Abstract
Bacteria lack an endoplasmic reticulum, a Golgi apparatus, and transport vesicles and yet are capable of sorting and delivering integral membrane proteins to particular sites within the cell with high precision. What is the pathway by which membrane proteins reach their proper subcellular destination in bacteria? We have addressed this question by using green fluorescent protein (GFP) fused to a polytopic membrane protein (SpoIVFB) that is involved in the process of sporulation in the bacterium Bacillus subtilis. SpoIVFB-GFP localizes to a region of the sporulating cell known as the outer forespore membrane, which is distinct from the cytoplasmic membrane. Experiments are presented that rule out a mechanism in which SpoIVFB-GFP localizes to all membranes but is selectively eliminated from the cytoplasmic membrane by proteolytic degradation and argue against a model in which SpoIVFB-GFP is selectively inserted into the outer forespore membrane. Instead, the results are most easily compatible with a model in which SpoIVFB-GFP achieves proper localization by insertion into the cytoplasmic membrane followed by diffusion to, and capture in, the outer forespore membrane. The possibility that diffusion and capture is a general feature of protein localization in bacteria is discussed.
Keywords: sporulation
Despite their small size and apparent simplicity, bacteria exhibit a high degree of subcellular organization in which proteins frequently localize to particular sites within the cell (1). Examples include the cell division protein FtsZ, which assembles into a cytokinetic ring at the mid cell of Escherichia coli (2); the chemotaxis receptor McpA, which localizes to the cell pole in Caulobacter crescentus (3); the membrane phosphatase SpoIIE, which localizes to the sporulation septum in Bacillus subtilis (4); and the actin polymerization protein IcsA, which localizes to the older of the two cell poles of Shigella flexneri (5). Here, we are concerned with the problem of how integral membrane proteins become localized within the bacterial cell. Unlike eukaryotes, bacteria lack organelles and membrane vesicles that sort and deliver membrane proteins to particular subcellular addresses. Thus, an outstanding question in the field of prokaryotic biology is the nature of the pathway(s) by which integral membrane proteins reach their proper destination inside the cell.
We envision three models for how bacterial membrane proteins become properly localized. In the first model, which we call targeted insertion, the protein is directly and selectively inserted into the membrane at the site where it will ultimately reside. This model has been proposed for the mechanism by which the outer membrane protein IcsA achieves proper localization at the old cell pole in Shigella (5, 6). The second model, which we call selective degradation, involves the random insertion of the protein into all membranes in the cell followed by proteolytic elimination of the protein from sites other than its proper destination. Selective degradation could be achieved by a protease with a spatially restricted distribution in the cell or by an alternative mechanism in which the protein is protected from degradation at its proper location by a protease that is distributed uniformly throughout the cell. In the third model, which we call diffusion and capture, the protein is inserted randomly into all membranes and becomes localized by diffusion to, and capture at, the site where it will ultimately reside.
An attractive system in which to address the question of how membrane proteins localize is the process of sporulation in B. subtilis. Sporulation takes place in two principal morphological stages (7). In the first stage, the sporulating cell (or sporangium) divides asymmetrically through the formation of a polar septum, which creates a small cell known as the forespore and a larger cell called the mother cell. Initially, the forespore and mother cell lie side-by-side, but in the second stage of development the forespore is engulfed by the mother cell. Engulfment is a phagocytic-like process in which the membranes of the polar septum migrate around the forespore, pinching it off as a free protoplast within the mother cell. Engulfment eventually creates two topologically distinct membranes within the mother cell: the cytoplasmic membrane, which delimits the periphery of the cell, and the mother cell membrane that surrounds the forespore (also known as the outer forespore membrane). Several proteins have been identified that localize specifically to the outer forespore membrane, one of the best-studied examples is the sporulation protein SpoIVFB (8–10).
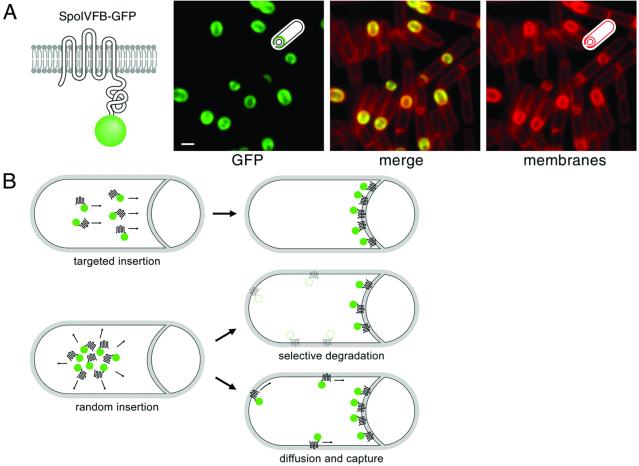
SpoIVFB is a proprotein processing enzyme that is involved in the activation of a transcription factor that acts at a late stage of development (11–14). SpoIVFB, which has six transmembrane segments (15), is synthesized in the mother cell during the process of engulfment (8). That SpoIVFB exhibits a highly specific pattern of subcellular localization is seen through the use of a functional fusion of the sporulation protein to the green fluorescent protein (GFP) (ref. 9; Fig. 1A). Fluorescence from SpoIVFB-GFP is coincident with the outer forespore membrane with little or no signal detectable at the cytoplasmic membrane (9, 10) (Fig. 1A). Proper localization of SpoIVFB-GFP depends on a second integral membrane protein (SpoIVFA) that resides in a complex with SpoIVFB-GFP (10). In the absence of SpoIVFA, SpoIVFB-GFP localizes uniformly to all membranes within the mother cell (10). SpoIVFA also localizes to the outer forespore membrane and does so in a manner that does not depend on SpoIVFB (10).
Figure 1.
The sporulation membrane protein SpoIVFB-GFP localizes to the mother-cell membrane that engulfs the forespore. (A) Model of the polytopic membrane protein SpoIVFB fused to GFP (green ball). Fluorescent micrograph of SpoIVFB-GFP from strain BDR497 at hour 2.5 of sporulation. SpoIVFB-GFP is synthesized in the mother-cell compartment and localizes to the mother-cell membrane that surrounds the forespore. Membranes (false-colored red) were labeled with TMA-DPH at the start of sporulation. The asymmetric septa and membranes engulfing the forespore stain more intensely because they are composed of two layers of membrane. (Bar = 1 μm.) (B) Three models for how SpoIVFB-GFP achieves proper localization to the engulfing septal membrane. In the first model, SpoIVFB-GFP is inserted directly into the septal membrane. In the second and third models, SpoIVFB-GFP is randomly inserted into all mother-cell membranes. The protein then achieves proper localization by selective degradation (dashed lines) in the cytoplasmic membrane. Alternatively, randomly inserted SpoIVFB-GFP diffuses and is captured in the engulfing septal membrane.
SpoIVFB-GFP could achieve proper localization to the outer forespore membrane by any of the three models we have presented (Fig. 1B). It could be inserted directly and selectively into the mother-cell membrane that surrounds the forespore. This targeted insertion could occur cotranslationally or posttranslationally as shown for simplicity in Fig. 1B. Alternatively, SpoIVFB-GFP could be inserted randomly into all mother-cell membranes and then selectively degraded from the cytoplasmic membrane. Finally, SpoIVFB-GFP could be inserted into all membranes and diffuse to the engulfing mother-cell membrane (before engulfment is complete) and then become captured in the outer forespore membrane.
With SpoIVFB-GFP as a model polytopic membrane protein and taking advantage of the requirement for SpoIVFA in the localization of SpoIVFB-GFP, we attempted to distinguish among the three models for how proper localization of an integral membrane protein is achieved. The results of our analysis are most easily compatible with a diffusion-and-capture model in which SpoIVFB-GFP is inserted into the cytoplasmic membrane from which it diffuses to, and is captured in, the membranes that surround the forespore during the process of engulfment. A further and unexpected finding to emerge from our investigation is that the outer forespore membrane is apparently refractory to membrane protein insertion. Hence, the only pathway by which SpoIVFB-GFP localizes to the outer forespore membrane may be by diffusion from the cytoplasmic membrane before the completion of engulfment.
Materials and Methods
General methods were as described (10). Sporulation was induced by the resuspension method in Sterlini–Mandelstam medium (16). All B. subtilis strains were derived from prototrophic strain PY79 (17) and are listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. See Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org, for detailed experimental procedures.
Results
SpoIVFB-GFP Is Stable in the Absence of SpoIVFA.
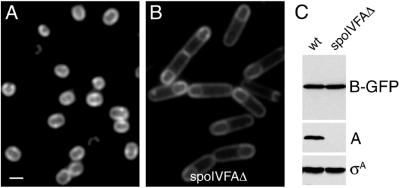
One model for how the subcellular localization of SpoIVFB-GFP is achieved is that the fusion protein is initially inserted into all membranes of the mother cell but is selectively eliminated from the cytoplasmic membrane by proteolytic degradation (Fig. 1B). We have shown that proper localization of SpoIVFB-GFP requires SpoIVFA, which exists in a complex with SpoIVFB-GFP in the engulfing septal membrane (10). In the absence of SpoIVFA, SpoIVFB-GFP is distributed uniformly in all membranes of the mother cell (Fig. 2, compare A and B). Because SpoIVFA is itself restricted to the engulfing forespore membrane, a model in which SpoIVFA is responsible for degrading SpoIVFB in the cytoplasmic membrane seems implausible. Conversely, however, SpoIVFA could be responsible for protecting SpoIVFB-GFP in the septal membrane from degradation by uniformly distributed protease. Such a protection-from-proteolysis model is, however, inconsistent with the results of fluorescence microscopy. If SpoIVFA were to protect SpoIVFB-GFP from degradation in the septal membrane, then, in the absence of SpoIVFA, SpoIVFB-GFP should appear mislocalized but be substantially eliminated from both the septal membrane and the cytoplasmic membrane. Instead, the absence of SpoIVFA causes SpoIVFB-GFP to be unlocalized but does not noticeably diminish the overall intensity of the SpoIVFB-GFP signal (Fig. 2B).
Figure 2.
SpoIVFB-GFP does not achieve proper localization by selective degradation. (A) SpoIVFB-GFP localized to the mother-cell membrane that surrounds the forespore in the presence of SpoIVFA. SpoIVFB-GFP from strain BDR497 was visualized by fluorescence microscopy at hour 2.5 of sporulation. (B) In the absence of SpoIVFA, SpoIVFB-GFP was uniformly distributed in all membranes. SpoIVFB-GFP from strain BDR493 was visualized by fluorescence microscopy at hour 2.5 of sporulation. (Bar = 1 μm.) (C) Immunoblots of whole-cell extracts from the same cultures used for microscopy in A and B. Lysates were from hour 2.5 of sporulation and were analyzed with polyclonal Abs that recognize the GFP moiety of SpoIVFB-GFP (B-GFP), SpoIVFA (A), and σA. The anti-σA immunoblot served as a control for loading.
To obtain a more quantitative assessment of the possible effect of SpoIVFA on SpoIVFB-GFP levels, we monitored levels of the GFP-tagged protein by immunoblot analysis of whole-cell lysates from wild-type and SpoIVFA mutant cells undergoing sporulation. In confirmation and extension of the results of fluorescence microscopy, SpoIVFA had no measurable effect on the total level of SpoIVFB-GFP (Fig. 2C). Immunoblots of serially diluted extracts confirmed that detection of SpoIVFB-GFP was in the linear range (data not shown). In sum, these results argue against a localization model in which SpoIVFA protects SpoIVFB-GFP from proteolysis in the engulfing forespore membrane. These results are also inconsistent with yet an additional variation on selective degradation in which SpoIVFA confers on SpoIVFB-GFP sensitivity to a protease that is restricted to the cytoplasmic membrane. If this were the case, then the levels of SpoIVFB-GFP should have been higher in the absence of SpoIVFA than in its presence, which was not the case (Fig. 2C). We conclude that the subcellular localization of SpoIVFB-GFP is not a result of its selective degradation in the cytoplasmic membrane.
SpoIVFB-GFP Can Diffuse from the Cytoplasmic Membrane and Be Captured in the Engulfing Septal Membrane.
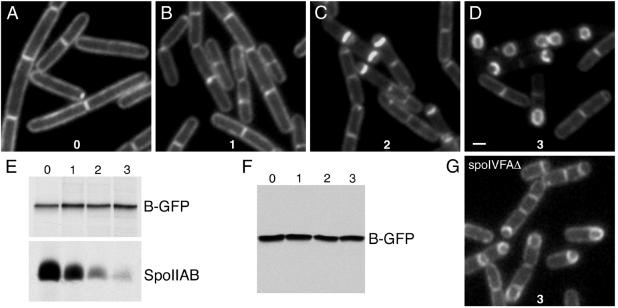
In an alternative model, SpoIVFB-GFP is initially inserted into both the cytoplasmic and septal membranes in the mother cell and then is captured in the engulfing septal membrane (Fig. 1B). According to this diffusion-and-capture model, SpoIVFB-GFP molecules that had been inserted into the cytoplasmic membrane would reach the engulfing septal membranes by diffusion. To investigate this model, we took advantage of the observation that SpoIVFB-GFP localizes uniformly to all membranes in cells that had been engineered to synthesize the fusion protein during growth (Fig. 3A). For these experiments we fused the gene encoding SpoIVFB-GFP to a xylose-inducible promoter (PxylA) and inserted it in single copy at a nonessential locus in the chromosome. When grown in the presence of xylose, SpoIVFB-GFP localized uniformly to all membranes (Fig. 3A). In the absence of xylose, the SpoIVFB-GFP signal was virtually undetectable both during growth and sporulation (data not shown).
Figure 3.
SpoIVFB-GFP is capable of diffusion and capture in the engulfing septal membrane. (A–D) The fate of uniformly distributed SpoIVFB-GFP (synthesized during growth) through the first 3 h of sporulation. BDR647, which contains a xylose-inducible promoter fused to the gene encoding SpoIVFB-GFP, was grown in the presence of xylose. After the initiation of sporulation, xylose was removed and the localization of SpoIVFB-GFP was monitored by fluorescence microscopy (see Materials and Methods). Time (in hours) after the start of sporulation is indicated in each image. (Bar = 1 μm.) (E) Pulse-chase analysis of SpoIVFB-GFP (B-GFP) and SpoIIAB synthesized at the start of sporulation. Sporulating cultures BDR524 and QPB550 were pulsed-labeled 30 min after the start of sporulation (see Materials and Methods). Time (in hours) after the addition of unlabeled methionine is indicated above the protein gels. (F) Immunoblot of whole-cell extracts from cells undergoing sporulation. Lysates were prepared from strain BDR647 treated identically to the culture in A–D and were analyzed with polyclonal Abs that recognize the GFP moiety of SpoIVFB-GFP (B-GFP). Time (in hours) after the start of sporulation is indicated above the immunoblot. (G) The fate of uniformly distributed SpoIVFB-GFP in the absence of SpoIVFA at hour 3 of sporulation. SpoIVFB-GFP was synthesized during growth in strain BDR526 and treated identically to BDR647 as described in A–D.
To determine whether SpoIVFB-GFP, which was initially present in the cytoplasmic membrane during growth, could diffuse into the engulfing septal membrane during sporulation, we grew the cells in the presence of xylose and then removed the inducer at the start of sporulation (see Materials and Methods). The fate of SpoIVFB-GFP molecules that had been produced during growth was monitored by fluorescence microscopy over the course of sporulation (Fig. 3 A–D). The results show that at the onset of engulfment the fusion protein began to accumulate in the septal membranes (Fig. 3C). By the time engulfment was near completion SpoIVFB-GFP was virtually absent from the cytoplasmic membrane and had apparently redistributed to the forespore membranes (Fig. 3D). These data are consistent with the idea that SpoIVFB-GFP can diffuse and be captured in the engulfing septal membrane.
A possible complication in our interpretation of the experiment of Fig. 3 is that the forespore is surrounded by two layers of membrane. One membrane, the inner forespore membrane, is derived from the forespore itself whereas the other, the outer forespore membrane, is derived from the mother-cell membrane that engulfs the forespore. Because SpoIVFB-GFP had been synthesized before asymmetric division, the fusion protein would have been present in the membranes of both the mother cell and forespore. It was therefore possible that the apparent localization of SpoIVFB-GFP around the forespore (Fig. 3D) was an artifact resulting from the presence of SpoIVFB-GFP in both membranes. To address this possibility, we repeated the experiment in a SpoIVFA mutant in which SpoIVFB-GFP fails to localize. As expected, the SpoIVFB-GFP signal surrounding the forespore was approximately twice as intense as the signal in the cytoplasmic membrane (Fig. 3G). However, SpoIVFB-GFP, which was virtually undetectable in the cytoplasmic membrane of wild-type sporangia on completion of engulfment, was readily detectable in the cytoplasmic membrane of the SpoIVFA mutant sporangia (Fig. 3, compare D and G). Thus, these results are consistent with the idea that SpoIVFB-GFP molecules that were originally present in the cytoplasmic membrane of growing cells had redistributed with high selectivity to the membranes that surround the forespore.
An alternative interpretation of the experiment of Fig. 3 is that the removal of xylose at the start of sporulation had not shut off the synthesis of the SpoIVFB-GFP completely and that the accumulation of SpoIVFB-GFP around the forespore was actually the result of targeted insertion of newly synthesized molecules into the septal membranes. According to this view, SpoIVFB-GFP molecules that had been synthesized during growth were eliminated by proteolysis during the course of sporulation and newly synthesized molecules were inserted directly into the septal membranes. If the SpoIVFB-GFP molecules that accumulated around the forespore during engulfment were in fact the same molecules that had been present in the cytoplasmic membrane at the start of sporulation, then the fusion protein must have been stable over the course of the experiment. To investigate the stability of SpoIVFB-GFP, we determined the half-life of the fusion protein synthesized at the beginning of sporulation by a pulse-chase experiment (see Materials and Methods). Consistent with the idea that SpoIVFB-GFP is stable during sporulation, radioactively labeled SpoIVFB-GFP was not measurably depleted during the chase over the course of the first 3 h of sporulation (Fig. 3E). As a control, radioactively labeled SpoIIAB, a sporulation protein that is known to be unstable when its binding partners (SpoIIAA and σF) are absent (18), was largely eliminated during the first 3 h of sporulation in a parallel pulse-chase experiment (Fig. 3E).
Knowing that SpoIVFB-GFP is stable, we further investigated the possibility that the xylose-inducible promoter had not been shut off after the removal of inducer by asking whether the levels of the fusion protein continued to increase during sporulation. SpoIVFB-GFP levels were monitored by immunoblot analysis. Consistent with the idea that synthesis of SpoIVFB-GFP was shut off when xylose was removed, the level of SpoIVFB-GFP remained approximately constant during the first 3 h of sporulation (Fig. 3F).
In toto, these data indicate that SpoIVFB-GFP molecules that had been synthesized during growth and that were initially localized to the cytoplasmic membrane of vegetative cells were the same molecules that eventually localized to the outer forespore membrane during sporulation. We conclude that there is no barrier to diffusion between the cytoplasmic and septal membranes during sporulation and that SpoIVFB-GFP is capable of diffusing to, and being captured in, the engulfing septal membrane.
SpoIVFB-GFP Is Inserted into the Cytoplasmic Membrane During Sporulation.
Thus far, we have shown that SpoIVFB-GFP that is initially present in the cytoplasmic membrane (in cells engineered to synthesize the fusion protein during growth) is capable of diffusion to, and capture in, the engulfing septal membrane. However, we could not exclude the possibility that normally during the course of sporulation SpoIVFB-GFP is directly and selectively inserted into the engulfing septal membranes by targeted insertion (Fig. 1B).
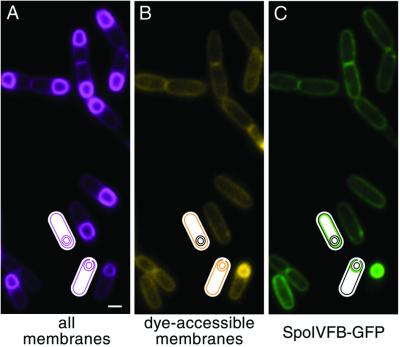
To distinguish between these possibilities, we took advantage of a unique feature of endospore formation. After completion of engulfment the mother-cell membrane that surrounds the forespore fuses with itself so as to pinch off the forespore as a free protoplast within the mother cell, in effect creating a cell within a cell (7). This terminal step in the phagocytic-like process of engulfment is referred to as membrane fusion (19). As a consequence of membrane fusion the mother-cell membrane that surrounds the forespore becomes topologically distinct from the cytoplasmic membrane. Importantly, after completion of engulfment, integral membrane proteins should no longer be capable of freely diffusing between the cytoplasmic and outer forespore membranes. Membrane fusion can be monitored with the lypophylic fluorescent dye FM 4-64, which cannot cross the lipid bilayer (19, 20). If FM 4-64 is added before the completion of engulfment the fluorescent dye is able to access all membranes of the sporangium: the cytoplasmic membrane, the mother-cell membrane that is engulfing the forespore, and the forespore membrane (Fig. 4B). However, if FM4-64 is added after membrane fusion, the dye can no longer access the two membranes that surround the forespore. Thus, in sporangia that have been treated with FM4-64 after membrane fusion, only the cytoplasmic membrane is labeled (Fig. 4B). The membranes that surround the forespore can, however, be visualized in these sporangia with a second membrane dye (TMA-DPH) that has distinct spectral properties from FM4-64 if the second membrane dye is added at the start of sporulation (Fig. 4A).
Figure 4.
SpoIVFB-GFP is inserted into the cytoplasmic membrane during sporulation. Strain BDR524, which contains a xylose-inducible promoter, fused to the gene encoding SpoIVFB-GFP was sporulated in the absence of xylose. After ∼80% of the sporulating cells had undergone membrane fusion, xylose was added to induce the synthesis of SpoIVFB-GFP. Labeled membranes and SpoIVFB-GFP were visualized by fluorescence microscopy 30 min after induction. (A) All membranes (false-colored magenta) were labeled with the TMA-DPH added at the start of sporulation. (B) To determine which cells had undergone membrane fusion, dye-accessible membranes (false-colored yellow) were visualized with FM 4-64 added just before visualization. (C) SpoIVFB-GFP synthesized after membrane fusion localized to the cytoplasmic membrane. (Bar = 1 μm.)
To distinguish between the targeted insertion and the diffusion-and-capture models, we investigated the fate of SpoIVFB-GFP molecules that were synthesized after completion of engulfment. If SpoIVFB-GFP achieves proper localization by targeted insertion, then the protein synthesized after membrane fusion should be properly localized to the outer forespore membrane. However, if SpoIVFB-GFP achieves proper localization by diffusion and capture, then SpoIVFB-GFP should be present in both the outer forespore membrane and the cytoplasmic membrane. The reason is that SpoIVFB-GFP inserted into the cytoplasmic membrane after engulfment would not be able to diffuse to the outer forespore membrane.
To carry out this experiment, we induced expression of the gene encoding SpoIVFB-GFP after the completion of engulfment with the xylose-inducible promoter fusion described previously. In the course of our analysis we discovered that this promoter was not active (and could not be induced) in the forespore compartment at this late stage of sporulation. Expression of the gene encoding GFP fused to the xylose-inducible promoter produced only a weak fluorescent signal in the forespore compartment (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). This fortuitous result simplified our analysis, because we were interested only in SpoIVFB-GFP synthesized in the mother-cell compartment. Cells harboring the inducible gene fusion were sporulated in the absence of inducer, and membrane fusion was monitored every 30 min with the membrane dyes FM4-64 and TMA-DPH. At hour 3 when ∼80% of the sporulating cells had topologically distinct membranes, synthesis of SpoIVFB-GFP was induced by the addition of xylose. The localization of SpoIVFB-GFP was determined 30 min later by fluorescence microscopy.
Surprisingly, in 87% (n = 234) of the sporangia that had completed engulfment, SpoIVFB-GFP was present exclusively in the cytoplasmic membrane of the mother-cell compartment (Fig. 4C). In the remaining 13% of the sporangia fluorescence from SpoIVFB-GFP was observed in both the cytoplasmic membrane and the outer forespore membranes. The simplest explanation for these results is that SpoIVFB-GFP is inserted only into the cytoplasmic membrane of sporangia that have completed engulfment and that the minority category in which fluorescence was observed in both the mother-cell and forespore membranes represented sporangia that had not completed engulfment at the time xylose was added but underwent membrane fusion during the 30-min period of induction. In this view, SpoIVFB-GFP that is made before membrane fusion is inserted into the cytoplasmic membrane from which it diffuses to, and is captured in, the outer forespore membrane. Meanwhile, protein that is synthesized after membrane fusion is inserted in, and remains restricted to, the cytoplasmic membrane. Several sporangia were also observed that had not completed engulfment by the end of the induction period (Fig. 4B) and in which SpoIVFB-GFP was properly localized to the outer forespore membrane (Fig. 4C). In these sporangia all of the SpoIVFB-GFP would have been able to freely diffuse from the cytoplasmic to the engulfing septal membrane.
The lack of SpoIVFB-GFP insertion into the mother-cell membrane that surrounds the forespore was unexpected and prompted a series of control experiments. It was possible that insertion of SpoIVFB-GFP into the outer forespore membrane was prevented by the presence of a nascent spore coat, which is assembled around the outer forespore membrane during engulfment. To address this possibility we repeated the xylose induction experiment in SpoIVA (not to be confused with SpoIVFA) and SpoVM mutants in which proper assembly of the protein coat is blocked (21–23). Once again, SpoIVFB-GFP was found exclusively in the cytoplasmic membrane in a high proportion of mutant sporangia that had completed engulfment (data not shown). Thus, it is unlikely that the spore coat was responsible for preventing insertion of SpoIVFB-GFP into the outer forespore membrane. Insertion exclusively in the cytoplasmic membrane was also observed after induction of SpoIVFB-GFP synthesis in sporangia mutant for the mother-cell transcription factors SpoIIID, which is active at a late stage of engulfment (24), or σK, which is active after engulfment (25) (data not shown). Thus, the block to insertion of SpoIVFB-GFP into the mother-cell membrane that surrounds the forespore did not require new gene expression before or after the completion of engulfment and conceivably already existed before engulfment.
In toto, these data are consistent with the idea that SpoIVFB-GFP achieves proper localization during sporulation by insertion into the cytoplasmic mother-cell membrane followed by diffusion and capture in the engulfing septal membrane.
Discussion
Eukaryotes have a sophisticated system for ensuring that integral membrane proteins reach their proper subcellular location. Sorting signals on membrane proteins direct them to specific transport vesicles, which then deliver the proteins to the apical or basolateral plasma membrane or to the membranes of a particular intracellular organelle (26). Here, we have investigated the mechanism by which a bacterial membrane protein (SpoIVFB-GFP) reaches its proper destination in the absence of a conspicuous sorting machine.
We ruled out models based on selective degradation, because the mislocalization of SpoIVFB-GFP was not accompanied by a change in the levels of the sporulation protein (Fig. 2C) as would be predicted by such models. Next, we demonstrated that there is no barrier to diffusion between the cytoplasmic and engulfing septal membranes. Uniformly distributed SpoIVFB-GFP present at the start of sporulation (in cells that had been engineered to synthesize the protein prematurely) could diffuse to, and be captured in, the septal membrane during engulfment (Fig. 3 A–D). This result is of particular interest because a barrier to diffusion of integral membrane proteins between the mother and daughter cells of the budding yeast Saccharomyces cerevisiae has been found to exist at the mother-bud neck (27). Finally, we found that SpoIVFB-GFP was inserted into the cytoplasmic membrane during sporulation when synthesized after the completion of engulfment (Fig. 4C) (in cells in which the synthesis of the protein could be delayed beyond its normal time of induction). In toto, these results are most easily compatible with a model in which SpoIVFB-GFP reaches its destination by a pathway in which it is initially inserted into the cytoplasmic membrane followed by diffusion to, and capture in, the septal membranes during the process of engulfment.
In the final step of this process, SpoIVFB-GFP is retained in the engulfing septal membrane by interaction with its partner protein SpoIVFA (10). This finding raises the question of the basis for the restricted localization of SpoIVFA in the engulfing septal membrane. SpoIVFA localizes properly in the absence of SpoIVFB (10), and a similar analysis with cells engineered to express GFP-SpoIVFA prematurely suggests that it too localizes by a diffusion-and-capture mechanism. At present we do not know what feature of the septal membrane is responsible for capturing SpoIVFA, but we have reported that a domain of SpoIVFA that is located in the space between the double membranes that surround the forespore shares homology with proteins involved in peptidoglycan remodeling (10). Thus, it is possible that SpoIVFA is captured in the engulfing septal membrane through an interaction with a unique feature of the peptidoglycan that is sandwiched between the two membranes of the sporulation septum.
The rate of diffusion of integral membrane proteins in the lipid bilayer has not been determined in bacteria. However, lateral membrane protein diffusion in eukaryotes has been found to be ∼0.3 μm2/sec (28). If the rate of diffusion is similar in B. subtilis, which is only 2–4 μm in length, then the time it would take a molecule of SpoIVFB-GFP to diffuse from the cytoplasmic membrane into the septal membrane would be less than a minute. Thus, a diffusion-and-capture mechanism is easily compatible with the strikingly restricted pattern of localization of SpoIVFB-GFP and the apparent absence of the protein in the cytoplasmic membrane.
The only other integral membrane protein whose mechanism of localization has been explored is the outer membrane protein IcsA from the Gram-negative bacterium S. flexneri, which localizes to the old pole of the cell. In contrast to the diffusion-and-capture mechanism that we have proposed for SpoIVFB-GFP, analysis of IcsA localization is most consistent with a targeted-insertion model. A GFP fusion to IcsA lacking both its signal sequence and outer membrane translocation domain is still capable of localizing to the cell pole (6). This result suggests that the nascent IcsA protein localizes to the cell pole on the cytoplasmic face of the inner membrane before its translocation into the periplasm and subsequent insertion in the outer membrane. It will be of interest to see whether both the diffusion-and-capture mechanism that we have invoked for SpoIVFB-GFP and the targeted-insertion mechanism proposed for IcsA prove to be general features of the localization pathways for bacterial membrane proteins.
Although our data favor a diffusion-and-capture model, we cannot rule out the possibility that SpoIVFB-GFP becomes localized in the outer forespore membrane by targeted insertion during the process of engulfment and that this pathway is shut off after the completion of engulfment. If this was the case, then we would expect that some gene that is responsible for blocking targeted insertion must be switched on during or just after engulfment. If so, such a gene would be expected to be under the control of the transcription factor SpoIIID, which appears in the mother cell at a late stage in engulfment (24), or σK, which becomes active shortly after the completion of engulfment (25). Importantly, neither SpoIIID nor σK mutants relieved the block to insertion into the outer forespore membrane observed after the completion of engulfment. Thus, we think the block to insertion exists during the process of engulfment and conceivably before its commencement. Because the mother-cell membrane that engulfs the forespore is derived from the septal membrane, this interpretation raises the intriguing possibility that integral membrane proteins are not, in general, inserted directly into the septum and only localize there by diffusion and capture.
The finding that SpoIVFB-GFP is exclusively inserted into the cytoplasmic membrane of the mother cell after completion of engulfment was unexpected and prompted us to determine whether this block was unique to SpoIVFB-GFP. We examined the polytopic membrane protein MalF of E. coli (29) fused to GFP, and it too localized exclusively to the cytoplasmic membrane in the majority of cells that had completed engulfment when its synthesis was induced after membrane fusion (data not shown). Thus, it is possible that the block to insertion into the outer forespore membrane is a general feature of all membrane proteins synthesized in the mother cell after engulfment.
Consistent with this idea, the mother-cell transcription factors σK and GerE, which are active only after the completion of engulfment (7), principally govern the synthesis of soluble proteins that are incorporated into the spore coat (30). Only two genes under the control of these late-acting transcription factors are known that encode proteins with predicted transmembrane segments. These are CotV and Csk22 (YobW) (31, 32). CotV is thought to reside on the spore coat, and it has been suggested that its hydrophobic segments might not be inserted into the outer forespore membrane but instead face outward, contributing to the hydrophobic surface of the spore coat (33). Csk22 has not been characterized but is predicted to have five transmembrane segments and is therefore likely to be a polytopic membrane protein. Based on our analysis of SpoIVFB-GFP and MalF-GFP, we predict that Csk22 (and any other integral membrane protein synthesized under the control of σK or GerE) will be found to localize exclusively to the cytoplasmic membrane.
Supplementary Material
Acknowledgments
We thank P. Stragier, G. Guidotti, R. Hampton, J. Theriot, and members of the Losick lab for valuable discussions throughout the course of this work; L. Shapiro and E. Judd for critical reading of the manuscript; S. Ben-Yehuda for assistance with the microscopy; M. Fujita (Harvard University) for anti-σA antiserum and strain MF240; C. van Ooij (Harvard University) for strain CV1103; and R. Hellmiss for help with digital art. This work was supported by National Institute of Health Grant GM18568 (to R.M.L.). D.Z.R. was supported in part by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship (DRG-1514).
Abbreviation
- GFP
green fluorescent protein
References
- 1.Shapiro L, Losick R. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 2.Lutkenhaus J, Addinall S G. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 3.Alley M R, Maddock J R, Shapiro L. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 4.Arigoni F, Pogliano K, Webb C D, Stragier P, Losick R. Science. 1995;270:637–640. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- 5.Steinhauer J, Agha R, Pham T, Varga A W, Goldberg M B. Mol Microbiol. 1999;32:367–377. doi: 10.1046/j.1365-2958.1999.01356.x. [DOI] [PubMed] [Google Scholar]
- 6.Charles M, Perez M, Kobil J H, Goldberg M B. Proc Natl Acad Sci USA. 2001;98:9871–9876. doi: 10.1073/pnas.171310498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stragier P, Losick R. Annu Rev Genet. 1996;30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 8.Cutting S, Roels S, Losick R. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 9.Resnekov O, Alper S, Losick R. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 10.Rudner D Z, Losick R. Genes Dev. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudner D Z, Fawcett P, Losick R. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Resnekov O, Losick R. Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y T, Kroos L. J Bacteriol. 2000;182:3305–3309. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 15.Green D H, Cutting S M. J Bacteriol. 2000;182:278–285. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harwood C R, Cutting S M. Molecular Biological Methods for Bacillus. New York: Wiley; 1990. [Google Scholar]
- 17.Youngman P J, Perkins J B, Losick R. Proc Natl Acad Sci USA. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan Q, Garsin D A, Losick R. Mol Cell. 2001;8:873–883. doi: 10.1016/s1097-2765(01)00362-8. [DOI] [PubMed] [Google Scholar]
- 19.Sharp M D, Pogliano K. Proc Natl Acad Sci USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pogliano J, Osborne N, Sharp M D, Abanes-De Mello A, Perez A, Sun Y L, Pogliano K. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price K D, Losick R. J Bacteriol. 1999;181:781–790. doi: 10.1128/jb.181.3.781-790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roels S, Driks A, Losick R. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin P A, Fan N, Ricca E, Driks A, Losick R, Cutting S. Mol Microbiol. 1993;9:761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 24.Kroos L, Zhang B, Ichikawa H, Yu Y T. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 25.Rudner D Z, Losick R. Dev Cell. 2001;1:733–742. doi: 10.1016/s1534-5807(01)00094-6. [DOI] [PubMed] [Google Scholar]
- 26.Hunziker W, Mellman I. Semin Cell Biol. 1991;2:397–410. [PubMed] [Google Scholar]
- 27.Takizawa P A, DeRisi J L, Wilhelm J E, Vale R D. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 28.Jacobson K, Ishihara A, Inman R. Annu Rev Physiol. 1987;49:163–175. doi: 10.1146/annurev.ph.49.030187.001115. [DOI] [PubMed] [Google Scholar]
- 29.Boyd D, Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piggot P J, Losick R. In: Bacillus subtilis and Its Closest Relatives: From Genes to Cells. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 483–517. [Google Scholar]
- 31.Henriques A O, Bryan E M, Beall B W, Moran C P., Jr J Bacteriol. 1997;179:389–398. doi: 10.1128/jb.179.2.389-398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Ichikawa H, Halberg R, Kroos L, Aronson A I. J Mol Biol. 1994;240:405–415. doi: 10.1006/jmbi.1994.1456. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Fitz-James P C, Aronson A I. J Bacteriol. 1993;175:3757–3766. doi: 10.1128/jb.175.12.3757-3766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.