Abstract
Posttranslational modifications of histone tails regulate chromatin structure and transcription. Here we present global analyses of histone acetylation and histone H3 Lys 4 methylation patterns in yeast. We observe a significant correlation between acetylation of histones H3 and H4 in promoter regions and transcriptional activity. In contrast, we find that dimethylation of histone H3 Lys 4 in coding regions correlates with transcriptional activity. The histone methyltransferase Set1 is required to maintain expression of these active, promoter-acetylated, and coding region-methylated genes. Global comparisons reveal that genomic regions deacetylated by the yeast enzymes Rpd3 and Hda1 overlap extensively with Lys 4 hypo- but not hypermethylated regions. In the context of recent studies showing that Lys 4 methylation precludes histone deacetylase recruitment, we conclude that Set1 facilitates transcription, in part, by protecting active coding regions from deacetylation.
In eukaryotes, DNA and histone proteins are organized into nucleosomes, which, in turn, form the higher-ordered structure of chromatin. The amino-terminal tails of histone proteins are subject to posttranslational modifications, including acetylation and methylation, which recruit downstream regulatory factors, influence chromatin structure, and are critical determinants of transcription (1, 2). Acetylation, which occurs at specific lysine residues in these tails, is generally associated with transcriptional activity (3). In contrast, methylation of tail lysines and arginines has alternately been linked to activation and repression, depending on the residue modified (4).
Because histone proteins are highly conserved from yeast to humans, Saccharomyces cerevisiae (budding yeast) has been widely exploited as a model system for the study of chromatin. The yeast histone deacetylase (HDAC) Rpd3 is recruited by Ume6 to promoters containing URS1 sites where it deacetylates locally and represses transcription (5, 6). Yeast Hda1 is recruited along with the Tup1/Ssn6 complex to deacetylate the ENA1 promoter (7). mRNA expression profiles reveal global influences of Rpd3 and Hda1 on genes involved in cell cycle regulation and carbohydrate metabolism, respectively (8). In addition to targeted deacetylation, these enzymes also act on large genomic regions in an untargeted fashion (9).
Set1, a homolog of human MLL and Drosophila trithorax, has recently been identified as the yeast histone H3 Lys 4 methyltransferase (10, 11). Lys 4 methylation has been linked to transcriptional activity in several organisms, and, in S. cerevisiae, the PPH3 gene is Lys 4 methylated and activated by Set1 (12–15; R.S. and T.K., unpublished data). However, Set1 is also required for transcriptional silencing at the silent mating-type locus, the rDNA locus, and at telomeres (10, 16).
Chromatin immunoprecipitation (chIP) has been combined with DNA microarray technology to localize transcription factors, silencing proteins, and chromatin remodelers genomewide (17–21). By identifying novel gene targets and by documenting a broad role for the RSC remodeling complex in transcriptional activation and repression, these studies validate the global approach. To obtain a global perspective on the physiologic roles of histone modifications we have analyzed acetylation and methylation patterns genomewide in S. cerevisiae. Our analysis confirms and extends existing models of acetylation and methylation and suggests that histone modifications in coding regions, as well as promoters, play a general role in transcriptional regulation.
Materials and Methods
Yeast Strains.
The strains used are listed in Table 1. An rpd3 mutant was generated by homologous recombination replacing the entire RPD3 gene with a HIS3 marker by using plasmid pFA6a-His3MX6 and standard methodology (23). Clones were selected on CSM-HIS plates and verified by PCR. Primers 5′-ACAATTGCGCCATACAAAACATTCGTGGCTACAACTCGATATCCGTGCAGG GTCGACGGATCCCCGGGTT-3′ and 5′-GCACTTCTCATACACAATTGGATAGCGT CTTAAGTGCCTTTTATTCACTTTCGATGAATTCGAGCTCGTT-3′ were used to generate the rpd3∷HIS3 strain.
Table 1.
Yeast strains
chIP.
Immunoprecipitation of DNA associated with modified histones was carried out as described (24, 25). Briefly, yeast cultures (strain UCC1001) grown in yeast extract/peptone/dextrose to a density of ∼2 × 107 cells per ml were cross-linked in 1% formaldehyde for 15 min, washed twice in PBS, and lysed with glass beads. The resulting extract was sonicated to fragment chromatin (4 × 30 sec burst/30 sec rest with a Branson Sonifier 250 at 70% duty, power 3) and centrifuged for 15 min at 14,000 × g. Chromatin was immunoprecipitated with Abs against acetylated histone H3 (Lys 9 and 14), acetylated histone H4 (Lys 5, 8, 12, and 16), or dimethylated histone H3 (Lys 4) (Upstate Biotechnology, Lake Placid, NY). Immunoprecipitated (enriched) sample and whole-cell extract (unenriched) were incubated in TE (10 mM Tris/1 mM EDTA, pH 8.0) plus 1.0% SDS and 150 mM NaCl overnight at 65°C to reverse cross-links. DNA was purified by proteinase K treatment, phenol/chloroform extraction, ethanol precipitation, and incubation with RNase.
DNA Amplification and Labeling.
Immunoprecipitated DNA (from ≈2 × 108 cells) and whole-cell extract DNA were amplified in parallel by random-primer PCR as described (19) incorporating amino-allyl dUTP. Amplified DNA was purified with a QIAquick PCR purification kit (Qiagen, Chatsworth, CA) and fluorescently labeled by incubation with monofunctional reactive Cy5 (chIP sample) or Cy3 (whole-cell extract) dye as described (26). Before hybridization, Cy5- and Cy3-labeled samples were purified with a QIAquick PCR kit and combined.
Deacetylation chIP Experiments.
For the Rpd3 and Hda1 deacetylation chIP data sets, DNA associated with H4Ac (Rpd3 experiment) or H3Ac (Hda1 experiment) was immunoprecipitated in parallel from mutant and wild type, amplified, labeled (Cy5 for mutant; Cy3 for wild type), and combined as above.
Microarray Preparation.
Intergenic regions (6,438) were amplified by PCR (Research Genetics, Huntsville, AL) as described (17, 18). ORFs were amplified from a set of 6,218 plasmids by using universal primers as described (27). These sets of intergenics and ORFs, which together encompass >95% of the yeast genome, were printed on separate slides, hydrated, and snap-dried as described (27).
Hybridization, Washing, and Scanning.
Mixed Cy5/Cy3-labeled probe was allowed to hybridize to microarrays for 12–14 h at 60°C. After hybridization, microarrays were washed as described (27) and scanned with a GenePix 4000A scanner with genepix pro software (Axon Instruments, Foster City, CA).
Microarray Data Processing.
The following chIP microarray data sets were analyzed in this article: H3Ac IP vs. whole-cell extract; H4Ac IP vs. whole-cell extract; H3 Lys 4 dimethyl IP vs. whole-cell extract; H4Ac IP (rpd3Δ) vs. H4Ac IP (wild type); and H3Ac IP (hda1Δ) vs. H3Ac IP (wild type).
Each microarray data set was determined from two independent chIP experiments and hybridizations. Composite Cy5:Cy3 ratios were calculated from weighted averages by using a single-array error model described in ref. 28. Overall correlations between duplicate experiments were ∼80% for all experiments except the hda1Δ vs. wild-type chIP data sets, which correlate at ∼50%. This lower correlation is a consequence of the more subtle influence of HDA1 deletion on acetylation and transcription and is not, in itself, a reflection of greater experimental variation. All data sets were log-transformed and zero-centered before further analysis. Complete data sets are available at www.schreiber.chem.harvard.edu.
Comparisons of H3 Acetylation and Lys 4 Methylation.
chIP microarray experiments comparing H3 Lys 4 methylation with H3 acetylation were carried out in duplicate as follows. Equal quantities of amplified DNA from an H3 Lys 4 dimethyl chIP and an H3 acetyl chIP were labeled with Cy5 and Cy3 dyes, respectively. Labeled samples were combined, and equal volumes were hybridized to ORF and intergenic slides. On the same slides (two identical microarrays were printed on each slide), probe containing equal quantities of Cy5- and Cy3-labeled whole-cell extract was hybridized to verify equal dye labeling and intensity. After hybridization, both slides were scanned with identical parameters. After quantitation, the ratio between H3 Lys 4 methylation and H3 acetylation in coding regions relative to intergenic regions was determined by dividing the average Cy5:Cy3 ratio over all ORFs by the average Cy5:Cy3 ratio over all intergenics.
mRNA Expression Profiles.
The mRNA expression profiles rpd3Δ vs. wild type, hda1Δ vs. wild type, and set1Δ vs. wild type (15) were analyzed in this article.
mRNA expression profiles were determined by conventional cDNA microarray analysis, in duplicate, as described (26, 27, 29). Briefly, mRNA isolated from mutant or parental wild-type cells grown to a density of 2 × 107 cells per ml was reverse-transcribed incorporating amino-allyl dUTP. The resulting cDNAs were labeled (Cy5 for mutant; Cy3 for wild type), combined, and hybridized to microarrays containing >95% of ORFs. Microarrays were scanned, spots were quantified, and composite ratios were determined as described in ref. 28.
Transcriptional Activity Data Set.
mRNA from wild-type (BY4741) yeast grown in yeast extract/peptone/dextrose to a density of ∼2 × 107 cells per ml was extracted and analyzed on a GeneChip array (Affymetrix, Santa Clara, CA), in duplicate as described (8). The GeneChip “average difference” measurements reflect absolute cellular mRNA levels and were used as surrogates for transcriptional activity with the caveat that mRNA levels are, to a certain extent, also a function of transcript half-life (30). Transcripts with absolute average difference measurements of 20 or less (3.7% of transcripts) were judged “not detected” and the corresponding genes were excluded from further analysis. For correlation analysis transcriptional measurements were log-transformed and zero-centered.
Correlation Analysis.
Statistical analysis was carried out with s-plus statistical software (Insightful, Seattle). Correlation coefficients that assess the linear association between transcriptional activity and coding region acetylation or methylation were computed between the log-transformed transcriptional activity data set and the log-transformed Cy5:Cy3 ratios for the set of all candidate ORFs. Similarly, correlation coefficients between transcriptional activity and promoter acetylation or methylation were computed between the log-transformed transcriptional activity and the log-transformed Cy5:Cy3 ratios for the adjacent, upstream intergenic regions. Confidence intervals of 95% for the correlation coefficients were assigned by bootstrapping methods. Correlations termed “significant” in the text have P values <1 × 10−3 based on permutation analysis.
Statistical Significance of Gene List Overlaps.
The lists of hypermethylated, hypomethylated, and deacetylated chromatin regions contain elements that are significantly different from background. To assess the statistical significance of observed overlap between two lists, a hypergeometric probability model was applied. Specifically, the null hypothesis of no association implies that the two lists were independently selected without replacement from the total set of intergenics and ORFs. The P values given describe the extent to which the observed overlap exceeds that expected under the null hypothesis (31).
Conventional chIP Analysis.
Conventional chIP was carried out with the immunoprecipitation protocol described previously. chIP DNA and whole-cell extract (input) were subjected to 23 cycles of quantitative PCR in the presence of two primer pairs designed to amplify a sequence in the promoter, and a sequence in the coding region, of a specific gene. Amplified DNA was visualized on polyacrylamide gel stained with 0.5% ethidium bromide and quantified with the Alpha DigiDoc System Gel Documentation System (Alpha Innotech, San Leandro, CA). The following primer pairs were used for conventional chIP analysis.
SUB2 coding region.
5′-ATGCCAGACGTTAAGACAGCAGTC-3′ and 5′-GTTATCTTAGCTTCATCATCGACG-3′.
SUB2 promoter.
5′-GAAGATTCGCGTTACCCTTACTCG-3′ and 5′-AGCTAGTTATACCGATGAGAATAG-3′.
MNN9 coding region.
5′-CCCTCGGGAATTGATTGAATTGGG-3′ and 5′-GGTCATGTCTTGAATTAAAGATGG-3′.
MNN9 promoter.
5′-TAGACACGACAGCATTGGTTTGTC-3′ and 5′-TCTTCTTACTTTCACAAAGTGCGG-3′.
YHM2 coding region.
5′-GTCCCACAATCTTCTTGGAGTGTG-3′ and 5′-ACCACGGTAAAGACCCTTTAGACC-3′.
YHM2 promoter.
5′-GCCTCTTGCATGGATCTTACCTTC-3′ and 5′-TCGGGCTTTTCCTAGCTTTATTTG-3′. Conventional chIP analysis of the rDNA used described NTS primers (32). Duplex PCR reactions comparing the rDNA locus and the SUB2 coding region in whole-cell extract and Lys 4 dimethyl chIP used a 25 times greater concentration of SUB2 primers than NTS primers. This ratio was necessary to amplify both fragments to equal concentrations from whole-cell extract, presumably because of the higher representation of the repeated rDNA in the template.
Results
Yeast Heterochromatin Is Hypoacetylated and Hypomethylated (Lys 4).
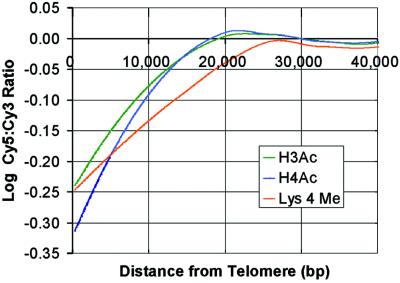
chIP followed by hybridization to microarrays was used to determine genomewide patterns of histone modifications. chIP microarray data sets for H3 acetylation, H4 acetylation, and H3 Lys 4 dimethylation each contain approximately 12,000 Cy5:Cy3 ratios that reflect the relative acetylation or methylation status of the corresponding genomic region. In Fig. 1, relative acetylation levels are plotted with respect to distance from chromosomal ends. On average, microarray features that correspond to telomere-proximal regions exhibit Cy5:Cy3 ratios significantly lower than the global average, indicating that associated histones are hypoacetylated. This finding is consistent with conventional studies that have found telomeres to be hypoacetylated relative to active genes (33). We also find that histone H3 Lys 4 is hypomethylated in these regions, consistent with a recent report that the S. pombe silent mating-type region lacks H3 Lys 4 methylation (14). Global analysis further indicates that other silenced regions, including the silent mating-type and rDNA loci, are also hypomethylated at Lys 4. Although previous studies have shown that the silent rDNA locus exhibits some degree of Lys 4 methylation (10), we used conventional chip to confirm that this locus is hypomethylated with respect to an active gene (Fig. 3C).
Figure 1.
An in-depth analysis of histone acetylation and methylation patterns at telomeres confirms and extends prior studies. Histone acetylation (H3 and H4) and methylation (H3 Lys 4) is plotted with respect to distance from the end of the chromosome and smoothed with a Lowess algorithm. Each curve represents data from ∼1,100 microarray elements corresponding to ends of the 16 yeast chromosomes. The Cy5:Cy3 ratios for telomere-proximal elements are significantly below the genome average (represented by the zero on the y axis), indicating that histones at telomeres are hypoacetylated and Lys 4 is hypomethylated.
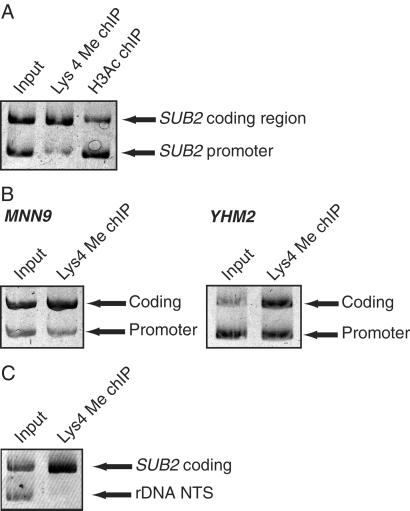
Figure 3.
(A) The ability of chIP microarray analysis to reflect relative levels of acetylation and methylation in promoters and coding regions was verified by conventional chIP experiments. SUB2 is a transcriptionally active gene that is, according to chIP microarray analysis, hypermethylated in its coding region and hyperacetylated in its promoter. Conventional chIP and quantitative PCR (shown) confirms that the SUB2 coding region is enriched 4.1-fold in a Lys 4 dimethyl chIP (with respect to the SUB2 promoter), and that the SUB2 promoter is enriched 2.8-fold in an H3 acetyl chIP (with respect to the SUB2 coding region). (B) Like SUB2, MNN9 and YHM2 are active genes found by chIP microarray analysis to be coding region-hypermethylated at Lys 4. Conventional chIP verifies both findings. The coding region of the transcriptionally active gene MNN9 is enriched 2.4-fold in the Lys 4 dimethyl chIP, relative to its promoter. Similarly, the coding region of the transcriptionally active YHM2 gene is enriched 2.0-fold relative to its promoter. These findings also verify the ability of chIP microarray to resolve between promoters and coding regions. (C) Although the rDNA locus may exhibit some degree of Lys 4 methylation, this locus is Lys 4 hypomethylated relative to the active SUB2 gene.
Histone Modifications Correlate with Transcriptional Activity.
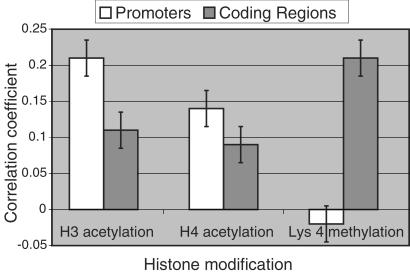
The DNA microarray elements used in this analysis correspond either to gene coding regions (ORFs) or to intergenic regions that contain the gene promoters. Hence, the chIP data sets contain separate measurements of promoter and coding region acetylation for each yeast gene. Histone acetylation has been genetically and biochemically linked to transcriptional activity (3). To investigate this association globally, we compared our chIP data sets to a measure of transcriptional activity. Transcriptional activity for every gene in yeast was approximated from cellular mRNA expression levels determined with an Affymetrix GeneChip array. From here on, we use the term “transcriptional activity” to reflect these measurements of mRNA expression levels, with the caveat that mRNA levels are, to a certain extent, also a function of transcript half-life. Genomewide correlation analysis reveals a significant positive association between the transcriptional activity of a gene and the acetylation of its promoter. Specifically, the correlation between transcriptional activity and acetylation status is 21.1% for histone H3 and 13.8% for histone H4. To a lesser extent, transcriptional activity also correlates with the acetylation levels of histones H3 and H4 within gene coding regions (Fig. 2).
Figure 2.
Transcriptional activity correlates with histone acetylation in promoters and coding regions, but with histone H3 Lys 4 dimethylation in coding regions. Correlations between transcriptional activity and acetylation and methylation status of corresponding promoters and coding regions were calculated from chIP microarray data sets and expression data. Error bars represent 95% confidence intervals determined by bootstrap analysis. Because these correlations are not perfect, we anticipate that the modification status of certain genes will differ from genomewide trends.
Although less well understood than acetylation, histone H3 Lys 4 methylation has been linked to transcriptional activity in Tetrahymena, S. pombe, and chicken (12–14). Nevertheless, we do not observe a global correlation between transcriptional activity and histone H3 Lys 4 dimethylation in promoter regions. Instead, we find that Lys 4 dimethylation in coding regions correlates with transcription. Statistical analysis confirms that this association is significant, with a genomewide correlation of 21.0% (Fig. 2). Although our analysis does not detect a global correlation between promoter Lys 4 methylation and transcription, we cannot rule out the possibility that promoter Lys 4 methylation plays some role in transcriptional regulation.
We used conventional chIP methodology to verify a subset of the chip microarray data and to confirm that this technique has sufficient resolution to differentiate between promoters and coding regions. According to chIP microarray, the transcriptionally active SUB2 gene is hyperacetylated within its promoter and hypermethylated (Lys 4) within its coding region. Conventional chIP followed by quantitative PCR with specific primer pairs (Fig. 3A) confirms that the SUB2 coding region is enriched 4.1-fold in a Lys 4 dimethyl chIP (with respect to the SUB2 promoter) and that the SUB2 promoter is enriched 2.8-fold in an H3 acetyl chIP (with respect to the SUB2 coding region). The hypermethylated status of two additional coding regions, MNN9 and YHM2, was also verified by conventional methods (Fig. 3B).
Global Distributions of Histone Modifications.
The observation that transcriptional activity correlates with histone acetylation in promoters but with Lys 4 methylation in coding regions led us to consider the global distribution of these modifications among promoters and coding regions. The absolute distribution of a particular modification cannot be determined with chIP microarray methodology. However, by directly hybridizing H3 Lys 4 dimethyl chIP DNA against H3 acetyl chIP DNA we assessed the relative distribution of these modifications over all coding and intergenic regions. The ratio between Lys 4 methylation and H3 acetylation is 1.6-fold greater in coding regions than in intergenic regions. This finding suggests that, on average, Lys 4 methylation is more common in coding regions, that H3 acetylation is more common in the promoter-containing intergenic regions, or that both phenomena are true.
Set1 Facilitates Transcription.
The Set1 protein has recently been identified as the primary histone H3 Lys 4 methyltransferase in yeast (10, 11). To determine whether methylation by Set1 is generally required for transcriptional activity, we examined an mRNA expression profile of the set1Δ mutant (15). Genomewide comparison of the set1Δ profile and the transcriptional activity data set reveals a 37.1% correlation between transcriptional activity and dependence on Set1 for expression. In other words, Set1 facilitates transcription of active genes in a manner that is proportional to their absolute transcriptional activity.
Genomewide Histone Deacetylation.
Biochemical studies suggest that a function of Lys 4 methylation may be to preclude recruitment of HDACs by the histone H3 tail. To investigate a relationship between Lys 4 methylation and histone deacetylation in vivo we examined the influence of the yeast HDACs Rpd3 and Hda1 on histone acetylation levels genomewide. chIP microarray data sets that reflect the influence of these enzymes on acetylation were generated by hybridizing DNA associated with acetylated histones in an HDAC mutant (Cy5-labeled) with DNA associated with acetylated histones in wild-type yeast (Cy3-labeled). In these experiments, microarray features that correspond to chromatin deacetylated by Rpd3 (or Hda1) have Cy5:Cy3 ratios significantly greater than 1 (i.e., these regions are more acetylated in the mutant). A list of gene promoters deacetylated by Rpd3 was collated from the Rpd3 chIP data set and includes the 150 promoter-containing intergenic regions with the highest Cy5:Cy3 ratios. Prior studies have demonstrated that the transcription factor Ume6 recruits Rpd3 to deacetylate the promoters of repressed genes (5, 6). Consistent with this paradigm, sequence analysis of the 150 deacetylated gene promoters reveals that 32 contain Ume6 binding sites. This overlap is highly significant with a P value of 9.2 × 10−15. There is also statistically significant overlap between genes whose promoters are deacetylated by Rpd3 (based on the rpd3Δ vs. wild-type chIP microarray) and genes that are repressed by Rpd3 (based on the rpd3Δ vs. wild-type mRNA expression profile) with a P value of 4.0 × 10−7 (8). An analogous set of experiments reveals significant overlap between genes whose promoters are deacetylated by Hda1 (histone H3) and genes repressed by Hda1 with a P value of 2.4 × 10−32. We conclude that our HDAC mutant vs. wild-type chIP experiments reflect the in vivo functions of these enzymes.
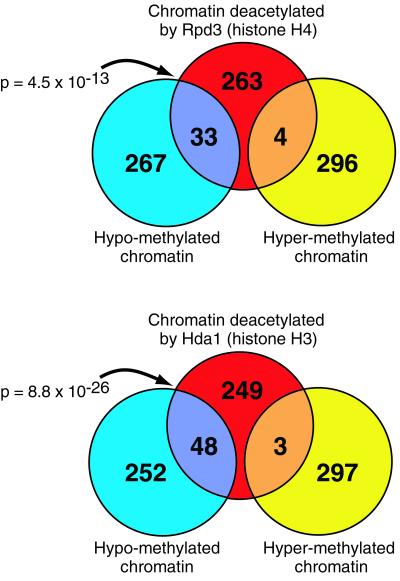
Lys 4 Hypomethylated and Deacetylated Chromatin Overlap.
Lists of hypo- and hypermethylated genomic regions were collated from the H3 Lys 4 dimethyl chIP data set and contain the 300 features with the lowest and highest Cy5:Cy3 ratios, respectively. Lists of deacetylated regions were collated from the Rpd3 and Hda1 chIP data sets and contain the 300 features with the highest Cy5:Cy3 ratios. These lists, which correspond in size to approximately the 95th percentile, were kept at exactly 300 to facilitate statistical analysis of list overlaps.
These lists contain elements that are significantly different from background. A large overlap between such lists indicates a possible causal association. To investigate a relationship between Lys 4 methylation and deacetylation, the histone H3 Lys 4 dimethyl chIP data set was compared with the Rpd3 and Hda1 chIP data sets. Lists of hypo- and hypermethylated chromatin regions were compared with lists of chromatin regions deacetylated by Rpd3 or Hda1. We find extensive, statistically significant overlap between hypomethylated chromatin and chromatin deacetylated by these enzymes (Fig. 4). In contrast, we find almost no overlap between hypermethylated chromatin and chromatin deacetylated by these enzymes.
Figure 4.
Chromatin regions methylated at histone H3 Lys 4 are not deacetylated by Rpd3 or Hda1. Lists of H3 Lys 4 hypomethylated and hypermethylated chromatin regions were collated from chIP microarray analyses, and compared with lists of chromatin regions deacetylated by Rpd3 or Hda1. There is extensive overlap between deacetylated regions and hypomethylated regions. In contrast, there is essentially no overlap between deacetylated regions and hypermethylated regions. These findings and those presented in Fig. 2 are consistent with a model in which methylation of histone H3 Lys 4 protects active coding regions from deacetylation.
Discussion
To obtain a global perspective on histone acetylation and Lys 4 methylation patterns in yeast, we have combined chIP with DNA microarray technology. Confirming and extending prior conventional studies (14, 33), we find that yeast heterochromatin is hypoacetylated and H3 Lys 4 hypomethylated relative to the genome average. A detailed comparison of modification patterns at telomeres indicates that, whereas hypoacetylation extends ∼15 kb from chromosome ends, Lys 4 hypomethylation extends ∼25 kb (Fig. 1). We note a parallel to the influences of Sir2 and Set1 on telomeric genes. That is, whereas repression by the deacetylase Sir2 extends ∼8 kb from chromosome ends (34, 35), repression by the Lys 4 methyltransferase Set1 extends ∼20 kb (B.E.B. and S.L.S., unpublished data).
Sir2 localizes to silent loci and presumably represses transcription directly by deacetylating chromatin (19, 32). The observation that Set1 also represses transcription at silent loci raises the possibility that Lys 4 methylation might repress transcription in certain cases. However, our observation that silent loci are relatively hypomethylated at Lys 4 suggests that the influence of Set1 on these regions is indirect and does not support a role for Lys 4 methylation in repression.
To investigate functions of acetylation and methylation within active chromatin, we examined the association between these modifications and transcriptional activity. We find that histone acetylation in promoters and, to a lesser extent, in coding regions, correlates globally with transcriptional activity. Like acetylation, H3 Lys 4 methylation has been generally linked to transcription (12–15). However, in contrast to acetylation, we find primarily that Lys 4 dimethylation in coding regions correlates globally with transcriptional activity (Fig. 2). The preferential association of transcription with promoter acetylation and coding region methylation led us to examine the relative distributions of these modifications. A direct comparison of H3 acetylation and H3 Lys 4 dimethylation indeed suggests that, relative to acetylation, Lys 4 dimethylation is more common in coding regions.
To characterize further the physiologic function of Lys 4 methylation, we examined the mRNA expression profile of a set1Δ mutant. By comparing this data set with the transcriptional activity data set we discovered that Set1 facilitates transcription of active genes in a manner that is proportional to their absolute transcriptional activity. This finding suggests that Set1-mediated methylation of histone H3 Lys 4 in coding regions of active genes is necessary to maintain their expression. We speculate that coding region methylation facilitates RNA polymerase II elongation through chromatin.
Efficient transcriptional elongation in vitro depends on histone acetylation (36). Furthermore, deacetylation by the yeast enzymes Rpd3 and Hda1 has been found to extend into coding regions (9). To investigate a relationship between Lys 4 methylation and deacetylation in vivo, we compared the histone H3 Lys 4 dimethyl chIP data set with the Rpd3 and Hda1 chIP data sets. We find extensive, statistically significant overlap between hypomethylated chromatin and chromatin deacetylated by these enzymes (Fig. 4). In contrast, we find almost no overlap between hypermethylated chromatin and chromatin deacetylated by these enzymes. Although this correlative data does not prove causation, the finding that Lys 4 methylation precludes recruitment of the mammalian HDAC complex NuRD (37–40), in vitro (41, 42), leads us to favor a model in which methylation by Set1 facilitates transcription by preventing deacetylation of coding regions.
In conclusion, by analyzing all promoters and coding regions and determining their modification status, we observe that histone methylation at Lys 4 frequently occurs in coding regions to facilitate transcription. This observation is in contrast to acetylation, whose function has previously been found to be primarily on histones associated with promoter regions. Genes found to be hypermethylated do not overlap with genes deacetylated by HDACs, consistent with binding studies that show Lys 4 methylation occludes binding of HDACs. These results suggest that the mechanism by which Lys 4 methylation activates transcription may involve processes other than (or in addition to) recruitment of the basal transcriptional components and may involve effects on transcriptional elongation.
Acknowledgments
We thank Andrew Murray, Christina Cuomo, George Busby, and the Bauer Center for Genomics Research for resources and helpful discussion. We also thank V. Geli for the set1∷KAN strain. This research was supported by National Institute of General Medical Sciences Grant GM38627 (to S.L.S.). B.E.B. is supported by a Howard Hughes Medical Institute Physician Postdoctoral Fellowship. S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Abbreviations
- chIP
chromatin immunoprecipitation
- HDAC
histone deacetylase
References
- 1.Jenuwein T, Allis C D. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Turner B M. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Grunstein M. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 5.Kadosh D, Struhl K. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Nature (London) 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Suka N, Carlson M, Grunstein M. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein B E, Tong J K, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogelauer M, Wu J, Suka N, Grunstein M. Nature (London) 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 10.Briggs S D, Bryk M, Strahl B D, Cheung W L, Davie J K, Dent S Y, Winston F, Allis C D. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roguev A, Schaft D, Shevchenko A, Pijnappel W W, Wilm M, Aasland R, Stewart A F. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strahl B D, Ohba R, Cook R G, Allis C D. Proc Natl Acad Sci USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litt M D, Simpson M, Gaszner M, Allis C D, Felsenfeld G. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 14.Noma K, Allis C D, Grewal S I. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 15.Boggs B A, Cheung P, Heard E, Spector D L, Chinault A C, Allis C D. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 16.Nislow C, Ray E, Pillus L. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren B, Robert F, Wyrick J J, Aparicio O, Jennings E G, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 18.Iyer V R, Horak C E, Scafe C S, Botstein D, Snyder M, Brown P O. Nature (London) 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 19.Lieb J D, Liu X, Botstein D, Brown P O. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 20.Damelin M, Simon I, Moy T I, Wilson B, Komili S, Tempst P, Roth F P, Young R A, Cairns B R, Silver P A. Mol Cell. 2002;9:563–573. doi: 10.1016/s1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- 21.Ng H H, Robert F, Young R A, Struhl K. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 23.Longtine M S, McKenzie A, 3rd, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 24.Hecht A, Grunstein M. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- 25.Kuo M H, Allis C D. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 26.Carroll A S, Bishop A C, DeRisi J L, Shokat K M, O'Shea E K. Proc Natl Acad Sci USA. 2001;98:12578–12583. doi: 10.1073/pnas.211195798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 28.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, et al. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 29.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Gentile C, Hwang S Y, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 31.Tavazoie S, Hughes J D, Campbell M J, Cho R J, Church G M. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 32.Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy B K, Grunstein M, Gasser S M. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 34.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 35.Wyrick J J, Holstege F C, Jennings E G, Causton H C, Shore D, Grunstein M, Lander E S, Young R A. Nature (London) 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 36.Protacio R U, Li G, Lowary P T, Widom J. Mol Cell Biol. 2000;20:8866–8878. doi: 10.1128/mcb.20.23.8866-8878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Nature (London) 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 38.Wade P A, Jones P L, Vermaak D, Wolffe A P. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 39.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 41.Zegerman P, Canas B, Pappin D, Kouzarides T. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- 42.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis C D, Tempst P, Reinberg D. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]