min), and then separated on a Bio-gel P2 column as described above.
Cloning and Expression of Arabidopsis Candidate Genes in Pichia pastoris.
Full-length forms of the putative Arabidopsis glycosyltransferase genes (AtGTs) were PCR-amplified with a PRL2 cDNA library (15) or genomic DNA as template and a proofreading DNA polymerase TaKaRa Ex Taq (PANVERA). PCR reactions were performed in 50 μl final reaction volume containing 0.2 mM dNTPs, 0.5 μM of each primer, 1× ExTaq buffer, 0.1–0.5 mg of template, and 1.25 units of polymerase. Primers for full-length clones were designed from flanking coding regions according to the genomic sequence, and the 5′ oligonucleotides used contained a 6×His-tag in-frame with the start codon of the genes (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). The amplified fragments were first cloned into pGEM-T-Easy vector (Invitrogen) and their identity confirmed by DNA sequencing. EcoRI or NotI digestion released fragments that contained the tagged genes that were cloned then into the P. pastoris expression vector pPICZ versions B or A. Clones with the correct orientation were selected for each construct to use for Pichia transformation. X-33 Pichia cells were prepared for transformation by electroporation according to the manufacturer's recommendations (Bio-Rad). Transformants were selected on media containing Zeocin and confirmed by direct PCR amplification to have the gene of interest.
Xylosyltransferase Assays with Extracts from Pichia-Expressing Candidate Genes.
For each construct, 5 colonies were selected and grown in 10 ml of buffered glycerol-complex medium (BMGY) at 30°C until OD600 reached ≈2 (≈2 days). To induce the expression of candidate genes, Pichia cells were first harvested by centrifugation (3,000 × g, 5 min) and then resuspended in buffered methanol-complex medium (BMMY) to an OD600 ≈1 (≈30 ml) in a 250-ml baffled flask. A time course of the induction was conducted for up to 4 days by adding methanol every 24 h to the final concentration of 0.5% (vol/vol). The activity was tested on soluble fractions prepared by vortexing the Pichia cells in EB containing 0.4% Triton X-100 (8 × 30 sec) followed by centrifugation (14,000 × g, 10 min, 4°C). Assays were performed as above with 20–40 μl of enzyme.
Results
α-Xylosyltransferase Activity and Its Relationship to XyG Biosynthesis.
XyG biosynthesis from UDP-[14C]Xyl and UDP-Glc by pea membranes was performed as described (8, 10, 16). The identity of the radiolabeled products as XyG was confirmed by cellulase and/or Driselase degradation followed by high pH anion exchange chromatography analysis as described (14) (data not shown). Xylose incorporation did not occur in the absence of UDP-Glc, but preincubation of membranes with 1 mM UDP-Glc alone before incubation with 0.2 mM UDP-[14C]Xyl alone was found to give increased incorporation of 14C-labeled Xyl into an endogenous acceptor. Degradation of the radiolabeled products with cellulase yielded the heptasaccharide Glc4Xyl3 (XXXG; for nomenclature, see ref. 17) and the pentasaccharide Glc3Xyl2 (XXG; data not shown). The observation that incorporation from UDP-[14C]Xyl into a product resembling XyG was stimulated by preincubation with excess UDP-Glc led to the conclusion that the xylosyltransferase activity could use a preformed endogenous acceptor, in this case, one made by XyG β(1,4)glucan synthase. Thus, we attempted to solubilize the α-xylosyltransferase activity and test its activity toward exogenous acceptors.
Thirteen detergents were tested for their effects on XyG biosynthesis. The data indicated that for concentrations equal to or higher than 0.1% (wt/vol), all these detergents disrupted XyG polymer production, as judged by the reduction of 14C-labeled Xyl incorporation into 67% ethanol-insoluble material. However, 0.4% digitonin preserved ≈50% of the 14C-labeled Xyl incorporation (see Table 1). The radiolabeled products formed under these conditions were also analyzed by cellulase digestion and fractionation on a Bio-gel P2 column followed by high pH anion exchange chromatography characterization and were confirmed to be XyG (data not shown). From these data we concluded that XyG synthase activity progressively lost the ability to make the polymer in the presence of an increasing amount of detergent. The loss of the activity may be the result of (i) loss of activity of one or both transferases by denaturation or (ii) dissociation of the transferases from each other and/or from their acceptors. To test the second option, we added exogenous acceptor and monitored pea xylosyltransferase solubilization.
Requirements for Substrate Acceptors and Solubilization of Pea α-Xylosyltransferase.
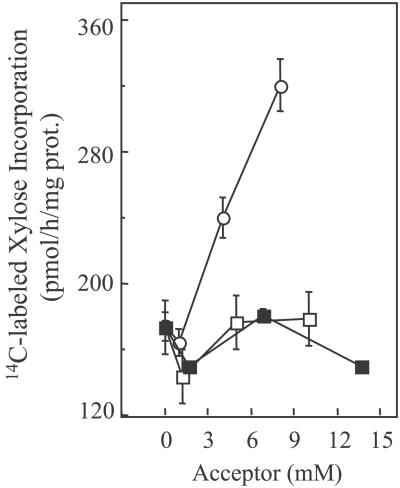
Assays to test the ability of cello-oligosaccharides to act as acceptors for xylosyltransferase activity were conducted with pea microsomes in the presence of 0.4% digitonin. Fig. 1 shows that 14C-labeled Xyl incorporation depends on the DP of the added oligosaccharides when present. Those having less than five Glc residues were not acceptor substrates, whereas substantial increases of 14C-labeled Xyl incorporation were observed with increasing amounts of added G5 (Fig. 1). Larger cello-oligosaccharides (DP >6) could not be tested because of their insolubility.
Figure 1.
Transfer of 14C-labeled xylose from UDP-[14C]Xyl to various acceptors by pea microsomal membranes treated with 0.4% digitonin. The reactions were stopped by addition of 4 volumes of water before addition of a strong basic ion exchange resin and centrifugation, as described in Materials and Methods. The supernatants containing the radiolabel incorporated into cellotriose (■), cellotetraose (□), and G5 (○) were counted. The values are the average of three assays, and the bars show SE.
To determine whether digitonin-solubilized as well as residual membrane-bound xylosyltransferase activity was able to transfer 14C-labeled Xyl into G5, we repeated the same incubations described above (Fig. 1) by using as enzyme the supernatant (solubilized) and pelleted (bound) fractions from microsomes treated with various concentrations of digitonin. As expected, the progressive loss of the ability of microsomal membranes to incorporate UDP-Glc and UDP-Xyl into XyG was correlated with the progressive increase of the G5-dependent xylosyltransferase activity in the soluble fraction (data not shown). Furthermore, the soluble xylosyltransferase had the same properties as presented in Fig. 1, namely, the ability to incorporate Xyl into G5 but not to cellotetraose or cellotriose (data not shown). These findings lead us to conclude that the pea α-xylosyltransferase activity is involved in XyG biosynthesis and can act independently from XyG β(1,4)glucan synthase.
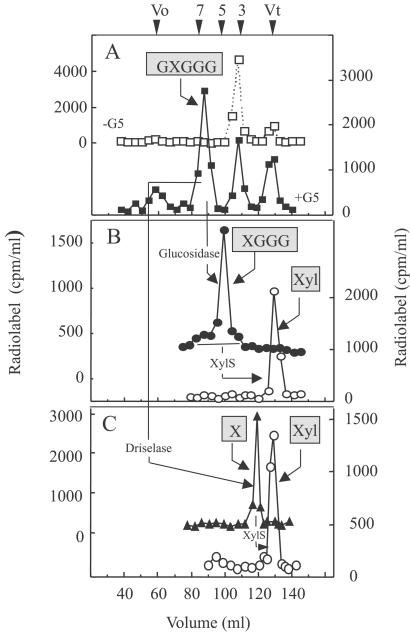
Characterization of the Radiolabeled Products Formed by Pea α-Xylosyltransferase from G5.
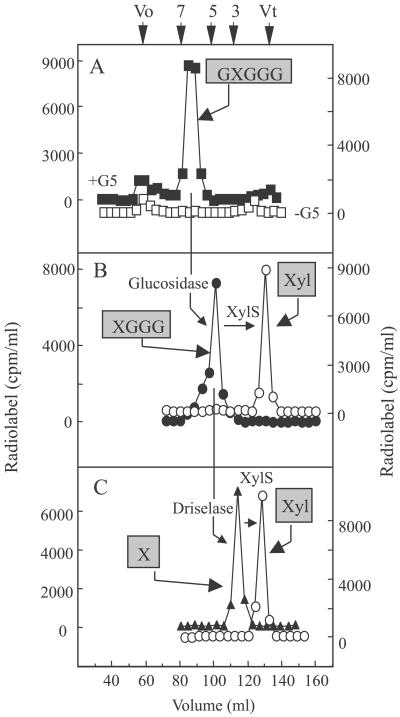
To characterize the radiolabeled products synthesized from G5 by the solubilized enzyme (1% digitonin), the reactions were scaled up 10–30 times and the products were fractionated on a Bio-gel P2 column (Fig. 2) both before and after treatment with various glycosidases. The elution profile of radiolabeled products (Fig. 2A) is consistent with a hexasaccharide; other radiolabeled products were also present in a control reaction (Fig. 2A), indicating that the hexasaccharide was the major product formed specifically when G5 was supplied to the reaction. When the radiolabeled hexasaccharide (Fig. 2A) was treated with XylS (12), very little 14C-labeled Xyl was released (data not shown). This result indicated that the hexasaccharide did not have a terminal isoprimeverose unit at the nonreducing end, because XylS is an exo-acting enzyme that releases xylose only from terminal isoprimeverose units (12). However, Driselase treatment of the radiolabeled hexasaccharide yielded radiolabeled disaccharide that coeluted with isoprimeverose, as expected from a product with an α-xylose residue somewhere along the chain of five Glc units (Fig. 2C). Moreover, the radiolabeled disaccharide was degraded by treatment with XylS, confirming that it contained an α-linked xylose (Fig. 2C).
Figure 2.
Characterization of radiolabeled products formed by digitonin-soluble pea enzyme in the presence of G5 and UDP-[14C]Xyl. (A) The products produced in the absence (□) or presence (■) of G5 were fractionated on a Bio-gel P2 column. The numbers along the top correspond to the size (DP) as estimated with malto-oligosaccharides (DP 2-7) and glucose as standards. (B) The products that eluted as hexasaccharides from A were treated with β-glucosidase (●), followed by XylS (○), then refractionation on a P2 column. (C) In another experiment, products the size of the hexasaccharides from A were digested with Driselase (▴) then with XylS (○) before separation on a P2 column. Reaction conditions for each treatment are described in Materials and Methods. The structure of the product shown in boxes presents the conclusions deduced from the product characterization (see text). Boxed X, isoprimeverose (17). Vo, void volume; Vt, total volume of the column.
When the radiolabeled hexasaccharide (Fig. 2A) was treated with β-glucosidase from Caldocellum saccharolyticum, which trims the unbranched Glc residues from the nonreducing end, the radiolabeled product shifted to a size expected of a pentasaccharide (90%), with small shoulders at hexasaccharide (6%) and tetrasaccharide (4%) (Fig. 2B). When these radiolabeled products were digested further with XylS, all of the radiolabel was released and eluted at the size expected of a monosaccharide. We conclude from these product characterization studies that the solubilized enzymes transferred one xylosyl residue from UDP-[14C]Xyl mainly to the second Glc residue from the nonreducing end of the G5, i.e., producing GXGGG, with minor amounts of XGGGG and GGXGG. This xylose was attached to glucose in an α-linkage, as would be expected in XyG.
Pea α-Xylosyltransferase Shares Molecular Mechanisms Similar to Fenugreek α(1,6)Galactosyltransferase.
The preliminary characterization of xylosyltransferase activity from peas led to the conclusion that it shared characteristics with a recently cloned α(1,6)galactosyltransferase from fenugreek (4). Common features include the formation of α(1,6)-linked branches on a linear β(1,4)glycan backbone. Most interestingly, when using exogenous acceptors both enzymes require five or more sugar residues (DP > 5). This similarity in molecular mechanisms suggested that the two proteins may share some amino acid sequence similarity. Therefore, the fenugreek gene was used to search the Arabidopsis genomic database. A cluster of 20 entries was identified and computer analysis of their sequences allowed us to narrow down the number of candidate genes to 7. All these Arabidopsis candidate genes are annotated as galactosyltransferases and are included in family 34, according to the CAZymes classification system (http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). This family includes genes encoding both α(1,6)- and α(1,2)galactosyltransferase from several species.
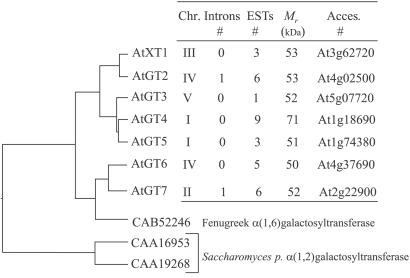
Fig. 3 summarizes some structural characteristics found in these putative AtGT proteins. All were predicted to have type-II membrane topology according to Kyte–Doolittle plots (18). The phylogenetic tree indicates that these proteins cluster into three groups, with A. thaliana xylosyltransferase 1 (AtXT1) (At3g62720) and AtGT2 (At4g02500) in one group sharing 85% identity at amino acid level. AtGT3 (At5g07720), AtGT4 (At1g18690), and AtGT5 (At1g74380) form a second subgroup sharing identity between 75–80% at the amino acid level. The third group is formed by AtGT6 (At4g37690) and AtGT7 (At2g22900) (75% identical). This group contained a third member, but the gene (At4g38310) is predicted to encode a 14-kDa polypeptide that does not contain a transmembrane domain or the DXD motif, which seems to be only a fragment of a glycosyltransferase. Whether this is a sequencing mistake or a real fragment needs to be determined. The third group also clusters with the fenugreek α(1,6)galactosyltransferase (≈53% identity to AtGT6 and AtGT7) (Fig. 3). The two α(1,2)galactosyltransferases from Schizosaccharomyces pombe form a distinct subgroup that shares low sequence identity (<15%) with the AtGTs. Despite this low sequence similarity, the alignment (clustal w) of all of the predicted proteins indicated a very conserved peptide region around a DXD motif, namely [E][FW][IVL][W][W][IVL][D][X][D][A][IVLM]. This motif occurs after a stretch of hydrophobic amino acids and is most likely to be involved in the stabilization/coordination of divalent cations in the binding of UDP-sugar (19). In this work we focused on six genes, namely AtXT1, AtGT2, AtGT3, AtGT4, AtGT5, and AtGT6.
Figure 3.
Structural characteristics and phylogenetic tree of AtXT1 and AtGT proteins. The molecular phylogenetic tree was based on the deduced amino acid sequences of AtXT1 and AtGTs and their alignment with the CLUSTAL W program. Lengths of lines indicate the relative distance between nodes. Accession numbers of each gene are indicated on the right. Because AtXT1 was confirmed to be a xylosyltransferase, we are using “AtXT” for its designation. Because computer predictions support the hypothesis that the other genes might be putative glycosyltransferases, we are using “AtGT” to designate them.
AtXT1 Has Cello-Oligosaccharide-Dependent α-Xylosyltransferase Activity.
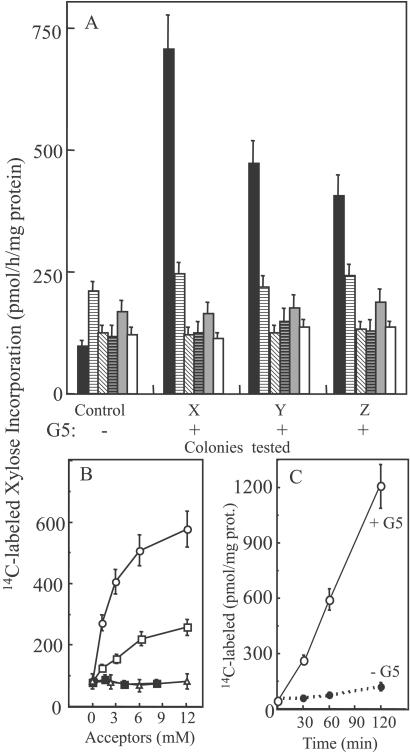
Full-length clones of these six genes were expressed in the methylotrophic yeast P. pastoris under the alcohol oxidase promoter. Screening for the α-xylosyltransferase activity was carried out with G5 as acceptor, UDP-[14C]Xyl, and 0.4% Triton X-100-solubilized proteins from yeast as the source of enzyme. Only the AtXT1 gene (At3g62720) product showed xylosyltransferase activity (Fig. 4A). To investigate its substrate requirement, other cello-oligosaccharides were tested as acceptors. Unlike the pea enzyme, AtXT1 was able to transfer the xylose onto an acceptor with a minimum of four Glc residues (Fig. 4B). Furthermore, the radiolabel incorporation is time-dependent, consistent with the kinetics of an enzyme-catalyzed reaction (Fig. 4C), and boiled enzyme lost its xylosyltransferase activity (data not shown). To investigate whether larger cello-oligosaccharides were good acceptors for AtXT1, we used the soluble cellulose derivatives, CMC, as substrate. The results indicated that the 14C-radiolabel incorporation increased with the time of incubation up to 4 h and was temperature-sensitive, consistent with an enzymatic reaction (data not shown).
Figure 4.
Cello-oligosaccharide-dependent α-xylosyltransferase activity from UDP-[14C]Xyl in detergent-soluble extracts from P. pastoris-expressing candidate genes. (A) Screening for activity in five colonies but only three colonies (X, Y, and Z) are shown for each clone: AtXT1 (■), AtGT2 (░⃞), AtGT3 (▧), AtGT4 (▤), AtGT5 (▩), and AtGT6 (□). The cells were grown under induction conditions (3- 4 days) before solubilization in EB containing 0.4% Triton X-100, and then the soluble fraction was used as the source of enzyme. (B) A single colony showing high activity with AtXT1 was tested for xylosyltransferase activity with cellobiose (▵), cellotriose (■), cellotetraose (□), and G5 (○). C shows the time-dependent incorporation of Xyl into G5 (6 mM). The values are the average of three assays, and the error bars show the SE.
When UDP-[14C]Glc or UDP-[14C]Gal were used as sugar donors for the transfer reaction in the presence of cello-oligosaccharides and AtXT1 or AtGTs, no radiolabel incorporation above the controls was observed under our conditions (data not shown). Also, AtXT1 and AtGTs did not catalyze 14C-labeled Gal incorporation from UDP-[14C]Gal into manno-oligosaccharide (data not shown), as was observed for fenugreek α(1,6)galactosyltransferase (4).
AtXT1 Produces Similar Radiolabeled Products from G5 as Pea α-Xylosyltransferase.
Reaction products obtained from G5 by AtXT1 were subjected to similar characterization procedures as described earlier for products produced by the pea enzyme. Transfer of 14C-labeled Xyl to G5 produced a radiolabeled product of the size of a hexasaccharide (Fig. 5A). Treatment of the product with β-glucosidase shifted its size to DP5 in Bio-gel P2 fractionation (Fig. 5B). α-Xylosidase digestion of this latter product released all of the radiolabel as free xylose (Fig. 5B). When the product from the β-glucosidase digestion was treated with Driselase, it released a radiolabeled product that eluted as disaccharide. Treatment of this latter product with α-xylosidase released a radiolabeled product that migrated as free xylose (Fig. 5C). Taking these data together, we concluded that AtXT1 transfers only one xylose residue to an α-linkage along the glucan chain, thereby producing the same product as the pea enzyme, namely GXGGG. Although a good incorporation of 14C-radiolabeled Xyl into CMC was obtained, the analysis of the resulting radiolabeled products was not carried out because of the presence of carboxymethyl groups, which interfered with hydrolysis by the various enzymes.
Figure 5.
Characterization of the radiolabeled products formed from G5 and UDP-[14C]Xyl by 0.4% Triton X-100 extract from Pichia cells expressing full-length AtXT1 (see Fig. 4C). (A) Products produced in the absence (□) or presence (■) of G5 were fractionated on a Bio-gel P2 column. (B) The radiolabeled product that eluted as hexasaccharide from A was treated with β-glucosidase (●) and then with Xyls (○). (C) Products the size of pentasaccharides were treated with Driselase (▴) and then with Xyls (○), as described in Materials and Methods and as in Fig. 2. The structure of the product shown in boxes presents the conclusions deduced from the product characterization (see text). Boxed X, isoprimeverose (17). Vo, void volume; Vt, total volume of the column.
Also, we observed that unlike fenugreek α(1,6)galactosyltransferase, the truncated AtXT1 protein failed to improve the activity. Surprisingly, the truncated AtXT1 lost the activity toward the G5 and showed substantial decrease of the activity for CMC (data not shown).
Discussion
An enzyme assay was developed to measure the activity of XyG xylosyltransferase that was solubilized from pea microsomal membranes. Preliminary characterization of the enzymatic properties of the XyG xylosyltransferase revealed that it had certain important properties in common with a galactosyltransferase that were described earlier (4). Taking advantage of the completed Arabidopsis genomic sequence, seven Arabidopsis genes with sequence similarity were identified that contained significant sequence similarity to galactomannan galactosyltransferase. Heterologous expression of six of these plant genes in the yeast P. pastoris led to the identification of one gene product that displayed α-xylosyltransferase activity.
Two lines of evidence support the hypothesis that the Arabidopsis α-xylosyltransferase is involved in XyG biosynthesis. First, it has acceptor substrate specificity very similar to the pea enzyme and what might be expected of an XyG xylosyltransferase. Both the pea and Arabidopsis enzymes are able to use G5 as acceptor; however, AtXT1 also was able to use cellotetraose as acceptor. Although it was not possible to test cello-oligosaccharides longer than 5 residues because of the insolubility of such oligosaccharides, the Arabidopsis enzyme was able to use CMC as substrate. Apparently, the presence of occasional substitutions on carbon-6 of the glucosyl residue did not prevent the xylosyltransferase from acting. The second, and most important, line of evidence supporting a role for these enzymes in XyG biosynthesis is that they produce the expected products. When G5 was used as acceptor, both the pea enzyme and AtXT1 produced mainly GXGGG.
One important question remains to be answered: How many xylosyltransferases are needed to make the core XyG? It is possible that only a single xylosyltransferase is needed. Although both the pea enzyme and AtXT1 add only one xylosyl residue, this could be attributed to the large excess of acceptor substrate in the reaction mixture. However, in this case, it is difficult to explain why the enzymes add xylose residue selectively on the penultimate Glc residues from the nonreducing end, producing mainly GXGGG. Because glycosyltransferases normally have high specificity we prefer the hypothesis that core XyG biosynthesis requires at least two or possibly three xylosyltransferases. In this case, it is tempting to speculate that the enzymes that add the second and third xylosyl residues may require the first xylosyl residue to be present. Detailed studies of acceptor specificity will be required to resolve these points. Regardless of whether one, two, or three enzymes are required, it is likely that they normally function in a complex that includes both a glucan synthase and the xylosyltransferases, therefore any conclusions based on in vitro assays with a single enzyme will need to be confirmed by other studies.
Given the hypothesis described above, it is tempting to speculate that some of the other Arabidopsis genes, AtGT2 to AtGT7, may encode xylosyltransferases with different specificity. For example, AtGT2 is 85% identical to AtXT1, but preliminary attempts to measure xylosyltransferase activity of the protein expressed in yeast cells give mixed results (unpublished observation). Thus, it seems possible that this gene may encode xylosyltransferase that cannot function independently and requires cooperative action from glucan synthase. Also, it is conceivable that AtGT2 does not act on unsubstituted glucan chains but functions to add the second and/or the third xylosyl residues to form XyG heptasaccharide XXXG, once the first xylose has been added. Efforts to explore these hypotheses are underway with functional and purified truncated proteins.
Supplementary Material
Acknowledgments
We thank Dr. Marco Moracci for providing XylS; Gordon Maclachlan, Prof. Emeritus (McGill University, Montreal) for his helpful discussion and comments; Ms. Karen Bird for editorial assistance; and CarboSource for the effort to make UDP-Xyl available (National Science Foundation Grant MCB 0090281). This work was supported by the Energy Biosciences Program at the U.S. Department of Energy and by National Science Foundation Plant Genome Program DBI 9975815.
Abbreviations
- AtXT1
A. thaliana xylosyltransferase 1
- AtGTs
A. thaliana glycosyltransferases
- G5
cellopentaose
- CMC
carboxymethyl-cellulose
- XyG
xyloglucan
- DP
degree of polymerization
- XylS
α-xylosidase from S. solfataricus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McCann M C, Roberts K. In: The Cytoskeletal Basis of Plant Growth and Form. Lloyd C W, editor. New York: Academic; 1991. pp. 109–120. [Google Scholar]
- 2.Carpita N C, Gibeaut D M. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 3.Perrin R M, DeRocher A E, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel N V, Keegstra K. Science. 1999;284:1976–1979. doi: 10.1126/science.284.5422.1976. [DOI] [PubMed] [Google Scholar]
- 4.Edwards M E, Dickson C A, Chengappa S, Sidebottom C, Gidley M J, Reid G. Plant J. 1999;19:691–697. doi: 10.1046/j.1365-313x.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- 5.Faik A, Bar-Peled M, DeRocher A E, Zeng W, Perrin R M, Wilkerson C, Raikhel N V, Keegstra K. J Biol Chem. 2000;275:15082–15089. doi: 10.1074/jbc.M000677200. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi T. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:139–168. [Google Scholar]
- 7.Vincken J P, York W S, Beldman G, Voragen A G J. Plant Physiol. 1997;114:9–13. doi: 10.1104/pp.114.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray P M. Biochim Biophys Acta. 1980;629:431–444. doi: 10.1016/0304-4165(80)90149-x. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi T, Matsuda K. J Biol Chem. 1981;256:11117–11122. [PubMed] [Google Scholar]
- 10.Gordon R, Maclachlan G. Plant Physiol. 1989;91:373–378. doi: 10.1104/pp.91.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brummell D A, Camirand A, Maclachlan G A. J Cell Sci. 1990;96:705–710. [Google Scholar]
- 12.Moracci M, Ponzamo B C, Trincone A, Fusco S, De Rosa M, Van der Oost J, Sensen C W, Charlebois R L, Rossi M. J Biol Chem. 2000;275:22082–22089. doi: 10.1074/jbc.M910392199. [DOI] [PubMed] [Google Scholar]
- 13.Love D R, Fisher R, Bergquis P L. Mol Gen Genet. 1988;213:84–92. doi: 10.1007/BF00333402. [DOI] [PubMed] [Google Scholar]
- 14.Faik A, Chileshe C, Sterling J, Maclachlan G. Plant Physiol. 1997;114:245–254. doi: 10.1104/pp.114.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman T, de Bruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whit A R, Xin Y, Pezeshk V. Biochem J. 1993;294:231–238. doi: 10.1042/bj2940231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry S C, York W S, Albersheim P, Darvill A, Hayashi T, Joseleau J-P, Kato Y, Lorences E P, Maclachlan G A, McNeil M, et al. Physiol Plant. 1993;83:1–3. [Google Scholar]
- 18.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Gastinel L N, Bignon C, Misra A K, Hindsgaul O, Shaper J H, Joziasse D H. EMBO J. 2001;20:638–649. doi: 10.1093/emboj/20.4.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.