Abstract
It has been believed that XIST RNA requires a discrete window in early development to initiate the series of chromatin-remodeling events that form the heterochromatic inactive X chromosome. Here we investigate four adult male HT-1080 fibrosarcoma cell lines expressing ectopic human XIST and demonstrate that these postdifferentiation cells can undergo chromosomal inactivation outside of any normal developmental context. All four clonal lines inactivated the transgene-containing autosome to varying degrees and with variable stability. One clone in particular consistently localized the ectopic XIST RNA to a discrete chromosome territory that exhibited striking hallmarks of inactivation, including long-range transcriptional inactivation. Results suggest that some postdifferentiation cell lines are capable of de novo chromosomal inactivation; however, long-term retention of autosomal inactivation was less common, which suggests that autosomal inactivation may confer a selective disadvantage. These results have fundamental significance for understanding genomic programming in early development.
Random inactivation of one of the two X chromosomes in mammalian females occurs early in embryogenesis to provide equivalent gene dosage between the sexes (reviewed in ref. 1; ref. 2). The inactivation process involves a number of changes in chromatin, producing greater condensation (evidenced by the heterochromatic Barr body), hypermethylation, late replication, hypoacetylation on core histone tails, and association with the unique histone macro H2A (reviewed in ref. 1). These chromosomal changes occur, before gastrulation, when facultative heterochromatin is first formed in previously totipotent cells of the early embryo.
The inactivation process is controlled by the XIST gene, which produces a polyadenylated RNA that does not encode a protein (3, 4). XIST is expressed exclusively from inactive X (Xi) and can initiate the X-inactivation process in developing mouse cells (5–7). The XIST RNA coats the inactive X chromosome in cis (8), triggering a cascade of events that synergistically produce the transcriptionally inactivated state. However, somatic cell hybrid studies provided evidence that the complete inactivation process may require the appropriate developmental context, because stable Xist transcripts localized without subsequent gene inactivation (9). In addition, Wutz and Jaenisch (10) showed that postdifferentiation embryonic stem (ES) cells lost the capacity to undergo inactivation in response to Xist RNA. Because of the apparent need for this developmental window, studies using ectopic XIST have focused on either developing mouse embryos or differentiating mouse ES cells. Although such studies have shown that just 15–35 kb of the mouse Xist gene are sufficient to trigger autosomal inactivation in these cells (7, 10), whether this is also the case for the human XIST gene has yet to be demonstrated in any model system. We have examined four transgenic clones carrying the full-length human XIST gene, stably integrated into a male human adult cancer cell line. This process avoids the potential complications of using interspecific hybrid cells and allows us to determine whether any hallmarks of X inactivation were seen after introduction of XIST. We show that these somatic fibrosarcoma cells are competent to respond to ectopic XIST RNA outside any normal developmental context.
Materials and Methods
Generation of Transgenic Clones and Cell Culture.
Transgene construct.
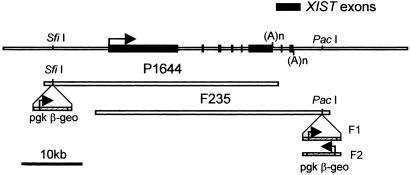
Two genomic XIST clones were isolated (P1644 and F235) from a Lawrist 4 human genomic library (Imperial Cancer Research Fund, London). Each genomic construct contained the entire 30-kb human XIST gene and approximately 10-kb flanking sequence (Fig. 1).
Figure 1.
XIST construct. (Top) Diagram represents genomic DNA spanning the human XIST gene. Cosmids P1644 and F235 contain the entire XIST gene (30 kb) and a β-geo selection marker. (Middle) P1644 includes ≈10 kb of 5′ flanking sequence, and (Bottom) F235 includes ≈10 kb of 3′ DNA. The F1–2 and F2–6 clones have the F235 construct, and the P1 clone has the P1644 construct. F1–2 has the β-geo marker in one orientation (F1) and the F2–6 line has the opposite orientation (F2).
Production and isolation of clones.
Cosmid clones were linearized and transfected into the male human sarcoma cell line HT-1080 (11) by using Lipofectin (Life Technologies, Gaithersburg, MD). Cells were cultured in 0.6 mg/ml G418 for 1–2 weeks (≈10 days). Colonies were propagated in media without G418. Three clones were isolated that expressed XIST RNA, two contained the F235 cosmid (F1–2 and F2–6) and one contained the P1644 cosmid (P1). All three expressed the β-geo gene as well, indicating that the entire cosmid had integrated. The F1–2 clone has the F1 construct, and the F2–6 clone has the F2 (see Fig. 1). Although initial Southern blot and fluorescence in situ hybridization (FISH) analysis suggested a high copy number (>100), and two separate integration sites, the cell cultures studied here have only one site of XIST expression and a much smaller DNA FISH signal, suggesting a copy number no higher than 5–10.
Cell culture and fixation.
The three HT-1080 clones were maintained in MEM (GIBCO/BRL) and 10% FCS. Some cells were grown in the presence of 300 ng/ml G418 for 1 week before FISH analysis to determine neomycin resistance. WI-38 human female fibroblast (control) cells were grown in basal media eagle (GIBCO/BRL) and 10% FCS. Our standard cell fixation protocol in 4% paraformaldehyde has been described (12).
RNA and DNA FISH.
Probes used were a 10-kb human XIST gene construct (XIST plasmid G1A) (9) and human Cot-1 DNA (GIBCO/BRL). The DNA probes (1 μg/reaction) were nick-translated by using biotin-11-dUTP or digoxigenin-16-dUTP (Roche Molecular Biochemicals). For whole-chromosome detection we used a chromosome library hybridization (biotinylated X-chromosome paint, Oncor) and XIST RNA detection (8). Our protocols for DNA and/or RNA FISH have been described (12, 13).
Antibody Detection and Replication Timing.
RNA hybridization was performed before α-acetylated histone H4 (Upstate Biotechnology, Lake Placid, NY) immunostaining. Cytokeratin immunostaining was performed before RNA hybridization by using a human pan-cytokeratin antibody (Sigma). RNase inhibitor was included in all antibody incubations to preserve RNAs. For replication timing assays, cells were incubated in the presence of 0.3 μM BrdUrd continuously for 4, 5, and 7 h and then were harvested and fixed by using standard cytogenetic techniques (14). Under these conditions, any mitotic cell that contains a signal would have incorporated label in late S phase (see Results).
Microscopy, Image Analysis, and Relative XIST RNA Analysis.
A Zeiss Axioplan microscope equipped with a triple-bandpass epifluorescence filter (Chroma Technology, Brattleboro, VT) was used for all imaging. Digital images were captured with a Photometrics series 200 charge-coupled device camera (Photometrics, Tucson, AZ). The ability of FISH signals to be used for relative RNA quantitation has been shown (15). Relative RNA levels were estimated after photographing approximately 25–35 individual cells of each cell line in each of 4–5 different experiments (P1 results were from only one experiment). Each channel, the 4′,6-diamidino-2-phenylindole (DAPI)/DNA (blue) and the XIST RNA (green), was photographed separately with a 16-bit digital image. All exposure times and fluorochromes were identical. Integrated intensities were measured for each nuclear territory, after subtraction of background fluorescence, by using metamorph (Universal Imaging, Media, PA). The mean integrated XIST fluorescence and SDs for each cell line were calculated, and the ratios of signal intensities between the cell lines were compared (see Results). ANOVA showed that the variation between the different experiments within each cell line was insignificant in comparison to the variation between cell lines (P ≪ 0.001), suggesting differences between lines were biological not technical.
Bromouridine-UTP (BrUTP) Transcription Assay.
Beginning with published protocols (16), several anti-BrdUrd antibodies and fixation conditions were tested for detection of all nascent transcription (BrUTP containing RNA). However, nucleolar labeling remained variable (further details are in additional Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org).
Results
XIST RNA Localizes and Produces a Distinct Barr Body in F2–6 Cells.
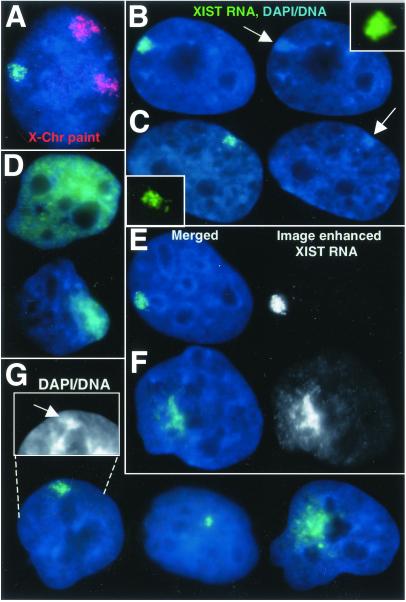
Three human clonal cell lines (F2–6, F1–2, and P1) derived from HT-1080 cells and expressing ectopic human XIST were investigated for XIST RNA localization by FISH, followed by the examination of other hallmarks of X inactivation. The frequency of cells expressing XIST for these experiments was 30% in F2–6, 40% in F1–2, and 72% in P1 and may reflect a decrease in expression during propagation. Although the original clones contained two integration sites (unpublished data), most cells (≈80% of expressing cells) studied here had only one focal site from which the RNA is detected. HT-1080 cells have an epithelium-like morphology and a near diploid (pseudodiploid) karyotype (11), but display many highly rearranged chromosomes. Although rearrangements of the chromosomes carrying the transgene made it difficult to identify the chromosomal origins in each line, it was too large to be the Y chromosome, and use of an X chromosome paint ruled out X chromosome involvement (see Fig. 2A).
Figure 2.
XIST RNA localization and Barr body formation. (A) The F2–6 TC chromosome is not the X chromosome. Ectopic XIST RNA (green), X chromosome paint (red). (B) F2–6 XIST RNA (green) and condensed DNA of a Barr body (arrow) (in 98% of expressing cells). (C) XIST RNA (green) and Xi Barr body (arrow) in normal female WI-38 cells. (D) Some F1–2 cells (5%) show large XIST RNA (green) drift. Most (70%) show diffuse cloud. (E and F) The XIST RNA (green) signal is enhanced equally for F1–2 and F2–6 cells (black and white images). XIST RNA drift can be seen in the more focal F1–2 cells (F). Equally enhanced F2–6 signal (E) shows little drift. (G) The P1 clone shows variable XIST localization: 74% are more localized, 20% have small spots, and 6% have large clouds. The more localized signal also shows some DNA condensation (arrow, Inset). (Magnification: ×2,000.)
In female nuclei, XIST RNA distribution is restricted to the interphase territory of the Xi (8). The XIST RNA distribution in 98% of the expressing F2–6 clones displays tight localization to a well-defined interphase region (Fig. 2B). This localization looks similar to normal female cells (Fig. 2C), as there is very little “drift” of the RNAs from the discretely bordered territories. In contrast, XIST RNA in the F1–2 clone does not localize well, with most (75%) cells showing unambiguous mislocalization (Fig. 2D). Even in 25% of F1–2 cells with more focal signal (Fig. 2F), close examination shows faint XIST RNA throughout the nucleoplasm. Equally enhanced images of F2–6 cells show very little dispersed signal (Fig. 2E). XIST RNA localization in the P1 clone is also variable (Fig. 2G); however, 74% show a somewhat localized signal.
The intense DAPI staining of the Barr body in human cells is a key hallmark of X inactivation. In 98% of expressing F2–6 cells, a prominent and distinct Barr body was readily apparent in these male cells (Fig. 2B). In fact, this Barr body appeared consistently more prominent than is often seen in the Xi in normal female WI38 fibroblasts (Fig. 2C). Interestingly, the F2–6 transgene-containing (TC) Barr body exhibits nuclear organization similar to that of the Xi in normal female cells, being localized to the heterochromatic compartment at the periphery of the nucleus (Fig. 2B) or the nucleolus (see Fig. 5B). Although such well-formed Barr bodies were not evident in the F1–2 cells, including cells with more “focal” XIST signals, most cells (72%) showed some DNA condensation (see Fig. 4C) coincident with the most concentrated XIST signal. The P1 line also had a significant fraction (42%) of cells that displayed a condensed region beneath the XIST signal (Fig. 2G) although they were not comparable to the obvious Barr bodies seen in either the WI-38 or F2–6 lines.
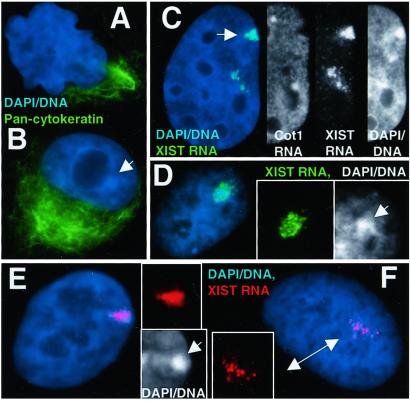
Figure 5.
State of differentiation and selection against inactivation. (A) F1–2 and (B) F2–6 cells contain cytoplasmic cytokeratins (green). A TC Barr body can be seen in the F2–6 cell (B) (arrow). (C) ≈1% of F2–6 cells exhibit two sites of XIST integration. Insets reveal only the top site (arrow) has a transcription hole, good RNA localization, and a clear Barr body. (D) Earlier passage F1–2 cells showed better localization of XIST RNA (green) and DNA condensation (arrow, DNA Inset). (E and F) L1.10.1 cells with localized XIST RNA (red) (E) and Barr bodies (arrow, DNA Inset). Localization was lost during propagation (F Inset) in all of the cells. (Magnifications: ×1,500, A, B, and D; ×2,000, C and F.)
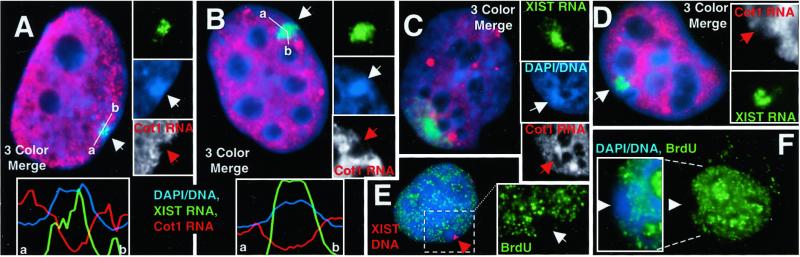
Figure 4.
Global transcription analysis. (A–D) Hybridization of Cot-1 DNA probe to predominantly intron containing heterogeneous nuclear RNA (red) and XIST RNA (green). Both the WI-38 normal female (A) and the F2–6 (B) lines show a hole (red arrows) in the Cot-1 signal coincident with the XIST RNA (green signal), and Barr body (arrow, blue insets). Fluorescence intensity plots for each signal across the area of the XIST localization (white line in merged image) illustrate the transcription holes coincident with the XIST signal and Barr bodies. The measurement line in the F2–6 cell (B) was curved to avoid the large Cot1 signal. Both F1–2 (C) and P1 (D) cells usually show diminished transcription (red arrows, Insets) under the brightest XIST signal. DNA condensation is sometimes associated with the transcription hole (C; arrow, blue insets). (E and F) F2–6 cells. BrUTP incorporation (green) into newly transcribed RNA shows a hole (white arrows) in the transcription signal coincident with the Barr body and the ectopic XIST gene (E; red arrow). (Magnifications: ×2,000, A–E; ×1,500, F.)
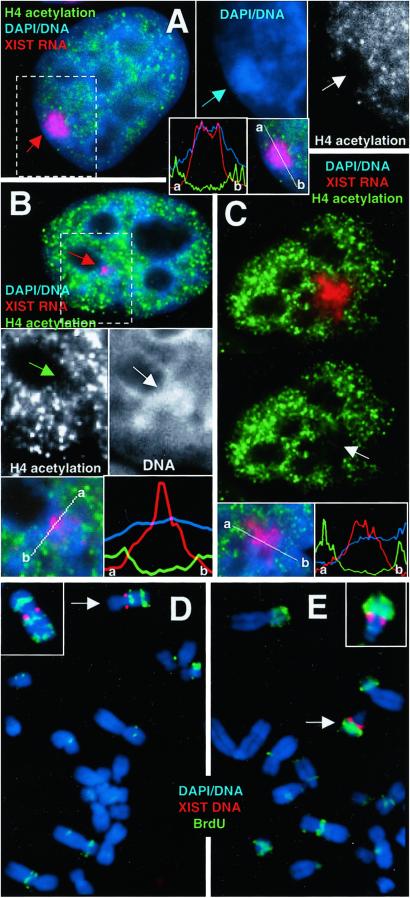
The F2–6 TC Autosome Is Hypoacetylated and Late Replicating.
The state of H4 acetylation was ascertained in interphase by using a double label of antiacetylated histone H4 antibody and XIST RNA FISH. The territory occupied by the XIST RNA/Barr body in the F2–6 clone exhibited little to no label for acetylated H4, producing an obvious “hole” in the overall nuclear staining (Fig. 3A). The fluorescence intensity plot of each signal shows that the overlapping peaks of XIST RNA and DNA are coincident with the hole in the acetylation signal. Both the F1–2 and P1 lines also contained some nuclei (≈60% and ≈30%, respectively), which exhibited a region of diminished acetylation coincident with the most concentrated area of XIST RNA (Fig. 3 B and C).
Figure 3.
Acetylation and late-replication analysis. (A–C) XIST RNA probe (red) and α-acetylated H4 antibody (green). (A) F2–6 cells reveal a hole (white arrow) in the acetylation over the XIST RNA (red arrow) and Barr body (blue arrow). Some diminished signal is seen in both P1 (B; green arrow) and F1–2 (C; arrow). Fluorescence intensity plots across the XIST signal (white line in merged Insets) illustrate the acetylation holes. Absence of acetylation in these cell lines sometimes corresponds to DNA condensation (B; white arrow). (D) F2–6 metaphase spread show XIST (red) and BrdUrd (green) (5 h BrdUrd sample). (Inset) The TC chromosome from a slightly earlier-replicating spread exhibiting a third late-replicating band on the p arm. The single copy endogenous XIST gene does not appear in these images because the signal intensity of the ectopic XIST is so much higher. (E) Some F1–2 cells (40%) show a late-replicating chromosome, (arrow). (Inset) An example of the same chromosome from another spread. (Magnifications: ×2,000, A–C; ×3,500, D and E.)
To determine whether the F2–6 TC chromosome is also late replicating, the cells were labeled continuously with BrdUrd 4, 5 and 7 h until fixation and visualization at metaphase. This process will label only late-replicating DNA. The predominant TC chromosome in the F2–6 cell line (78% of spreads) is a submetacentric C group chromosome, with the XIST integrated at the proximal region of the q arm. This autosome usually contained the latest-replicating DNA in the majority of spreads (68%). Particularly informative were spreads with very little label (representing the latest-replicating DNA; usually only telomeres and centromeres), indicating that large segments of the TC chromosome were among the last DNA in the cell to replicate (Fig. 3D). Although the whole chromosome did not replicate synchronously, very late-replicating segments were seen on both sides of the centromere and very distal to the XIST integration site, clearly indicating a long-range effect.
The replication results for the F1–2 line were variable, with the majority of TC chromosomes exhibiting early/middle replication (≈70%). However, a significant fraction displayed a late-replicating chromosome (≈40%) (Fig. 3E) (percentages do not equal 100% as some cells contained two TC chromosomes). Unlike the F2–6 and F1–2 lines, there was no obvious late-replicating chromosome in the P1 line, which showed mostly intermediate replication. Without knowing the homologous autosomal segments for the P1 TC chromosome, it remains possible that the inactivated TC chromosome is later replicating than its homolog.
Evidence of Local Transcriptional Silencing in the F2–6 Clone.
Because recent results demonstrate that replication timing is not strictly correlated with transcriptional inactivation (17), the most direct test for the final outcome of the inactivation process is to examine transcription more directly. Transcription of the neomycin resistance gene (neoR) located in the integrated cosmid, adjacent to the XIST gene, was assessed. When the F2–6 line was grown in the presence of 300 μg/ml G418 for 1 week, the number of XIST-expressing cells dropped to less than 1%, clearly indicating that the neoR gene had been inactivated. On the other hand, there was no decrease in expressing cells in the F1–2 line, indicating the neoR gene remained active. It is possible that the F2–6 neoR genes were inactivated independent of XIST-mediated inactivation (e.g., methylation); however, it would seem unlikely that only the tandem array of F2–6 neoR genes but not the accompanying XIST genes or the F1–2 neoR genes were affected.
Evidence for Long-Range Transcriptional Silencing Across the F2–6 TC Chromosome.
Although inactivation of the neoR gene in F2–6 cells suggests short-range transcriptional silencing, we wanted to develop an approach for assessing transcriptional status more broadly over the chromosome and avoiding the limitations of assaying only a few genes. In this case, analysis of other individual genes was particularly problematic because of the difficulty of identifying the highly rearranged chromosomes, as well as the difficulty of finding expressed gene probes detectable by FISH. Therefore, we developed an assay that could quickly and directly assess the transcriptional status across the interphase chromosome, using a separate, but related, approach to corroborate it.
To ascertain whether the TC Barr body is producing significant RNAs of any species, we hybridized with labeled Cot-1 fraction of genomic DNA (enriched for repeated sequences), which may broadly detect heterogeneous nuclear RNA, particularly intron-containing (pre)-mRNAs. Hybridization using the Cot-1 probe under nondenaturing conditions to prevent DNA hybridization produces a very bright granular fluorescence throughout the nucleoplasm (Fig. 4 A–D). This signal is not present over the Xi in normal WI-38 cells (Fig. 4A), demonstrating that this approach successfully identifies the inactive chromosome. Similarly, the F2–6 transcription signal is also absent from the territory defined by ectopic XIST RNA (Fig. 4B), coincident with the TC Barr body. The fluorescence intensity plot of each signal shows that the overlapping peaks of XIST RNA and DNA are coincident with the hole in the transcription signal. In fact, the absence of Cot-1 RNA over the F2–6 Barr body was often more pronounced than for WI-38 cells. Interestingly, both the F1–2 (Fig. 4C) and P1 (Fig. 4D) clones also showed some suggestion of transcriptional inhibition in the area of brightest XIST signal (63% and 42%, respectively).
To corroborate the above findings, we used an approach that is commonly used in studies of nuclear structure to assay the distribution of transcriptional activity. This procedure examines BrUTP incorporation into newly synthesized transcripts to visualize all nascent RNA (16, 18, 19). We tested several versions of these protocols to find that which gave the best labeling of both the nucleoplasm and nucleolus (Fig. 4F) (for examples of the other versions, see additional Materials and Methods, Results, and Figs. 6–8, which are published as supporting information on the PNAS web site). Sites of nascent transcription distributed broadly throughout the nucleus as punctate dots, but were excluded from the F2–6 TC Barr body (Fig. 4 E and F), as was previously suggested for the normal Xi (20). Although subsequent XIST DNA hybridization caused diminution of the transcription signal, particularly nucleolar signal (Fig. 5E), the transcription hole is still clearly visible.
Thus, we conclude that the F2–6 TC chromosome shows essentially the same degree of transcriptional inactivity as seen for the normal Xi in female cells, and the other two clones show partial inhibition.
Both the F1–2 and F2–6 Cells Express Epithelial Cell Differentiation Markers.
Because results suggest inactivation has occurred postdevelopment, we wanted to verify that these fibrosarcoma cells still expressed markers associated with a differentiated phenotype. Given the epithelial morphology of HT-1080 cells, we tested an antibody against cytokeratin proteins. The majority of cells in both lines (98% of parental HT-1080, 51% of F1–2, and 98% of F2–6) still exhibit normal levels of cytokeratin in the cytoplasm (Fig. 5 A and B), confirming that the original clones were differentiated adult cells. Interestingly, much of the F1–2 population shows loss of differentiation, with ≈60% displaying abnormal nuclear shape (Fig. 5A), and 37% exhibiting absence of cytokeratin expression (12% show very low expression), suggesting that loss of differentiation properties does not correlate with more efficient localization and inactivation.
Less Complete Inactivation Is Not Caused by Lower XIST RNA Levels.
We ascertained whether the differences between the clones were caused by insufficient levels of XIST RNA expression. Because of high variability between cells, we needed to estimate the XIST levels within individual cells in the population. We therefore measured the amount of XIST fluorescence detected in the nucleus of F1–2, F2–6, and WI-38 lines. These results suggest that on average, the F2–6 clone produces ≈3 times the amount of XIST signal seen in the normal female cultured cells, P1 ≈4.5 times, and the F1–2 line approximately 18 times more (Table 1). Although it appears that insufficient XIST expression is not correlated with the mislocalization, neither does excess RNA, because 40% of F1–2 cells contained relative RNA levels within the range of the highest F2–6 cells, yet still failed to localize the RNA properly.
Table 1.
Data summary
| Clone | XIST
|
Hallmarks
|
Other
|
||||
|---|---|---|---|---|---|---|---|
| XIST loc. | RNA level | Barr body | Trans. repress. | Late repl. | H4 Hypo-acetyl. | Diff. | |
| Female cells | + | 1× | + | + | + | + | + |
| GCL | + | ∼3× | + | + | + | + | + |
| BCL | − | ∼18× | +/− | +/− | +/− | +/− | + |
| P1 | +/− | ∼4.5× | +/− | +/− | +/− | +/− | □ |
| L1.10.1 | + | □ | + | □ | □ | □ | □ |
Column abbreviations: XIST loc., XIST localization; Trans. repress., transcriptional repression; Late repl., late replication; H4 Hypo-acetyl., H4 hypo-acetylation; and Diff., differentiation. Empty boxes represent analyses not determined for each cell line. Relative RNA estimates were obtained from ∼30 cells in four replica experiments, and the integrated intensity values (arbitrary units) obtained were F1–2 = 7.2 × 106 (SD 6 × 106), F2–6 = 1.2 × 106 (SD 0.7 × 106), WI-38 = 0.4 × 106 (SD 0.4 × 106), and P1 = 1.8 × 106. The RNA levels given are in relation to the amount seen in normal cells. +/− indicates that there was high variability for that hallmark in the population or that there were suggestions of hallmarks but they were not at the level seen in normal female cells.
Results Suggest Instability Selection Against Inactivation.
In a separate collaborative study on XIST and TSIX interactions (J. C. Chow, L.L.H., J.B.L., and C. J. Brown, unpublished work) another HT-1080 clone (L1.10.1) was generated as a control. This clone contains a P1 artificial chromosome with the full-length ectopic XIST gene. We briefly examined this clone in the context of this study to determine whether it also showed signs of inactivation. Initial observations were made on cells shortly after isolation (6–8 passages). These L1.10.1 cells (71%) showed highly localized XIST RNA and a well-defined Barr body, similar to the F2–6 line (Fig. 5E), suggesting a similar level of autosomal inactivation. However, after the cells had been propagated for further analyses and freezing (not more than 1 month), the phenotype of the population had clearly changed. The fraction of cells showing evidence of inactivation had decreased markedly and a more dispersed XIST localization had become the predominant phenotype (Fig. 5F).
We therefore re-examined slides of earlier passage F1–2 cells and established that before several months of culture (when most of the analyses were done) the F1–2 cells exhibited much more localized XIST RNA (Fig. 5D). In addition, the great variety in inactivation status (Table 1) and TC chromosome morphology in the F1–2 and P1 clones, as well as the loss of the second integration site in all three lines, further suggests that there have been changes and selection during propagation. Interestingly, there is a small fraction of the F2–6 population (≈1%) that still contains both integration sites, but they exhibit two different XIST localization phenotypes. The RNA from one appears localized and shows hallmarks of inactivation, whereas the other RNA is dispersed, with no associated Barr body (Fig. 5C). This finding, as well as the fact that some of these F2–6 cells now express very little XIST RNA from the second site, suggests that it was selectively lost, and now the only autosomal inactivation site preserved is one that can be stably maintained.
Discussion
As embryonic cells go from totipotent to the more restricted fate of postdifferentiation, chromatin is extensively remodeled to form a cell-type specific pattern of facultative heterochromatin. It was reasonable to expect that the competence to undergo these complex changes would be restricted to cells traversing a particular stage of development. However, this study establishes the precedent that a human XIST transgene can induce chromosomal inactivation in postdifferentiated human cells, outside this narrow developmental window. Because the cells studied here were derived from an adult human fibrosarcoma, we took measures to affirm that the cell lines studied still expressed clear postdifferentiation markers. In fact, the F2–6 line, which most clearly demonstrated inactivation, had the strongest and most consistent expression of the differentiation marker. Therefore, some postdifferentiation somatic cells still retain the potential for broad-scale formation of facultative heterochromatin. This finding means that the cells contain all of the necessary enzymatic functions to extensively remodel large blocks of DNA de novo, and that the genome of these cells remains in a state competent to respond to the relevant signals. In addition, this work demonstrates that in human cells a relatively small construct comprising essentially only the human XIST gene can induce inactivation of an autosome in cis, which had been previously shown only for the mouse Xist gene. Because working in human embryonic stem cells can be prohibitive and using human XIST in mouse cells has produced variable results, possibly caused by species differences (21–25), results here suggest a valuable alternative approach to studying human XIST RNA's role during inactivation in human somatic cells. Lastly, we demonstrate the value of an approach for evaluating general transcriptional activity directly on an interphase chromosome.
Initiation of Chromosomal Inactivation in Postdifferentiation Cells.
Of the four clonal ectopic XIST lines studied, the TC autosome in the F2–6 clone most clearly inactivated, by all criteria and in a manner stable for many generations in culture (summarized in Table 1). Surprisingly, most hallmarks of inactivation in the F2–6 line were typically more prominent than the normal Xi in female fibroblasts.
Although the F2–6 clone demonstrated unmistakable signs of stable inactivation, the other three clones also exhibited some evidence of inactivation (see Table 1). In fact, none of the HT-1080 clones that expressed XIST showed a complete absence of autosomal inactivation. For example, although the majority of cells in both the F1–2 and P1 clones did not localize the XIST RNA properly, a significant portion of the population consistently demonstrated some evidence of inactivation hallmarks. In addition, the fourth HT-1080 clone had displayed well-localized XIST RNA and obvious Barr body formation. Thus, although only one clone exhibited stable inactivation, all clones showed some evidence of inactivation, which may have been more complete at one point (discussed below).
There have been cases in which postdevelopment somatic cells failed to initiate inactivation in response to XIST RNA localization, suggesting the necessity for the appropriate early developmental context (8, 10, 26). Although the clones used here were capable of initiating inactivation, we do not conclude that all adult cells are as competent to do so. Some adult cells may have significant potential to undergo inactivation; however, they may respond to XIST RNA more slowly or with less efficiency. For example, there may be difference(s) in levels of some enzymatic activity necessary for inactivation between mature and immature cells. Levels of different methyl transferase enzymes are lower in adult cells (reviewed in ref. 27), in which DNA methylation is shown to occur more slowly over many generations (28). Thus, fully differentiated cells may need to be maintained for a few generations with continuous XIST expression to allow new chromatin changes to be slowly incorporated by enzymes that may not be as efficient as embryonic enzymes. Our results provide some support for this idea, as our F2–6 cells were initially selected with G418 yet after many generations became sensitive to it.
This study also raises the interesting possibility that cancer cells may have a more plastic capacity to remodel and repress chromatin in response to XIST RNA that is closer to that of early embryonic cells. Interestingly however, it is the regression of X inactivation and the loss of facultative heterochromatin and DNA methylation that is more frequently linked to the neoplastic state (reviewed in ref. 11; refs. 29–33).
Autosomal Inactivation Is Selectively Lost in Most Clones.
Although inactivation appears to be a fairly frequent occurrence in the HT-1080 cell line, only the F2–6 clone shows stable, long-range inactivation of the autosomal chromatin. Results for autosomal inactivation may be complicated by selection and/or by a more tenuous association of XIST RNA with autosomal chromatin (for example, refs. 6 and 34–37). Several aspects of our results suggest that autosomal inactivation may commonly be selected against. The progressive loss of inactivation may be caused by genetic and/or epigenetic changes in the original clone that circumvents one or more parts of the inactivation process. Because there is a subfraction of cells that contained two phenotypically different transgene-XIST phenotypes (localized and nonlocalized) within the same cell, the effect must be a cis effect. However, the human antisense TSIX gene is unlikely to be involved, as the promoter, which is ≈27 kb 3′ of XIST, is not included in either the P1644 or F235 constructs (38). Therefore, the loss of the localization and/or inactivation hallmarks may reflect unknown epigenetic cis effects that influence XIST RNA's interaction with the chromosome, such as DNA sequence differences or protein modifications.
Assaying Global Transcriptional Silencing on an Unknown Autosome.
A final contribution of this study is the introduction of an assay to assess global transcriptional activity from an unknown chromosome, using assays that reveal newly synthesized nuclear RNA. We find that the most consistent and convenient assay for chromosomal silencing is localization of heterogeneous nuclear RNA by hybridization with a Cot-1 probe (further information on this or the BrUTP assays are found in additional Results, Materials and Methods, and Figs. 6–9, which are published as supporting information on the PNAS web site).
Conclusion
At least some postdifferentiation adult cells contain all of the necessary factors essential to undergo chromosomal inactivation, although the process may be less efficient. However, given sufficient time, complete chromosomal inactivation in a postdifferentiation somatic cell is possible. An important future direction will be to determine whether the plasticity of chromatin regulation exhibited by these HT-1080 cells can also be manifest by cells with no neoplastic history, using an inducible expression system such that selection against the inactivation phenotype is reduced.
Supplementary Material
Acknowledgments
We especially thank Carolyn Brown and Jennifer Chow for generously allowing us to include the L1.10.1 clone in this article. Additional thanks go to John McNeil for assistance with imaging, Christine Clemson for help with XIST literature, Lindsay Shopland and Kelly Smith for proofreading, and Laurie Lizotte for organizational and miscellaneous assistance. This project was supported by National Institutes of Health Grants GM53234, GM45441, and HD07439-09.
Abbreviations
- FISH
fluorescence in situ hybridization
- DAPI
4′,6-diamidino-2-phenylindole
- Xi
inactive X
- TC
transgene-containing
- BrUTP
bromouridine-UTP
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brockdorff N. Curr Opin Genet Dev. 1998;8:328–333. doi: 10.1016/s0959-437x(98)80090-7. [DOI] [PubMed] [Google Scholar]
- 2.Carrel L, Willard H F. Proc Natl Acad Sci USA. 1999;96:7364–7369. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong Y K, Ontiveros S D, Strauss W M. Mamm Genome. 2000;11:220–224. doi: 10.1007/s003350010040. [DOI] [PubMed] [Google Scholar]
- 4.Memili E, Hong Y K, Kim D H, Ontiveros S D, Strauss W M. Gene. 2001;266:131–137. doi: 10.1016/s0378-1119(01)00353-5. [DOI] [PubMed] [Google Scholar]
- 5.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Nature (London) 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 6.Lee J T, Jaenisch R. Nature (London) 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 7.Herzing L B K, Romer J T, Horn J M, Ashworth A. Nature (London) 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 8.Clemson C M, McNeil J A, Willard H F, Lawrence J B. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemson C M, Chow J C, Brown C J, Lawrence J B. J Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wutz A, Jaenisch R. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 11.Rasheed S, Nelson-Rees W A, Toth E M, Arnstein P, Gardner M B. Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 12.Johnson C V, Singer R H, Lawrence J B. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- 13.Tam R, Johnson C, Shopland L, McNeil J, Lawrence J B. Applications of RNA FISH For Visualizing Gene Expression and Nuclear Architecture. Oxford: Oxford Univ. Press; 2002. [Google Scholar]
- 14.Wydner K L, Bhattacharya S, Eckner R, Lawrence J B, Livingston D M. Genomics. 1995;30:395–396. [PubMed] [Google Scholar]
- 15.Johnson C, Primorac D, McKinstry M, McNeil J, Rowe D, Lawrence J B. J Cell Biol. 2000;150:417–432. doi: 10.1083/jcb.150.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei X, Somanathan S, Samarabandu J, Berezney R. J Cell Biol. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharp A, Robinson D O, Jacobs P. Hum Genet. 2001;109:295–302. doi: 10.1007/s004390100578. [DOI] [PubMed] [Google Scholar]
- 18.Jackson D A, Hassan A B, Errington R J, Cook P R. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dundr M, Raska I. Exp Cell Res. 1993;208:275–281. doi: 10.1006/excr.1993.1247. [DOI] [PubMed] [Google Scholar]
- 20.Verschure P J, van Der Kraan I, Manders E M, van Driel R. J Cell Biol. 1999;147:13–24. doi: 10.1083/jcb.147.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heard E, Kress C, Mongelard F, Courtier B, Rougeulle C, Ashworth A, Vourc'h C, Babinet C, Avner P. Hum Mol Genet. 1996;5:441–450. doi: 10.1093/hmg/5.4.441. [DOI] [PubMed] [Google Scholar]
- 22.Heard E, Mongelard F, Arnaud D, Chureau C, Vourc'h C, Avner P. Proc Natl Acad Sci USA. 1999;96:6841–6846. doi: 10.1073/pnas.96.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuura S, Episkopou V, Hamvas R, Brown S D. Hum Mol Genet. 1996;5:451–459. doi: 10.1093/hmg/5.4.451. [DOI] [PubMed] [Google Scholar]
- 24.Migeon B R, Kazi E, Haisley-Royster C, Hu J, Reeves R, Call L, Lawler A, Moore C S, Morrison H, Jeppesen P. Genomics. 1999;59:113–121. doi: 10.1006/geno.1999.5861. [DOI] [PubMed] [Google Scholar]
- 25.Migeon B R, Winter H, Kazi E, Chowdhury A K, Hughes A, Haisley-Royster C, Morrison H, Jeppesen P. Genomics. 2001;71:156–162. doi: 10.1006/geno.2000.6421. [DOI] [PubMed] [Google Scholar]
- 26.Surralles J, Natarajan A T. Cytogenet Cell Genet. 1998;82:58–66. doi: 10.1159/000015065. [DOI] [PubMed] [Google Scholar]
- 27.Turker M S. Semin Cancer Biol. 1999;9:329–337. doi: 10.1006/scbi.1999.0133. [DOI] [PubMed] [Google Scholar]
- 28.Graff J R, Herman J G, Myohanen S, Baylin S B, Vertino P M. J Biol Chem. 1997;272:22322–22329. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh S N, Shah P N. Cancer Genet Cytogenet. 1981;4:269–274. doi: 10.1016/0165-4608(81)90020-0. [DOI] [PubMed] [Google Scholar]
- 30. Takagi, N., Yoshida, M. A., Sugawara, O. & Sasaki, M. (1983) Cell 1053–1062. [DOI] [PubMed]
- 31.Klein C B, Costa M. Mutat Res. 1997;386:163–180. doi: 10.1016/s1383-5742(96)00052-x. [DOI] [PubMed] [Google Scholar]
- 32.Takagi N. Cancer Genet Cytogenet. 1997;93:48–55. doi: 10.1016/s0165-4608(96)00274-9. [DOI] [PubMed] [Google Scholar]
- 33.Miniou P, Jeanpierre M, Blanquet V, Sibella V, Bonneau D, Herbelin C, Fischer A, Niveleau A, Viegas-Pequignot E. Hum Mol Genet. 1994;3:2093–2102. doi: 10.1093/hmg/3.12.2093. [DOI] [PubMed] [Google Scholar]
- 34.Lee J T, Lu N, Han Y. Proc Natl Acad Sci USA. 1999;96:3836–3841. doi: 10.1073/pnas.96.7.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caiulo A, Bardoni B, Camerino G, Guioli S, Minelli A, Piantanida M, Crosato F, Dalla Fior T, Maraschio P. Hum Genet. 1989;84:51–54. doi: 10.1007/BF00210670. [DOI] [PubMed] [Google Scholar]
- 36.Brockdorff N, Duthie S M. Cell Mol Life Sci. 1998;54:104–112. doi: 10.1007/s000180050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Csankovszki G, Nagy A, Jaenisch R. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Migeon B R, Chowdhury A K, Dunston J A, McIntosh I. Am J Hum Genet. 2001;69:951–960. doi: 10.1086/324022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.