Abstract
During spermiogenesis (the maturation of spermatids into spermatozoa) in many vertebrate species, protamines replace histones to become the primary DNA-packaging protein.It has long been thought that this process is facilitated by the hyperacetylation of histone H4. However, the responsible histone acetyltransferase enzymes are yet to be identified. CDY is a human Y-chromosomal gene family expressed exclusively in the testis and implicated in male infertility. Its mouse homolog Cdyl, which is autosomal, is expressed abundantly in the testis. Proteins encoded by CDY and its homologs bear the “chromodomain,” a motif implicated in chromatin binding. Here, we show that (i) human CDY and mouse CDYL proteins exhibit histone acetyltransferase activity in vitro, with a strong preference for histone H4; (ii) expression of human CDY and mouse Cdyl genes during spermatogenesis correlates with the occurrence of H4 hyperacetylation; and (iii) CDY and CDYL proteins are localized to the nuclei of maturing spermatids where H4 hyperacetylation takes place. Taken together, these data link human CDY and mouse CDYL to the histone-to-protamine transition in mammalian spermiogenesis. This link offers a plausible mechanism to account for spermatogenic failure in patients bearing deletions of the CDY genes.
Keywords: histone acetylation‖spermiogenesis‖chromodomain‖infertility
The acetylation of N-terminal lysine residues in core histones has been implicated in three distinct cellular processes. The first is the deposition of free histones onto newly synthesized DNA (1). The second is the regulation of gene expression (reviewed in ref. 2). The third is the displacement of histones by transition proteins and protamines during vertebrate spermatogenesis (3–6). Various histone acetyltransferase (HAT) enzymes involved in the first two processes have been characterized. HATs involved in the third process have yet to be identified.
In mammals and many other vertebrates, dramatic chromatin remodeling occurs during spermiogenesis, whereby histones are displaced from chromatin, first by transition proteins and later by protamines (7). With protamines, DNA in mature spermatozoa is packaged into an extremely condensed, functionally inert configuration (8). Several lines of evidence led to the view that chromatin remodeling during spermiogenesis is facilitated by hyperacetylation of histone H4. First, studies of numerous vertebrate species have shown that H4 hyperacetylation correlates with the occurrence of the histone-to-protamine transition in spermatogenesis. Extensive H4 hyperacetylation occurs in the testes of species where histones are replaced by protamines during spermiogenesis (3–6), but not in species that completely retain somatic-type histones in mature spermatozoa (4, 9). Second, the timing of H4 hyperacetylation is consistent with its role in promoting histone displacement. Studies in the rat, for instance, demonstrated that H4 hyperacetylation during spermiogenesis immediately precedes the histone-to-protamine transition (5, 6). Finally, the postulated role of histone hyperacetylation in facilitating histone displacement is consistent with the general perception that histone acetylation results in a more open chromatin structure and hence easier access by regulatory proteins. Indeed, it has been observed in the trout that H4 hyperacetylation during spermiogenesis results in a highly relaxed chromatin configuration (4). An important inroad into understanding the regulation of chromatin remodeling during vertebrate spermatogenesis will be the identification of enzymes responsible for H4 hyperacetylation during the remodeling process.
We have previously isolated a group of very closely related genes on the human Y chromosome, which we named CDY for ChromoDomain Y (10, 11). An autosomal homolog of CDY was identified in humans (named CDYL for CDY Like) and in mice (named Cdyl) (11). Human CDYL and mouse Cdyl are orthologs, their protein products sharing 93% identity, whereas human CDYL and CDY are paralogs, their protein products sharing 63% identity. Comparative evolutionary studies showed that human CDY arose on the Y chromosome through retroposition of CDYL mRNA, followed by amplification on the Y (11). Mice lack CDY on the Y chromosome. The mouse autosomal Cdyl gene produces a 3.6-kb ubiquitous transcript and a highly abundant 2.8-kb testis-specific transcript (11). The presence of these two distinct transcripts suggests that mouse Cdyl may perform two functions, one housekeeping and the other spermatogenic. In humans, CDYL expresses only a ubiquitous transcript, whereas CDY is expressed exclusively in the testis (11). Thus, CDYL and CDY in humans appear to have undergone functional specialization, with CDYL retaining the housekeeping function and CDY assuming the spermatogenic function.
On the human Y, two distinct versions of the CDY genes, CDY1 and CDY2, have been characterized. These two versions differ slightly in sequence, and each correspond to multiple nearly identical copies of the gene (11, 12). There are two copies of CDY1 on the human Y chromosome. They map within AZFc, a region of the Y frequently deleted in infertile men who suffer from spermatogenic failure (11–14). This finding suggests that CDY may play an important role in spermatogenesis and that its deletion may contribute to spermatogenic defects in AZFc-deleted men.
In this study, we investigated the spermatogenic function of human CDY and mouse Cdyl genes. We showed that protein products of these genes possess in vitro HAT activity. We further demonstrated that the expression pattern and subcellular localization of the proteins are consistent with a role in mediating histone H4 hyperacetylation during spermatid maturation.
Materials and Methods
HAT Activity Assay.
DNA fragments corresponding to the ORFs of the human CDY1 minor transcript (11), human CDYL, and mouse Cdyl were cloned into the His-tagged prokaryotic expression vector pRSET (Invitrogen). (CDY1 is one of the two known CDY isoforms on the human Y chromosome.) Escherichia coli strain BL21(DE3) containing these plasmids was induced with isopropyl β-d-thiogalactoside. Soluble proteins were extracted in a buffer containing 300 mM NaCl, 1 mM DTT, 1 mM phenylmethysulfonyl fluoride, and 50 mM Tris⋅HCl, pH 8.0. Insoluble proteins in inclusion bodies were isolated in a buffer containing 8 M urea, 100 mM Na-phosphate, and 100 mM Tris⋅HCl, pH 8. Recombinant proteins were then purified on Ni-NTA-agarose (Qiagen) according to the vendor's protocol and tested for HAT activity by either the liquid assay or the in-gel assay as described (15, 16).
Northern Analysis.
Each lane on the Northern blot contained 10 μg of mouse testis total RNA. The blot was incubated with 32P body-labeled probes at 65°C in 0.5 M sodium phosphate buffer (pH 7) and 7% SDS and washed at 65°C in 0.1× SSC and 0.1% SDS before exposing to film.
Western Analysis.
Western blots were made with SDS-solubilized protein extracted from mouse and human testes. Anti-CDYL and anti-CDY sera were raised in rabbits against whole mouse CDYL and human CDY proteins (Research Genetics, Huntsville, AL). The mouse blot was incubated with anti-CDYL serum at a 1:200 dilution. The human blot was incubated with anti-CDY serum at a 1:100 dilution. Both mouse and human blots were then incubated with goat anti-rabbit-IgG Ab conjugated to horseradish peroxidase (Jackson ImmunoResearch; 1:500 dilution). Immunoreactive bands on the blot were visualized by using the enhanced chemiluminescence reagent (Amersham Pharmacia) following the vendor's protocol.
Microdissection of Mouse Seminiferous Tubules.
Seminiferous tubules were dissected under a transillumination dissection microscope into 1-mm segments as described (17). Each segment was squashed by a cover-slip onto a slide, prefixed in liquid nitrogen, and shortly in 94% ethanol, air dried, and stored. The slides were postfixed in 10% formalin before use.
Immunohistochemistry.
Mouse testis sections or seminiferous tubule squash preparations were incubated with anti-CDYL serum at a 1:100 dilution or anti-hyperacetylated histone H4 (AcH4) serum at a 1:200 dilution. Human testis sections were incubated with anti-CDY serum at a 1:100 dilution. Both mouse and human sections were then incubated with goat anti-rabbit-IgG Ab conjugated to horseradish peroxidase (Jackson ImmunoResearch; 1:200 dilution). Secondary Ab was visualized with the VECTASTAIN ABC System (Vector Laboratories) following the vendor's protocol.
Reverse Transcription–PCR Detection of mRNA.
Extraction of mRNA from human tissues was performed by using the QuickPrep Micro mRNA Purification kit (Amersham Pharmacia). cDNA was made from mRNA by using the Marathon cDNA Amplification kit (CLONTECH). PCR was performed with 30 cycles under the following conditions: 94°C for 45 sec, 60°C for 45 sec, and 72°C for 45 sec. PCR primers for human genes were as follows: CDYL, forward GTACATCTCCGTTCATGGATG and reverse CTGATAGCTTCTGCCATTTTAG; DAZL, forward ATCCTCCTCCACCACAGTTTCAGA and reverse GTGGGCCATTTCCAGAGGG; PRM1, forward AGGTACAGATGCTGTCGCAG and reverse CATTGTTCCTTAGCAGGCTC; and CDY (able to amplify both CDY1 and CDY2), forward GGACTAGGTGCATCCATCC and reverse CAACAGAGGTATTTTTAACATGG.
Results and Discussion
Human CDY and Its Homologs Encode HATs.
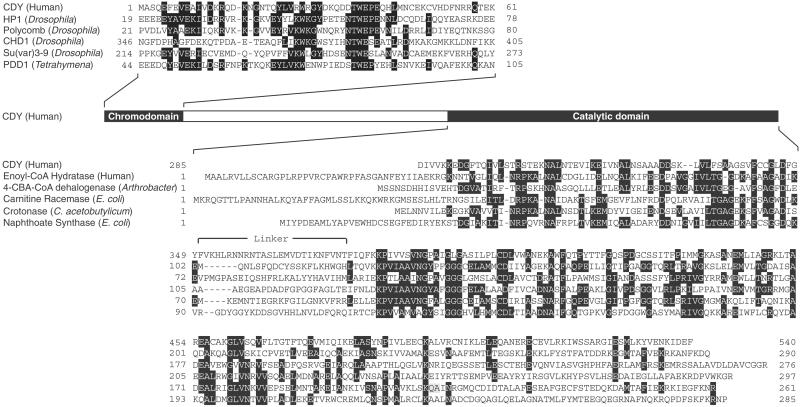
A homology search has previously revealed two functional domains in proteins encoded by human CDY, human CDYL, and mouse Cdyl (11). The N-terminal regions of the proteins contain a chromodomain, a sequence motif that has been implicated in chromatin binding (18). The C-terminal regions of the proteins contain a catalytic domain found in many CoA-dependent acylation enzymes (19) (Fig. 1). The presence of these two domains suggests that the proteins may be CoA-dependent chromatin-acylating enzymes. Such an enzyme could be a HAT.
Figure 1.
Amino acid sequence alignments of the two functional domains of CDY with their respective homologs. CDY protein sequence is deduced from the major transcript of CDY1, a version of CDY on the human Y chromosome (11). Numbers at the beginning and end of each sequence segment are start and end positions of the segment with respect to the initiator methionine. Consensus residues shared by three or more proteins are shaded.
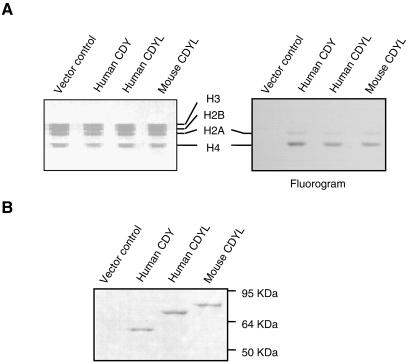
To address the possibility that CDY and its homologs encode HATs, recombinant proteins were produced and tested for HAT activity. First, we performed a liquid assay where recombinant proteins were incubated with all four core histones (H2A, H2B, H3, and H4) in the presence of acetyl-CoA (carrier of the acetyl moiety). We found that all recombinant proteins specifically acetylated H4 and H2A, with H4 being the strongly preferred substrate (Fig. 2A).
Figure 2.
HAT activity of recombinant human CDY, human CDYL, and mouse CDYL proteins. (A) Liquid assay demonstrating HAT activity of recombinant proteins. Bacterially expressed recombinant proteins were incubated with chicken erythrocyte core histones and [3H]acetyl-CoA. Reaction products were separated on an SDS/PAGE gel, stained with Coomassie blue to reveal all four core histone species (Left), and fluorographed to detect acetylated histone species (Right). (B) In-gel assay demonstrating HAT activity of recombinant proteins. Recombinant proteins were separated on an SDS/PAGE gel that preincorporated calf thymus histones. After electrophoresis, the gel was incubated with [3H]acetyl-CoA and fluorographed to detect HAT activity in the gel.
To confirm that the HAT activity detected by the liquid assay indeed came from recombinant proteins, an in-gel assay was performed. Purified recombinant proteins were separated on an SDS/PAGE gel, followed by an in-gel histone acetylation reaction. This assay demonstrated unequivocal HAT activity of the recombinant proteins (Fig. 2B). As a control for specificity, the in-gel assay was performed by using BSA instead of histones. This assay did not detect any nonspecific acetyltransferase activity (data not shown).
Mouse Cdyl Is Abundantly Expressed in Step 9–12 Spermatids.
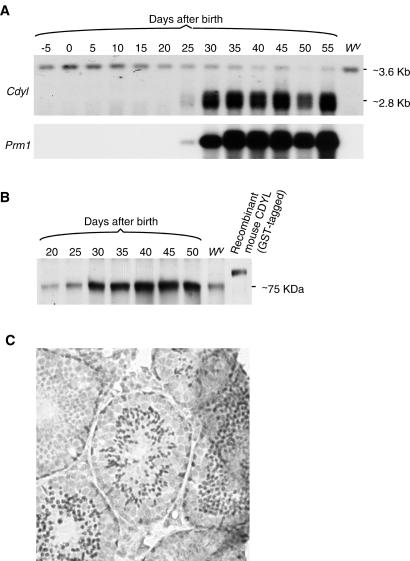
Based on the HAT activity of human CDY and mouse CDYL proteins and their testis expression, we hypothesized that they may mediate H4 hyperacetylation during spermiogenesis. A critical prediction of this hypothesis is a temporal correlation between CDY/CDYL expression and H4 hyperacetylation during spermiogenesis. We first analyzed the transcription of mouse Cdyl. Northern analysis of mouse testes collected at different developmental stages revealed two distinct Cdyl transcripts (Fig. 3A Upper). A long transcript, previously shown to be present in a wide range of tissues (11), is present in testes of all stages, as well as in germ-cell-deficient homozygous Wv testis (20). A short transcript, previously shown to be testis-specific (11), is abundantly expressed in the testis after day 25. This transcript is absent in the germ-cell-deficient homozygous Wv testis, indicating that it is produced only in germ cells. Based on morphology, mouse spermiogenesis can be divided into three major stages: round spermatids containing spherical nuclei, elongating spermatids where nuclear elongation is underway, and elongated spermatids with fully elongated nuclei (21). At day 25 of mouse postnatal development, germ cells in the first wave of spermatogenesis enter the late phase of the round-spermatid stage, and by day 30, germ cells reach the elongating-spermatid stage (22). By this developmental timeline, the Cdyl short transcript first appears in late-phase round spermatids at postnatal day 25 and becomes highly expressed in elongating spermatids at day 30. The same Northern blot also was probed for the expression of Prm1, the gene encoding Protamine 1. It showed that in the developing mouse testis, the temporal expression profile of Prm1 is essentially identical to that of the Cdyl short transcript (Fig. 3A). This is consistent with previous studies showing that Prm1 transcription begins in late-phase round spermatids and becomes highly abundant in elongating spermatids (23).
Figure 3.
Expression of mouse Cdyl in the testis. (A) Northern analysis showing abundant mouse Cdyl expression in germ cells after day 25. Total RNAs from wild-type testis were collected at 5-d intervals, from 5 d before birth to 55 d after birth, and homozygous Wv adult testis, which lacks germ cells (28), were blotted and hybridized with probes derived from the entire ORFs of mouse Cdyl cDNA (Upper) or mouse Prm1 cDNA (Lower). (B) Western analysis revealing dramatic increase in the abundance of mouse CDYL protein after day 25. Total proteins from wild-type testis were collected at at 5-d intervals, from 5 d before birth to 55 d after birth, and homozygous Wv adult testis (which lacks germ cells) were blotted and probed with anti-CDYL serum. Recombinant mouse CDYL protein tagged with glutathione S-transferase (GST) was included as positive control. (C) Immunohistochemistry revealing mouse CDYL immunoreactivity in elongating spermatids. Adult mouse testis was sectioned and stained with anti-CDYL serum. Adjacent seminiferous tubules in the section may contain germ cells of different spermatogenic stages, and only those tubules that contain elongating spermatids are immunoreactive.
To examine the expression of mouse CDYL protein, a Western blot made from testes of multiple developmental stages was probed with serum against mouse CDYL. A dramatic increase in CDYL protein level in the testis was seen after day 25 (Fig. 3B). Thus the temporal expression profile of mouse CDYL protein closely parallels transcription of the gene (compare Fig. 3 A Upper and B). Note that in Fig. 3B, even though the abundance of mouse CDYL protein is dramatically increased after day 25, this protein is also present in the testes of earlier developmental stages, as well as in germ-cell-deficient homozygous Wv testis. This is consistent with our previous interpretation that the two mouse Cdyl transcripts (the widely expressed 3.6-kb transcript and the testis-specific 2.8-kb one) differ only in their 3′-untranslated regions, and they thus encode the same protein (11).
To directly visualize cells that express the CDYL protein, mouse testis sections were immunostained with anti-CDYL serum. Intense staining was observed in a subset of germ cells that have the appearance of elongating spermatids, although precise staging of these cells was difficult (Fig. 3C).
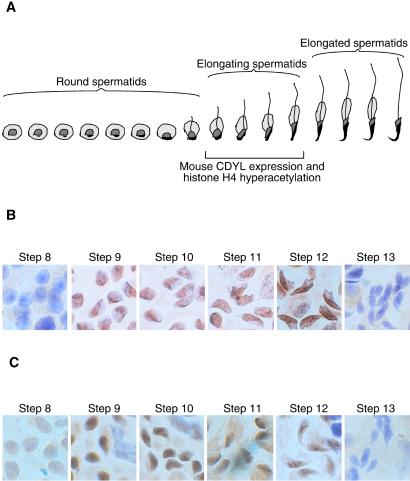
Developmental histologists have divided the three stages of mouse spermiogenesis (round spermatid, elongating spermatid, and elongated spermatid) further into 16 morphologically distinct steps (Fig. 4A) (21). To precisely map the expression of mouse CDYL protein within these 16 steps, seminiferous tubules from adult mice were separated into stage-specific segments by microdissection (17). This technique takes advantage of the fact that spermatogenesis initiates in sequential waves down the length of seminiferous tubules, such that adjacent segments of the tubule contain germ cells in consecutive stages of spermatogenesis. A contiguous series of microdissected tubule segments will thus represent germ cells in progressive stages of spermatogenesis. Squash preparations from contiguous segments were immunostained with anti-CDYL serum. This experiment revealed that mouse CDYL protein is abundantly expressed in the nuclei of step 9–12 spermatids (Fig. 4B), an observation consistent with the Northern and Western data.
Figure 4.
Staging CDYL protein expression and histone H4 hyperacetylation in mouse spermatids. (A) Schematic depiction of the 16 steps of mouse spermiogenesis and the time window where CDYL protein expression and histone H4 hyperacetylation were detected. (B) A series of mouse seminiferous tubule squash preparations containing spermatids of steps 8–13 was immunohistochemically stained with anti-CDYL serum. It reveals CDYL immunoreactivity in step 9–12 spermatids. (C) A parallel series of squash preparations were stained with serum against AcH4. It shows AcH4 immunoreactivity in step 9–12 spermatids.
Mouse CDYL Protein Expression During Spermatogenesis Coincides with Histone H4 Hyperacetylation.
Having staged mouse CDYL expression and demonstrated its nuclear localization, we sought to stage histone H4 hyperacetylation during mouse spermiogenesis. A previously developed antiserum that preferentially recognized AcH4 (24) was used to immunostain stage-specific seminiferous tubule squash preparations. The nuclei of step 9–12 spermatids exhibited the strongest AcH4-immunoreactivity (Fig. 4C). This result is in agreement with prior studies in the rat, where the same anti-AcH4 serum revealed the presence of H4 hyperacetylation in step 9–12 spermatids (rat spermatids of these steps are comparable in developmental maturity to mouse spermatids of corresponding steps) (5, 6). Thus, there exists a strong temporal correlation between the abundant expression of the CDYL protein in the nuclei of elongating spermatids and H4 hyperacetylation (Fig. 4).
Human CDY Is Expressed in Elongating Spermatids.
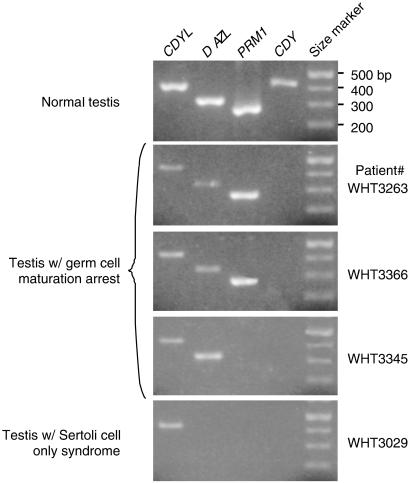
We also examined the expression profile of human CDY during spermatogenesis. An indirect approach was used to analyze the temporal expression of CDY mRNA in spermatogenic cells. Testis biopsy samples were obtained from four men with azoospermia (absence of sperm in semen), including three patients with germ cell maturation arrest and one patient with Sertoli cell only syndrome (complete absence of germ cells).
Reverse transcription–PCR analysis was performed on mRNAs extracted from these samples and on mRNA from a normal testis. First, we confirmed the histological evaluation of these patients by examining the presence or absence of two germ-cell-specific transcripts: DAZL, which is a marker for spermatogonia and spermatocytes (25), and PRM1, which is a marker for late-phase round spermatids and elongating spermatids (26) (Fig. 5). As expected, the Sertoli cell only syndrome sample (WHT3029), which lacked germ cells, was negative for both DAZL and PRM1 transcripts. Two of the maturation arrest samples (WHT3263 and WHT3366) contained both DAZL and PRM1 transcripts, indicating that spermatogenesis in these two patients arrested after the onset of PRM1 expression. The third maturation arrest sample (WHT3345) contained DAZL but not PRM1 transcript, showing that spermatogenesis in this patient arrested at a stage after DAZL expression but before PRM1 expression. Next, we examined CDY transcript in these samples, and found it to be absent from the Sertoli cell only syndrome sample as well as the three maturation arrest samples (Fig. 5). The absence of CDY transcript in these biopsy samples is not caused by deletions of CDY genomic loci, because these patients all appear to carry intact CDY genes. The most plausible explanation is that CDY is transcribed in a cell type that is absent from all four biopsy samples, i.e., late spermatids. This interpretation is consistent with a recent study showing that CDY1 is transcribed in late spermatogenic cells (27). Thus, the transcription profile of human CDY appears to closely parallel that of mouse Cdyl.
Figure 5.
Expression profile of human CDY mRNA in the testis. Reverse transcription–PCR detection of CDY mRNA in testis samples from a normal male, three males with germ cell maturation arrest, and one male with Sertoli cell only syndrome. The widely expressed CDYL (11) is used as a positive control for the reverse transcription–PCR assay. DAZL is used as a marker for spermatogonia and spermatocytes (25). PRM1 is used as a marker for late-phase round spermatids and elongating spermatids (26).
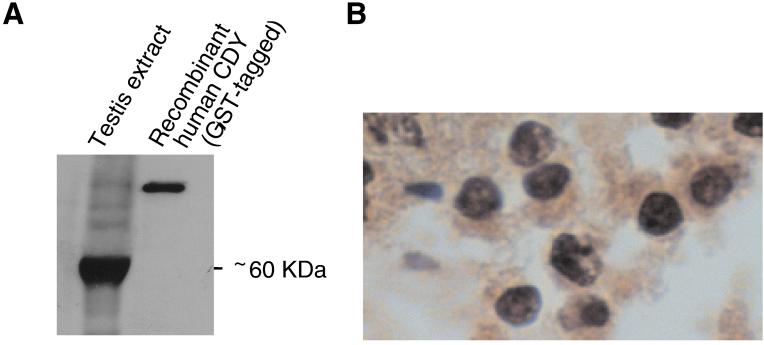
We then examined human CDY expression at the protein level. Western blot analysis with anti-CDY serum revealed an immunoreactive species in human testis extract of ≈60 kDa, the predicted size of CDY (Fig. 6A). When the same antiserum was used to probe human testis sections, spermatids exhibited strong immunoreactivity in their nuclei (Fig. 6B).
Figure 6.
Expression of human CDY protein in testis. (A) Western blot analysis with anti-CDY serum detecting abundant CDY protein in the testis. Recombinant human CDY protein tagged with glutathione S-transferase (GST) was included as positive control. (B) Immunohistochemical staining of human spermatids with anti-CDY serum (counterstained with hematoxylin).
Collectively, our biochemical, developmental, and histochemical data provide strong circumstantial evidence linking mouse CDYL and human CDY proteins with H4 hyperacetylation during spermiogenesis. The hypothesized function of CDY in chromatin remodeling provides a mechanistic rationale for how CDY deletion on the human Y chromosome might contribute to spermatogenic failure in AZFc-deleted men. In these men, CDY1 is deleted along with several other genes within the AZFc region (11–14). A major reduction in total CDY protein might interfere with the histone-to-protamine transition during spermiogenesis, thus contributing to overall spermatogenic failure. Finally, it is worth noting that as an enzyme specific to spermatids, CDY may be an ideal target for male contraceptive drugs.
Acknowledgments
We are grateful to the Cooperative Human Tissue Network, University of Pennsylvania Medical Center, and R. Oates for human testis tissues. We thank J. Borrow, A. Bortvin, L. G. Brown, S. L. Gilbert, M. L. Goodheart, S. R. Grimes, W. S. Kistler, K. C. Kleene, J. Lange, M. L. Meistrich, D. B. Menke, J. Potash, S. Roth, H. Skaletsky, and J. L. Workman for helpful discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health.
Abbreviations
- HAT
histone acetyltransferase
- AcH4
hyperacetylated histone H4
References
- 1.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis C D. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Oliva R, Mezquita C. Nucleic Acids Res. 1982;10:8049–8059. doi: 10.1093/nar/10.24.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen M E, Rattner J B, Dixon G H. Nucleic Acids Res. 1984;12:4575–4592. doi: 10.1093/nar/12.11.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimes S R, Jr, Henderson N. Exp Cell Res. 1984;152:91–97. doi: 10.1016/0014-4827(84)90232-5. [DOI] [PubMed] [Google Scholar]
- 6.Meistrich M L, Trostle-Weige P K, Lin R, Bhatnagar Y M, Allis C D. Mol Reprod Dev. 1992;31:170–181. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- 7.Wouters-Tyrou D, Martinage A, Chevaillier P, Sautiere P. Biochimie. 1998;80:117–128. doi: 10.1016/s0300-9084(98)80018-7. [DOI] [PubMed] [Google Scholar]
- 8.Balhorn R. J Cell Biol. 1982;93:298–305. doi: 10.1083/jcb.93.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy B P, Davies P L. J Biol Chem. 1980;255:2533–2539. [PubMed] [Google Scholar]
- 10.Lahn B T, Page D C. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 11.Lahn B T, Page D C. Nat Genet. 1999;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- 12.Kuroda-Kawaguchi T, Skaletsky H, Brown L G, Minx P J, Cordum H S, Waterston R H, Wilson R K, Silber S, Oates R, Rozen S, et al. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 13.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 14.Vogt P H, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Köhn F M, Schill W B, Farah S, Ramos C, et al. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 15.Candau R, Zhou J X, Allis C D, Berger S L. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brownell J E, Allis C D. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parvinen M, Hecht N B. Histochemistry. 1981;71:567–579. doi: 10.1007/BF00508382. [DOI] [PubMed] [Google Scholar]
- 18.Cavalli G, Paro R. Curr Opin Cell Biol. 1998;10:345–360. doi: 10.1016/s0955-0674(98)80011-2. [DOI] [PubMed] [Google Scholar]
- 19.Palosaari P M, Vihinen M, Mantsala P I, Alexson S E, Pihlajaniemi T, Hiltunen J K. J Biol Chem. 1991;266:10750–10753. [PubMed] [Google Scholar]
- 20.Nocka K, Tan J, Chin E, Chu T Y, Ray P, Traktman P, Besmer P. EMBO J. 1990;9:1805–1813. doi: 10.1002/j.1460-2075.1990.tb08305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell L D, Ettlin R A, Sinha Hikim A P, Clegg E D. Histological and Histopathological Evaluation of the Testis. Clearwater, FL: Cache River; 1990. [Google Scholar]
- 22.McCarrey J R. In: Cell and Molecular Biology of the Testis. Desjardins C, Ewing L L, editors. New York: Oxford Univ. Press; 1993. pp. 58–89. [Google Scholar]
- 23.Hecht N B, Bower P A, Waters S H, Yelick P C, Distel R J. Exp Cell Res. 1986;164:183–190. doi: 10.1016/0014-4827(86)90465-9. [DOI] [PubMed] [Google Scholar]
- 24.Lin R, Leone J W, Cook R G, Allis C D. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menke D B, Mutter G L, Page D C. Am J Hum Genet. 1997;60:237–241. [PMC free article] [PubMed] [Google Scholar]
- 26.Wykes S M, Nelson J E, Visscher D W, Djakiew D, Krawetz S A. DNA Cell Biol. 1995;14:155–161. doi: 10.1089/dna.1995.14.155. [DOI] [PubMed] [Google Scholar]
- 27.Kleiman S E, Lagziel A, Yogev L, Botchan A, Paz G, Yavetz H. Fertil Steril. 2001;75:166–173. doi: 10.1016/s0015-0282(00)01639-3. [DOI] [PubMed] [Google Scholar]
- 28.Mintz B, Russell E S. J Exp Zool. 1957;134:207–230. doi: 10.1002/jez.1401340202. [DOI] [PubMed] [Google Scholar]