Abstract
We demonstrate that two rhodopsins, identified from cDNA sequences, function as low- and high-light-intensity phototaxis receptors in the eukaryotic alga Chlamydomonas reinhardtii. Each of the receptors consists of an ≈300-residue seven-transmembrane helix domain with a retinal-binding pocket homologous to that of archaeal rhodopsins, followed by ≈400 residues of additional membrane-associated portion. The function of the two rhodopsins, Chlamydomonas sensory rhodopsins A and B (CSRA and CSRB), as phototaxis receptors is demonstrated by in vivo analysis of photoreceptor electrical currents and motility responses in transformants with RNA interference (RNAi) directed against each of the rhodopsin genes. The kinetics, fluence dependencies, and action spectra of the photoreceptor currents differ greatly in transformants in accord with the relative amounts of photoreceptor pigments expressed. The data show that CSRA has an absorption maximum near 510 nm and mediates a fast photoreceptor current that saturates at high light intensity. In contrast, CSRB absorbs maximally at 470 nm and generates a slow photoreceptor current saturating at low light intensity. The relative wavelength dependence of CSRA and CSRB activity in producing phototaxis responses matches precisely the wavelength dependence of the CSRA- and CSRB-generated currents, demonstrating that each receptor mediates phototaxis. The saturation of the two photoreceptor currents at different light fluence levels extends the range of light intensity to which the organism can respond. Further, at intensities where both operate, their light signals are integrated at the level of membrane depolarization caused by the two photoreceptor currents.
Keywords: retinal protein‖photoreceptor‖receptor currents‖signal transduction
Unicellular flagellate algae optimize their light environment by motility responses. Phototaxis (or oriented movement) guides them toward or away from a light source, whereas photophobic responses prevent their crossing a light/dark border (1). In Chlamydomonas reinhardtii these photomotility responses are mediated by retinal-containing receptor(s), as shown by retinal reconstitution studies in blind mutants (2–5). Moreover, it has been established that the native chromophore of the photoreceptor protein(s) is 6-s-trans all-trans-retinal, as in archaeal rhodopsins, and its all-trans/13-cis isomerization is required for triggering behavioral responses (3, 4, 6).
A complex photoreceptor apparatus is used to track the light source. The photoreceptor molecules appear to be localized in a small portion of the plasma membrane overlying the eyespot. Light absorption/reflection by the eyespot modulates the photoreceptor illumination during helical swimming if the helix axis does not coincide with the light direction (7). This modulated illumination serves as a signal for the correction of the swimming path.
A cascade of electrical phenomena plays a key role in the signal transduction. Photoexcitation of the receptor molecules results in the generation of photoreceptor currents, membrane depolarization, and, above a certain threshold of the stimulus intensity, activation of voltage-gated Ca2+ channels in the flagellar membrane (8, 9). These rapid electrical events bring about asymmetric motile responses of the two flagella during phototaxis (10, 11) and a brief alteration of the flagellar beating mode during the photophobic response (12).
Photoreceptor current generation is the earliest detected consequence of light absorption by the phototaxis-receptor molecules, occurring <3 μs after a laser flash (9, 13). The localization of the current to the eyespot region of the cell corresponds to the expected location of the pigments (10, 11), and its action spectrum measured in vivo matches that of phototaxis (8) and of the photophobic response (14). The sensitivity of the current to inhibitors (8, 10, 11) and its chromophore requirements studied in retinal-deficient mutants (15) confirm its role in photomovement. Thus, the photoreceptor current provides a direct probe for activity of the photoreceptors that mediate light-controlled swimming behavior in vivo.
As shown in Haematococcus (13), fluence-response curves for photocurrent amplitudes in flagellates are biphasic. Fitting with a sum of two saturating functions reveals a slower low-light-saturating current and faster high-light-saturating current (9–11, 16, 17), which were interpreted as being driven by either a single, or two different, photoreceptor proteins (18). Multiple photoreceptors have been considered, based on the complex shape of the photoreceptor-current action spectrum (8, 11, 14), and results from retinal-reconstitution studies (19).
For several years the most abundant protein in the eyespot membranes of Chlamydomonas and Volvox has been considered as the photoreceptor for photomotile responses in these algae (20, 21). However, recently the so-called “chlamyrhodopsin” (21) has been ruled out as the photoreceptor pigment for either phototaxis or the photophobic response in Chlamydomonas (22). Therefore, the identification of the genuine photomotility receptor(s) still posed a challenge (18, 23, 24).
Materials and Methods
Strains.
C. reinhardtii strain 495 (Chlamydomonas culture collection at St. Petersburg University, St. Petersburg, Russia) and its transformants were grown on TAP (Tris-acetate-phosphate medium; ref. 25) agar plates under continuous illumination at 30°C. Cells from 5- to 7-day plates were converted into gametes by overnight incubation in the nitrogen-deficient medium (NMM) (2) on a rotary shaker under illumination. To minimize electrical noise, the cells were transferred by centrifugation to a low ionic strength measuring medium containing 1 mM Tris⋅HCl buffer (pH 7.5) and 0.1 mM CaCl2 at a concentration of 1–2 × 107 cells per ml. Equal cell density in the samples was controlled by measurements of optical density at 800 nm.
Escherichia coli DH5α and XL1-Blue were used for cloning the genes from cDNA, and E. coli transformants were grown in LB (Luria–Bertani) medium in the presence of ampicillin (50 μg/ml) at 37°C. The full-length CSOA and CSOB‖ constructs were expressed in Pichia pastoris with the plasmid expression system used for bovine rhodopsin (26). The strains CC620 and CC621 (E. Harris, Chlamydomonas Genetics Center Collection, Duke University, Durham, NC) were used for the isolation of gametic autolysin used to remove cell walls for Chlamydomonas transformation (25).
Identification and Cloning of CSOA and CSOB.
Gene fragments with homology to the archaeal rhodopsin apoprotein genes were identified in the expressed sequence tag (EST) project of C. reinhardtii at the Kazusa DNA Research Institute (http://www.kazusa.or.jp/). The CSOA gene was cloned from a cDNA library of light-grown cells and the CSOB gene was cloned from cDNA of dark-grown cells in TAP medium. λ-cDNA libraries were purchased from the Chlamydomonas Genetics Center. After the positive λ clones were confirmed by Southern hybridization, ExAssist interference-resistant helper phage with SOLR strain (Stratagene) was used to clone the EcoRI/XhoI fragments into the pBluescript phagemid vector by in vivo excision. The size of the cDNA of CSOA and CSOB is 3.7 kb and 3.6 kb, respectively. After the termination codon of each gene, CSOA and CSOB had an additional 1.6-kb and 1.3-kb extension, respectively, before the poly(A) tail.
Construction of Plasmids Encoding Hairpin dsRNA for CSOA and CSOB.
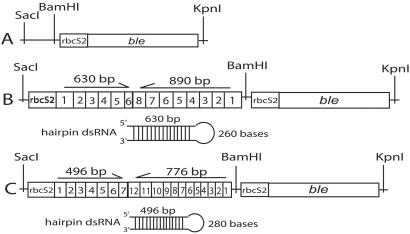
Restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were purchased from Promega, and Pfu DNA polymerase was purchased from Stratagene. Oligonucleotides were purchased from Sigma-Genosys (The Woodlands, TX), ampicillin from Sigma, and Zeocin from Invitrogen. The pSP124S plasmid (Fig. 1A) was used to construct hairpin A-RNAi (Fig. 1B) and B-RNAi (Fig. 1C). The rbcS2 promoter (27) and opsin genes were fused by recombinant PCR (28). The PCR was performed in 31 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 3 min, and the product was purified, digested with SacI and BamHI, and introduced into the pSP124S plasmid (29). Cloned PCR constructs were confirmed by DNA sequencing. The hairpin dsRNA constructs under the rbcS2 promoter in pSP124S were transformed by electroporation (30) into the C. reinhardtii strain 495 without carrier DNA, at an electrical-field strength of 1,375 V/cm in transformation medium of 40 mM sucrose in water. The bacterial phleomycin-resistance gene, ble, which is a dominant marker for nuclear transformation in the pSP124S plasmid, was used to directly select in a Zeocin plate (29). The cells were incubated on a TAP agar plate with Zeocin (10 μg/ml) for 7 days at 30°C. The transformant with pSP124S plasmid without RNAi constructs, which is resistant to Zeocin, was used as a control.
Figure 1.
Constructs of hairpin double-stranded (ds)RNAi. SacI/KpnI fragments of pSP124S (A), RNAi against CSOA (B), and RNAi against CSOB (C). Numbers within squares correspond to exons in the CSOA and CSOB genes. The rbcS2 sequence is a promoter for ribulose bisphosphate carboxylase/oxygenase in C. reinhardtii (27) and ble is the phleomycin-resistance gene (29).
Membrane Preparation.
The Chlamydomonas cells containing RNAi constructs were pelleted by centrifugation at 1,200 × g for 10 min. Membranes were isolated by sonication and suspended in 30 mM Tris, pH 7.0/30 mM NaCl/10% glycerol. To provide an epitope for detection of the full-length CSOA and CSOB in Pichia pastoris, hexahistidine tags were introduced at the C-terminal ends of the opsins (31). For detection in Chlamydomonas, anti-peptide antibody (Alpha Diagnostic, San Antonio, TX) was made against a synthetic peptide corresponding to the C-terminal 19 residues of CSOA (EMLQQLMSEINRLKNELGE).
Purified total membranes of Pichia and Chlamydomonas transformants were subjected to SDS/PAGE and immunoblotting using anti-CSOA-peptide antibody. The His-tagged CSOA in Pichia membrane was detected by anti-His-tag antibody (CLONTECH).
Photoelectric Measurements.
Flash-induced macroscopic electrical currents were recorded from a population of freely swimming cells as described (15, 16). The currents were detected by a pair of platinum electrodes immersed in a cell suspension and fed into a low-noise current amplifier 428 (Keithley). The signals were digitized and stored by using the DIGIDATA A1325 and PCLAMP 8.2 programs (both from Axon Instruments, Foster City, CA). Curves were fit by the ORIGIN 6.1 program (OriginLab, Northampton, MA). A photoflash Sunpak auto 383 (ToCAD America, Parsippany, NJ) with 0.14-ms rise time in combination with heat-absorbing and broad- and narrow-band interference filters was used as a light source. Flash energy was attenuated by neutral density filters and measured by a calibrated photodiode.
Two forms of the macroscopic current measurement were used. In the nonoriented mode, a spatially narrow flash oriented along the line connecting the two electrodes excites a suspension of randomly oriented cells. The cells with their eyespots toward the light source generate larger photoreceptor currents than those with their eyespots away from it, which gives rise to a difference signal.
In the second modification of the measurement (the preoriented mode), the excitation flash is applied perpendicularly to the line connecting the electrodes. Under these conditions, no electrical signals can be detected in a nonoriented suspension because asymmetric currents have zero projection on the line connecting the electrodes. However, when the cells are preoriented with continuous light along the line connecting the electrodes, the directions of their photoreceptor currents coincide, so that the electrodes pick up a sum of their projections on the direction of the orienting light. For more detailed description of the suspension assay, see Sineshchekov and Govorunova (32).
Phototaxis Measurements.
Phototaxis was measured by recording photoelectric currents in the preoriented mode (see above). The magnitude of the photoelectric signal recorded in this mode depends only on the degree of phototactic orientation of the cells, provided that the amplitude of the transmembrane currents generated by individual cells in response to the equal test flashes does not change (15, 16). The degree of phototactic orientation was assessed by measuring the amplitude of the photoelectric signal elicited by a white test flash after 10 s of continuous illumination (which was sufficient to reach a steady-state level of orientation) with orienting light of a desired spectral composition and intensity.
Results
Two Rhodopsins Are Encoded in the C. reinhardtii Genome.
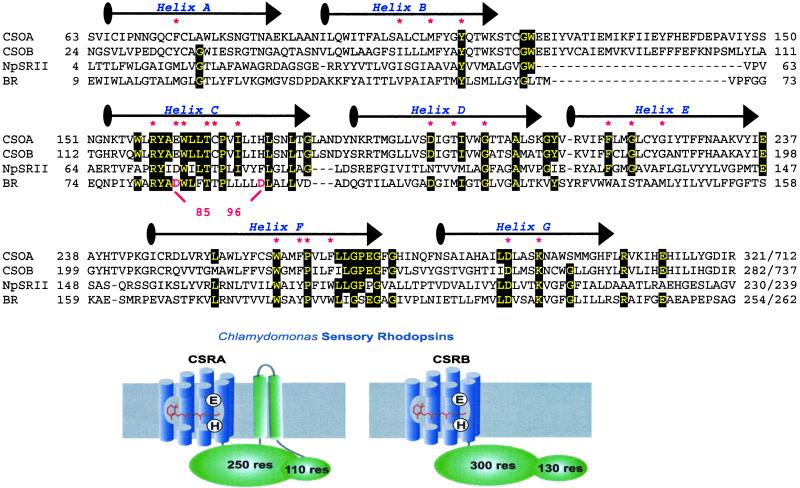
Based on cDNA fragments homologous to the archaeal rhodopsin apoprotein (“opsin”) genes, identified in the EST project of C. reinhardtii at Kazusa DNA Research Institute, we cloned the complete genes from a cDNA library as well as from C. reinhardtii genomic DNA. These putative opsin genes are predicted to encode 712- and 737-residue archaeal-type [type 1 (24)] rhodopsin apoproteins CSOA and CSOB (Chlamydomonas Sensory Opsins A and B, so named based on their sensory role demonstrated here), respectively. The N-terminal ≈300 residues of the rhodopsin homologs exhibit high identity in the most conserved regions in archaeal rhodopsins, namely helices C, F, and G (Fig. 2) followed by ≈400 residues of additional membrane-associated portion as shown in the cartoon in Fig. 2. In the case of CSOA, hydropathy plots predict two transmembrane helices. The CSOB sequence does not show clearly predicted transmembrane regions, although the 400-residue C-terminal domains of both CSOA and CSOB are membrane-associated when expressed in E. coli (data not shown).
Figure 2.
Primary sequences of the rhodopsin domains of sensory opsins from C. reinhardtii (CSOA and CSOB) aligned with sensory rhodopsin II (NpsopII) from Natronobacterium pharaonis, and bacteriorhodopsin (BR) from Halobacterium salinarum. Conserved residues are marked with black boxes and residues predicted to be in the retinal-binding pocket are marked with a red asterisk. The drawing of CSRA and CSRB is based on known microbial rhodopsin structures and secondary structure predictions.
Residues in contact with the retinal chromophore according to the crystal structure of the phototaxis receptor sensory rhodopsin II (NpSRII) (33) and bacteriorhodopsin (BR) (34), including the lysine that forms a Schiff base with retinal, are conserved (Fig. 2, marked with a red asterisk). A unique feature of the Chlamydomonas sensory rhodopsins is a Glu residue in place of the Asp counterion to the protonated Schiff base seen in other type-1 rhodopsins (Asp-85 in BR). The carboxylate proton donor specific to proton pumps (Asp-96 in BR), which is a noncarboxylate residue in known archaeal sensory rhodopsins, is replaced with a His residue. The C-terminal 110 residues of the rhodopsins blast-match to domain D of synapsin, which is regulated in synapsin by Ca2+/calmodulin-dependent protein kinases (35). This region contains several Ser/Thr which are predicted to be phosphorylation sites (36). The Chlamydomonas genome contains a single copy of each of the two opsin homologs according to our Southern hybridization with CSOA and CSOB genes against total genomic DNA of Chlamydomonas (data not shown). There are 14 introns in CSOA and 19 introns in CSOB. Ten introns are incorporated at the same positions in CSOA and CSOB.
Hairpin dsRNAi Suppression of Rhodopsin Expression.
Interference with the expression of specific genes by dsRNA is a powerful tool in many organisms, including Caenorhabditis elegans (37), Drosophila (38), Trypanosoma (39), and plants (40). Because efficient targeted gene disruption by homologous recombination is not available for Chlamydomonas nuclear genes (41), we used the hairpin dsRNAi technique (22, 39) to suppress the expression of the CSOA and CSOB genes (Fig. 1). To test for a functional role of the CSOA and CSOB rhodopsins in the photosensory control of motility, we prepared transformants directed against CSOA expression (A-RNAi), and separate transformants against CSOB expression (B-RNAi), and measured CSOA expression, photoreceptor currents, and photomotility responses.
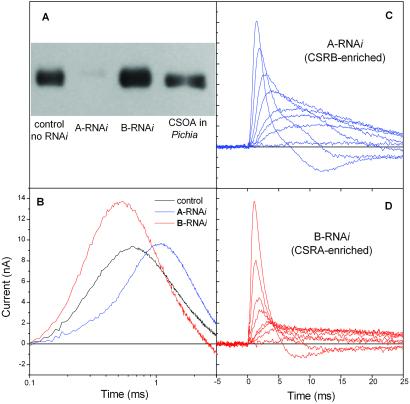
Immunoblots with anti-CSOA antibody showed that in A-RNAi transformants the amount of CSOA is greatly reduced (less than 5%), whereas in B-RNAi cells the CSOA amount is significantly increased (Fig. 3A). Given the high degree of homology between CSOA and CSOB, we expect the corresponding effects on CSOB, i.e., the amount of CSOB would be decreased in B-RNAi and increased in A-RNAi transformants. This expectation was confirmed by the electrophysiological evidence (see below).
Figure 3.
(A) Comparison of CSOA expression in C. reinhardtii strain 495 transformant with pSP124S (lane 1), 495 with A-RNAi (lane 2), 495 with B-RNAi (lane 3), and CSOA expressed in Pichia pastoris (lane 4). Antibody against CSOA C-terminal peptide was used in this immunoblot. The molecular mass of CSOA is 76–80 kDa on SDS/PAGE. (B) Photoreceptor currents in control cells, and A-RNAi and B-RNAi transformants. Currents were recorded under identical conditions in suspensions of nonoriented cells by unilateral excitation with a broad-band (350–550 nm) flash. (C and D) Sets of photoelectric currents excited by flashes with incrementally decreasing energy (approximately 60% steps) in A-RNAi (CSRB-enriched) and B-RNAi (CSRA-enriched) transformants, respectively.
CSRA Mediates the Faster High-Light-Saturating Current and CSRB Mediates the Slower Low-Light-Saturating Current.
Transformation with either A- or B-RNAi constructs specifically alters the kinetics of the photoreceptor currents compared with the wild-type control (C. reinhardtii strain 495 transformed with pSP124S plasmid). The main kinetic parameters of the signals are clearly different in the two transformants (Fig. 3 B–D). In A-RNAi cells, which have greatly reduced amounts of CSRA (Fig. 3A) the photocurrent signals are much slower; i.e., the initial slope of the current rise is decreased and the peak time and the decay time are increased (Fig. 3 B–D). In contrast, in B-RNAi cells, which have an increased amount of CSRA, the signals are accelerated, showing a greater rise slope as well as shorter rise time and decay time. Our interpretation is that CSRA generates the fast component of the photoreceptor current, and CSRB generates the slow component of photoreceptor current.
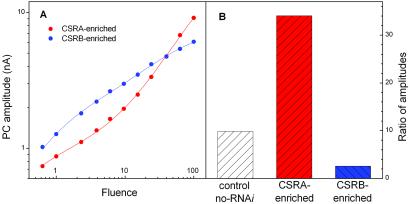
Analysis of fluence-response dependence of the current in the A-RNAi and B-RNAi transformants confirms that the two rhodopsins generate different components of the photoreceptor current (Fig. 4). In wild-type cells fitting fluence-response curves with a sum of two hyperbolic functions reveals a component of the current that saturates at low fluence and a second component that saturates at high fluence (9, 11, 17, 18). We examined the behavior of these two components in the RNAi transformants. Fluence-response curves for the amplitude of the overall photoreceptor current excited by broad-band (350–550 nm) flashes in the RNAi transformants and control cells were fitted by a sum of two hyperbolic functions as in the previously reported work. The results of the fits are presented in Fig. 4B. In control cells the 10:1 ratio of the amplitudes for high-fluence-saturating and low-fluence-saturating components corresponds to that previously reported. In A-RNAi transformants (i.e., CSRB-enriched) the amplitude of the high-saturating component decreases and the amplitude of the low-saturating component increases, giving rise to an ≈4-fold decrease in their ratio (2.5:1). In contrast, the ratio of high- and low-saturating amplitudes in CSRA-enriched cells is increased by >3-fold (34:1). Our interpretation is that the current generated by CSRA saturates at high light intensity, and that generated by CSRB saturates at low light intensity. Furthermore, this analysis of the currents in the transformants unambiguously shows that the high-light-saturating phase of the curve indeed corresponds to the fast current component, whereas the low-light-saturating phase corresponds to the slow component, as was proposed on the basis of kinetic analysis (9).
Figure 4.
(A) Fluence-response curves for the amplitude of the photoreceptor currents in CSRA-enriched and CSRB-enriched cells. (B) Ratio of the amplitudes of the high-light-saturating to low-light-saturating functions derived from fits of fluence-response curves for controls, CSRA-enriched, and CSRB-enriched cells. Experimental data were fit by the sum of two hyperbolae.
CSRB Absorption Is Shifted to the Blue of CSRA Absorption.
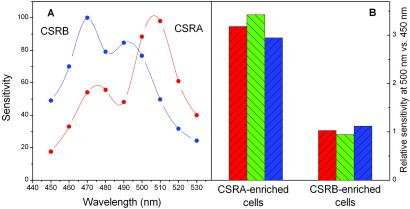
Action spectra for the photoreceptor-current amplitude in CSRA-enriched and CSRB-enriched cells reveal a clear difference between the absorption spectra of CSRA and CSRB and exclude the possibility of secondary effects of transformation on the sensory-transduction chain (Fig. 5A). In CSRA-enriched cells, the action-spectrum maximum is between 505 and 510 nm with a shoulder at 475 nm. In CSRB-enriched (CSRA-deficient) cells, maximum sensitivity was observed at 470 nm with a secondary minor band at 495–500 nm. The secondary bands in both spectra are likely to be due to residual amounts of the other (i.e., RNAi-suppressed) pigment. Their slight apparent spectral shifts toward the main bands are expected from the summation with the spectra of the stronger peaks.
Figure 5.
(A) Action spectra for photoreceptor currents in CSRA-enriched and CSRB-enriched cells. Sensitivity is the relative effectiveness of monochromatic light determined from fluence-response curves as the reciprocal of the fluence that gives a photoreceptor-current amplitude equal to that at a reference wavelength (500 nm for CSRA and 470 nm for CSRB), expressed in percent of the reference value. Spectra were measured at fluences of ≈1 μE/m2 (1 μE = 1 μmol of photons). Reproducibility was better than 90%. (B) Relative effectiveness of a spectral band absorbed predominantly by CSRA (480–520 nm) compared with that absorbed predominantly by CSRB (430–470 nm) in the generation of photoreceptor currents measured in the nonoriented mode (red columns), preoriented mode (green columns), and in the phototaxis-motility response (blue columns) in CSRA- and CSRB-enriched cells. Relative effectiveness was determined from fluence-response curves as a reciprocal of fluence (for photocurrents) and fluence rate (for phototaxis) giving equal responses. (See text for details.)
The appearance of macroscopic photocurrents in nonoriented cell suspensions is because of the directional sensitivity of the photoreceptor apparatus (see Materials and Methods). Directional sensitivity arises from the absorption and reflection properties of the highly pigmented eyespot, the primary shading device for the photoreceptor proteins. Therefore, the shift between the photoreceptor-current action spectra of the two pigments could in principle be because of differences in absorption/reflection of the eyespot. Comparison of the spectral dependence of the photoreceptor current in nonoriented cells with that in preoriented cells excludes this possibility. We measured the efficiency of photocurrent generation from light in two nonoverlapping spectral bands, one predominantly absorbed by CSRA (480–520 nm) and the other by CSRB (430–470 nm). In nonoriented suspensions, an increase in shading efficiency would increase the amplitude of the electric signal because the difference between “illuminated” and “shaded” receptors gives rise to the current. In contrast, in preoriented suspensions, asymmetric currents generated by all cells have the same direction because of their orientation along the line between the measuring electrodes and thus are summed to give the measured current. In this case any shading would decrease the light-induced electrical signal. Fig. 5B shows that the difference in relative efficiency of the two spectral bands in the CSRA-enriched versus CSRB-enriched cells is the same in both modes of measurement (red and green columns). Therefore, the difference between the action spectra of the two transformants (Fig. 5A) reflects the difference between the absorption spectra of the two pigments, and is not because of differences in screening.
Both CSRA and CSRB Mediate Phototaxis Motility Responses.
Both CSRA-enriched and CSRB-enriched cells exhibit phototaxis, as assessed by computerized motility analysis (data not shown). For quantitative determination of the degree of phototactic orientation of cells we used a simple, highly sensitive, and reproducible assay based on photocurrent measurement (see Materials and Methods). To test whether both or only one of the pigments mediates the motility behavior, we measured phototaxis orientation to light in two different spectral regions, one preferentially absorbed by CSRA and the other by CSRB. Cells were oriented along the line connecting the two electrodes with continuous light of the same short-wavelength and long-wavelength spectral bands as those used for the photocurrent measurements. The degree of cell orientation was determined with a test flash of constant fluence applied after 10 s of continuous illumination with the short- or long-wavelength light at 90°. The amplitude of the electric response measured under these conditions is directly proportional to the extent of cell orientation (15, 16). The relative phototaxis effectiveness of each photostimulus was derived from fluence-response curves as the reciprocal of the light intensity yielding a criterion extent of cell orientation. The relative phototaxis effectiveness of long-wavelength and short-wavelength light was different in A-RNAi and B-RNAi transformants, and exactly corresponded to the relative efficiencies of these spectral bands in generation of the photoreceptor current (Fig. 5B). We conclude that both CSRA and CSRB mediate phototactic orientation through the generation of their respective photoreceptor currents.
Discussion
In the 1970s and early 1980s four rhodopsins were discovered in the cytoplasmic membrane of the archaeon Halobacterium salinarum: the light-driven ion pumps bacteriorhodopsin and halorhodopsin, and the phototaxis receptors sensory rhodopsin I and II. Genome projects on a number of microbes have more recently revealed archaeal rhodopsin homologs in the other two domains of life as well, namely Eubacteria and Eucarya. Organisms containing these homologs live in such diverse environments as soil, freshwater, and ocean waters, and they include a broad range of microbial life, including proteobacteria, cyanobacteria, fungi, and algae (24). The Chlamydomonas rhodopsins provide an example of evolution fusing the seven-helix microbial rhodopsin motif with other domains (Fig. 2).
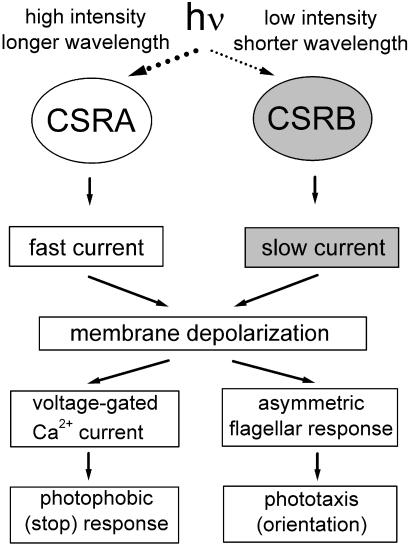
Our study of the RNAi transformants demonstrates that CSRA and CSRB function as photoreceptors for phototaxis responses in Chlamydomonas. Photoexcitation of either rhodopsin results in the generation of a photoreceptor current (Fig. 6). CSRA generates a faster photoreceptor current that saturates at high light intensities, and CSRB generates a slower (delayed) current, saturating at much lower intensities. Superposition of the CSRA- and CSRB-generated currents gives rise to the complex electrical response described in wild-type cells (9, 13). Analysis of the currents in RNAi transformants, which allowed assigning specific currents to CSRA and CSRB, has made clear the relationship between the kinetic components of the current and the saturation phases of the fluence-response curve, which was difficult to resolve in the wild type (18).
Figure 6.
Scheme of light signal transduction for photomotility responses initiated by the two sensory rhodopsin pigments in Chlamydomonas.
The two newly identified rhodopsin pigments have different absorption spectra, as revealed by measurement of the photoreceptor-current action spectra in the RNAi transformants. CSRB has a maximum absorption at 470 nm, and the absorption maximum of CSRA is shifted at least 35 nm to longer wavelengths. The action spectrum of the photoreceptor current measured in nonoriented suspensions may be blue-shifted relative to that of the receptor-absorption spectrum, because the screening by the eyespot drops sharply above 500 nm. Hence, the actual position of the absorption maximum of CSRA may be at a longer wavelength than the action spectra indicate. This notion is supported by the fact that the slope of the rise of the photoreceptor current continues to increase above 510 nm, whereas the amplitude of the photoreceptor current falls because of the decreasing directional sensitivity.
The action spectra maxima of photomotility responses in Chlamydomonas reported earlier vary from 490 to 503 nm (42, 43). These values are between the absorption maxima of CSRA and CSRB, and their variation under different experimental conditions is consistent with our finding that in the wild-type cells two spectrally different receptors, CSRA and CSRB, mediate motile responses. The residues in the predicted retinal-binding pockets of CSRA and CSRB are identical, indicating that differences in shells of residues surrounding the pocket are responsible for tuning their spectra, as has been found for NpSRII (33, 44).
Both photoreceptor pigments mediate phototaxis orientation. However, their relative contribution changes over the dynamic range of this response, because the two respective photoreceptor currents differ in their light sensitivity. CSRB operates at low light intensities, and absorption of one quantum leads to the transfer of at least hundreds of charges through the cell membrane, as was estimated earlier from single-cell measurements in Haematococcus pluvialis (10, 11). Sigmoidal kinetics of the CSRB-generated current and its low light saturation suggest that the transduction chain responsible for amplification of its signal includes diffusional, substrate-limited biochemical steps (10, 11).
The relative contribution of CSRA to the photoreceptor current increases with increasing stimulus intensity, because it saturates at higher fluences than the CSRB-mediated current. At high light intensities the involvement of amplification steps is less critical. The fast kinetics of the CSRA-generated current and its high light saturation may indicate a mechanism of CSRA-current generation different from that of CSRB. Possibly CSRA molecules can form ion channels themselves or are in a tight complex with other proteins, consistent with recent models (9, 18). Although the mechanisms of current generation by CSRA and CSRB may differ, the sensory signals from the two photoreceptors integrate at the level of membrane depolarization brought about by the two currents they produce. The saturation of the two photoreceptor currents at different light fluences extends the range of light intensity to which the organism can respond.
It has been shown that photophobic responses appear as a result of the opening of voltage-dependent Ca2+ channels when the depolarization of the plasma membrane caused by photoreceptor currents exceeds a threshold level (11, 45). In this view photophobic responses would result from stronger stimulation of the same receptors causing phototaxis orientation, as was proposed from the effects of retinal analogs and illumination background on phototaxis and on photophobic responses (46). Consistent with this model, we observed photophobic responses in both A-RNAi and B-RNAi transformants with spectral sensitivity similar to that for photoreceptor currents and phototaxis (data not shown).
Heterogeneous photoreceptor systems with different pigments covering low and high light-intensity ranges are known in photomorphogenesis and phototropism (47, 48). Different rhodopsins (although localized in separate cells) provide the low-intensity (night) and high-intensity (color) vision in higher animals. Multiple receptors with different spectral properties are known to mediate photobehavior in haloarchaea (49) and color vision in animals (50). The evolution of two photoreceptors for photomotile responses of Chlamydomonas further substantiates the importance of multipigment receptor systems in light intensity adaptation and color discrimination in microbes, plants, and animals.
Acknowledgments
We thank Elizabeth Harris for providing Chlamydomonas strains and cDNA libraries, Saul Purton for pSP124S plasmid, and Valerie Kliesing for technical assistance. This work was supported by National Science Foundation Grant 0091287 (to J.L.S.).
Abbreviations
- EST
expressed sequence tag
- RNAi
RNA interference
- dsRNAi
double-stranded RNAi
- NpSRII
phototaxis receptor sensory rhodopsin II
- BR
bacteriorhodopsin
Footnotes
Data deposition: The cDNA and genomic sequences reported in this paper have been deposited in the GenBank database [accession nos. AF508965 and AF508967 (CSOA) and AF508966 and AF508968 (CSOB)].
See commentary on page 8463.
CSOA and CSOB are opsin-encoding genes. CSRA and CSRB refer to the Chlamydomonas sensory rhodopsins A and B, respectively [the term “rhodopsin” designates an apoprotein (“opsin”) bound to its retinal chromophore]; CSOA and CSOB are the apoproteins (“opsins”) of CSRA and CSRB, respectively.
References
- 1.Witman G B. Trends Cell Biol. 1993;3:403–408. doi: 10.1016/0962-8924(93)90091-e. [DOI] [PubMed] [Google Scholar]
- 2.Foster K W, Saranak J, Patel N, Zarilli G, Okabe M, Kline T, Nakanishi K. Nature (London) 1984;311:756–759. doi: 10.1038/311756a0. [DOI] [PubMed] [Google Scholar]
- 3.Hegemann P, Gartner W, Uhl R. Biophys J. 1991;60:1477–1489. doi: 10.1016/S0006-3495(91)82183-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawson M A, Zacks D N, Derguini F, Nakanishi K, Spudich J L. Biophys J. 1991;60:1490–1498. doi: 10.1016/S0006-3495(91)82184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi T, Yoshihara K, Watanabe M, Kubota M, Johnson R, Derguini F, Nakanishi K. Biochem Biophys Res Commun. 1991;178:1273–1279. doi: 10.1016/0006-291x(91)91031-7. [DOI] [PubMed] [Google Scholar]
- 6.Sakamoto M, Wada A, Akai A, Ito M, Goshima T, Takahashi T. FEBS Lett. 1998;434:335–338. doi: 10.1016/s0014-5793(98)01004-7. [DOI] [PubMed] [Google Scholar]
- 7.Foster K W, Smyth R D. Microbiol Rev. 1980;44:572–630. doi: 10.1128/mr.44.4.572-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litvin F F, Sineshchekov O A, Sineshchekov V A. Nature (London) 1978;271:476–478. doi: 10.1038/271476a0. [DOI] [PubMed] [Google Scholar]
- 9.Sineshchekov O A, Govorunova E V. Trends Plant Sci. 1999;4:58–63. doi: 10.1016/s1360-1385(98)01370-3. [DOI] [PubMed] [Google Scholar]
- 10.Sineshchekov O A. In: Light in Biology and Medicine. Douglas R H, editor. New York: Plenum; 1991. pp. 523–532. [Google Scholar]
- 11.Sineshchekov O A. In: Biophysics of Photoreceptors and Photomovements in Microorganisms. Lenci F, editor. New York: Plenum; 1991. pp. 191–202. [Google Scholar]
- 12.Schmidt J A, Eckert R. Nature (London) 1976;262:713–715. doi: 10.1038/262713a0. [DOI] [PubMed] [Google Scholar]
- 13.Sineshchekov O A, Litvin F F, Keszthelyi L. Biophys J. 1990;57:33–39. doi: 10.1016/S0006-3495(90)82504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sineshchekov O A, Litvin F F. In: Molecular Mechanisms of Biological Action of Optical Irradiation. Rubin A B, editor. Moscow: Nauka; 1988. pp. 213–226. [Google Scholar]
- 15.Sineshchekov O A, Govorunova E G, Der A, Keszthelyi L, Nultsch W. Biophys J. 1994;66:2073–2084. doi: 10.1016/S0006-3495(94)81002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sineshchekov O A, Govorunova E G, Der A, Keszthelyi L, Nultsch W. J Photochem Photobiol. 1992;13:119–134. [Google Scholar]
- 17.Ehlenbeck S, Gradmann D, Braun F J, Hegemann P. Biophys J. 2002;82:740–751. doi: 10.1016/S0006-3495(02)75436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sineshchekov O A, Govorunova E G. Biochemistry (Moscow) 2001;66:1300–1310. doi: 10.1023/a:1013191504508. [DOI] [PubMed] [Google Scholar]
- 19.Zacks D N, Derguini F, Nakanishi K, Spudich J L. Biophys J. 1993;65:508–518. doi: 10.1016/S0006-3495(93)81067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegemann P. Planta. 1997;203:265–274. doi: 10.1007/s004250050191. [DOI] [PubMed] [Google Scholar]
- 21.Deininger W, Kroger P, Hegemann U, Lottspeich F, Hegemann P. EMBO J. 1995;14:5849–5858. doi: 10.1002/j.1460-2075.1995.tb00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuhrmann M, Stahlberg A, Govorunova E, Rank S, Hegemann P. J Cell Sci. 2001;114:3857–3863. doi: 10.1242/jcs.114.21.3857. [DOI] [PubMed] [Google Scholar]
- 23.Hegemann P, Fuhrmann M, Kateriya S. J Phycol. 2001;37:668–676. [Google Scholar]
- 24.Spudich J L, Yang C S, Jung K H, Spudich E N. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 25.Harris E H. Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic; 1989. [DOI] [PubMed] [Google Scholar]
- 26.Abdulaev N G, Popp M P, Smith W C, Ridge K D. Protein Expression Purif. 1997;10:61–69. doi: 10.1006/prep.1996.0704. [DOI] [PubMed] [Google Scholar]
- 27.Goldschmidt-Clermont M, Rahire M. J Mol Biol. 1986;191:421–432. doi: 10.1016/0022-2836(86)90137-3. [DOI] [PubMed] [Google Scholar]
- 28.Higuchi R. In: PCR Protocol: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 177–183. [Google Scholar]
- 29.Stevens D R, Rochaix J D, Purton S. Mol Gen Genet. 1996;251:23–30. doi: 10.1007/BF02174340. [DOI] [PubMed] [Google Scholar]
- 30.Shimogawara K, Fujiwara S, Grossman A, Usuda H. Genetics. 1998;148:1821–1828. doi: 10.1093/genetics/148.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bieszke J A, Spudich E N, Scott K L, Borkovich K A, Spudich J L. Biochemistry. 1999;38:14138–14145. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- 32.Sineshchekov O A, Govorunova E G. Photomovements. In: Haeder D P, Lebert M, editors. Comprehensive Series in Photosciences. Vol. 1. Amsterdam: Elsevier; 2001. pp. 245–280. [Google Scholar]
- 33.Luecke H, Schobert B, Lanyi J K, Spudich E N, Spudich J L. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luecke H, Schobert B, Richter H T, Cartailler J P, Lanyi J K. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 35.Valtorta F, Benfenati F, Greengard P. J Biol Chem. 1992;267:7195–7198. [PubMed] [Google Scholar]
- 36.Blom N, Gammeltoft S, Brunak S. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 37.Montgomery M K, Xu S, Fire A. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennerdell J R, Carthew R W. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 39.Ngo H, Tschudi C, Gull K, Ullu E. Proc Natl Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gura T. Nature (London) 2000;404:804–808. doi: 10.1038/35009245. [DOI] [PubMed] [Google Scholar]
- 41.Pazour G J, Witman G B. Methods. 2000;22:285–298. doi: 10.1006/meth.2000.1081. [DOI] [PubMed] [Google Scholar]
- 42.Uhl R, Hegemann P. Biophys J. 1990;58:1295–1302. doi: 10.1016/S0006-3495(90)82469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nultsch W, Throm G, von Rimscha I. Arch Microbiol. 1971;80:351–369. [Google Scholar]
- 44.Ren L, Martin C H, Wise K J, Gillespie N B, Luecke H, Lanyi J K, Spudich J L, Birge R R. Biochemistry. 2001;40:13906–13914. doi: 10.1021/bi0116487. [DOI] [PubMed] [Google Scholar]
- 45.Beck C, Uhl R. J Cell Biol. 1994;125:1119–1125. doi: 10.1083/jcb.125.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zacks D N, Spudich J L. Cell Motil Cytoskeleton. 1994;29:225–230. doi: 10.1002/cm.970290305. [DOI] [PubMed] [Google Scholar]
- 47.Sakai T, Kagawa T, Kasahara M, Swartz T E, Christie J M, Briggs W R, Wada M, Okada K. Proc Natl Acad Sci USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casal J J. Photochem Photobiol. 2000;71:1–11. doi: 10.1562/0031-8655(2000)071<0001:pcppii>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 49.Hoff W D, Jung K H, Spudich J L. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 50.Yokoyama S. Prog Retin Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]