Abstract
The replacement histone H2A.Z is variously reported as being linked to gene expression and preventing the spread of heterochromatin in yeast, or concentrated at heterochromatin in mammals. To resolve this apparent dichotomy, affinity-purified antibodies against the N-terminal region of H2A.Z, in both a triacetylated and non-acetylated state, are used in native chromatin immmuno-precipitation experiments with mononucleosomes from three chicken cell types. The hyperacetylated species concentrates at the 5′ end of active genes, both tissue specific and housekeeping but is absent from inactive genes, while the unacetylated form is absent from both active and inactive genes. A concentration of H2A.Z is also found at insulators under circumstances implying a link to barrier activity but not to enhancer blocking. Although acetylated H2A.Z is widespread throughout the interphase genome, at mitosis its acetylation is erased, the unmodified form remaining. Thus, although H2A.Z may operate as an epigenetic marker for active genes, its N-terminal acetylation does not.
INTRODUCTION
The inclusion of histone variants (‘replacement histones’) substituting for canonical histones may act as epigenetic information for the transmission of specific chromatin states, similarly to histone modifications or DNA methylation (1). The H2A.Z variant has a substantially altered sequence from canonical H2A and is highly conserved in higher eukaryotes, including Drosophila, though the Saccharomyces cerevisiae and Tetrahymena thermophila sequences are notably different, including the N-terminal domain. At a sub-set of genes in S.cerevisiae, H2A.Z (Htz1) fulfils a role related to the combination of SWI/SNF and SAGA, i.e. the remodelling of chromatin prior to transcription. This implies participation in gene activation, and mapping of H2A.Z at yeast genes showed a preferential location not on the genes themselves but in adjacent intergenic regions (2). Another activity of H2A.Z in S.cerevisiae is its link to preventing the spread of heterochromatin (3), and loss of H2A.Z leads to partial de-repression of HMR, the silent mating locus (4) and it is present at and close to yeast telomeres (5). In T.thermophila, an association with transcription was implied by the presence of H2A.Z in the transcriptionally active macronucleus but not in the inactive micronucleus (6). In mice, lack of H2A.Z is lethal at an early stage of embryogenesis (7) where it is enriched at pericentric heterochromatin (8), and in mammalian cell lines H2A.Z forms foci with HP1α at heterochromatic domains (9). These observations imply a structural role in higher eukaryotes but do not exclude a function in gene expression. Thus, while the proposed role for H2A.Z in yeast and protozoa is an involvement in transcription and barrier activity, in vertebrates participation in the formation of heterochromatin has been shown but there is no evidence for a role in transcription.
MATERIALS AND METHODS
Preparation and affinity purification of antibodies
The peptide AcAGGKAGKDSGKAKAKAC and its tri-acetyl counterpart acetylated at K4, K7 and K11 were coupled to KLH and used as immunogens in sheep. Total Ig was purified from serum using caprylic acid and then affinity purified antibodies isolated using a column of CPG beads carrying covalently attached immunogen peptides (10). SDS–PAGE and AUT–western analysis of the specificity and effectiveness of the affinity purified antibodies was carried out as described previously (11).
Preparation of nucleosomes and their immunoselection (nChIP)
Micrococcal nuclease digestion of nuclei, i.e. not crosslinked, followed by separation on exponential sucrose gradients were used to prepare ‘native’ nucleosomes from four chicken cell types: 15-day chicken embryo erythrocytes (15DCE), the HD37 erythroblast cell line, 10-day chicken brain (10DCB) tissue and the HD11 macrophage cell line. These were then immuno-precipitated as described previously (10). Antibody-Bound nucleosomes were retained on Protein G-agarose beads for sheep antibodies (H2A.Z and AcH2A.Z) and Protein A-Sepharose for rabbit antibodies (AcH4, AcH3 and AcH2B). The purified DNA from the Input, Unbound and Bound nucleosomes was UV quantified and using chicken genomic DNA as standard, analysed for sequence content by quantitative real-time PCR using Taqman probes on an ABI Prism 7900. The amplicon lengths varied between 68 and 113 bp and the results for each amplicon were expressed as the ratio B/I (which represents the absolute fold enrichment achieved by the ChIP) or, where a depletion occurred, as I/B (which represents the absolute fold depletion), all as described in ref. (11). Error bars represent standard deviations of triplicate measurements. Duplicate ChIPs with the anti-AcH2A.Z antibodies showed the same pattern of enrichments with a 15–20% variation in the absolute values. All the figures give data from single ChIPs.
RESULTS AND DISCUSSION
Antibody characterization
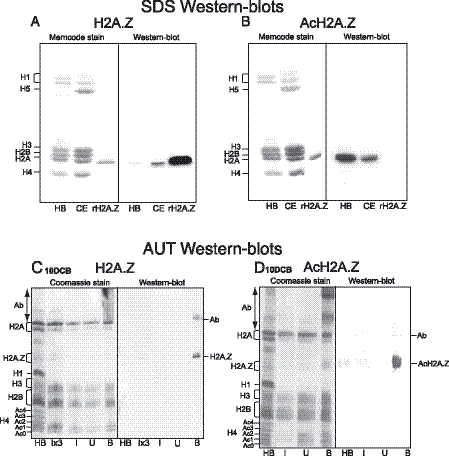
A synthetic 16-residue N-terminal peptide from chicken H2A.Z and also its tri-acetyl counterpart were used to raise and affinity purify antibodies. First, the SDS–western blots in Figure 1 show that no histone species other than the H2A fraction are recognized by either antibody. Figure 1A shows that anti-H2A.Z (i.e. antibodies to the unmodified protein) strongly recognizes recombinant H2A.Z, an H2A component in chicken erythrocyte (CE) histones and weakly a component in histones from butyrate-treated HeLa cells (HB). The low signal in HB is a result of most H2A.Z being driven up to acetylated species by histone deacetylase (HDAC) inhibition. Figure 1B shows that anti-AcH2A.Z fails to recognize the recombinant protein, i.e. non-acetylated H2A.Z, but strongly recognizes an H2A species in the HB histones, in consequence of butyrate treatment.
Figure 1.
(A and B) Characterization of anti-H2A.Z and anti-AcH2A.Z affinity purified antibodies. HB, butyrate-treated HeLa histones; CE, 15-day embryo chicken erythrocyte histones; rH2A.Z, recombinant H2A.Z. (C and D) Characterization of proteins from ChIPs using these affinity purified antibodies with nucleosomes from 10-day chicken brain tissue (10DCB). I, U, B: Input, Unbound, Bound histones; Ab, carry-over products of primary antibodies.
To demonstrate the effectiveness of the antibodies in ChIP experiments and provide further characterization, nucleosomes from several tissues were used in ChIP experiments with both antibodies. Figure 1C and D show acetic acid/urea/Triton (AUT) gels and corresponding western blots comparing proteins in the antibody-Bound chromatins with those of the Input and Unbound fractions from 10DCB tissue. For both antibodies the westerns exhibit only very weak H2A.Z signals in the Input lanes, emphasising the low levels of H2A.Z in this tissue. By comparing Bound with Input lanes, the westerns make clear that enrichment has occurred of a single H2A.Z species in the case of anti-H2A.Z, while for anti-AcH2A.Z several acetylated species are enriched in the Bound. The fastest-migrating H2A.Z species in the Bound lane of Figure 1D cannot be the unacetylated form since Figure 1B demonstrates that the anti-AcH2A.Z antibodies do not recognize unmodified H2A.Z (rH2A.Z); it follows that H2A.Z carries several acetyl groups. The Supplementary Data contain AUT gels/westerns of proteins from ChIPs performed using nucleosomes from 15DCE and from chicken HD11 cells, a macrophage line. These also make it clear that only a single H2A.Z species is enriched in the Bound proteins when the anti-H2A.Z antibodies are used and three species of AcH2A.Z when the anti-AcH2A.Z antibodies are used for the ChIPs.
The AUT PAGE–westerns also demonstrate that neither antibody recognizes the canonical H2A species: this follows from lack of H2A staining in the I and U lanes. Furthermore, absence of H2A signal in the HB lane also shows that acetylated forms of H2A are not recognized, nor are acetylated species of the other core histones. The AUT PAGE–westerns thus show that both antibodies are effective in ChIP experiments.
H2A.Z mapping
To decide whether H2A.Z is a feature of active genes in the chicken, nChIP experiments were performed using nucleosomes from HD37 cells, a chicken erythroblast line, 15DCE and 10DCB with both affinity-purified antibodies; assessment of sequence content was by real-time PCR, and high-resolution distribution maps were thereby generated. Table 1 gives the transcriptional status of the genes studied in the three cell types.
Table 1.
Transcriptional status of the genes studied in the three cell types
| Cell type | β-globin locus | Folate receptor | Lysozyme | Gas41 | Carbonic anhydrase | GAPDH | Vimentin | Insulin |
|---|---|---|---|---|---|---|---|---|
| 15DCE | Active | Inactive | Inactive | Active | Active | Active | Active | Inactive |
| HD37 | Inactive | Active | Inactive | Active | Active | Active | Active | Inactive |
| 10DCB | Inactive | Inactive | Inactive | Active | Inactive | Active | Active | Inactive |
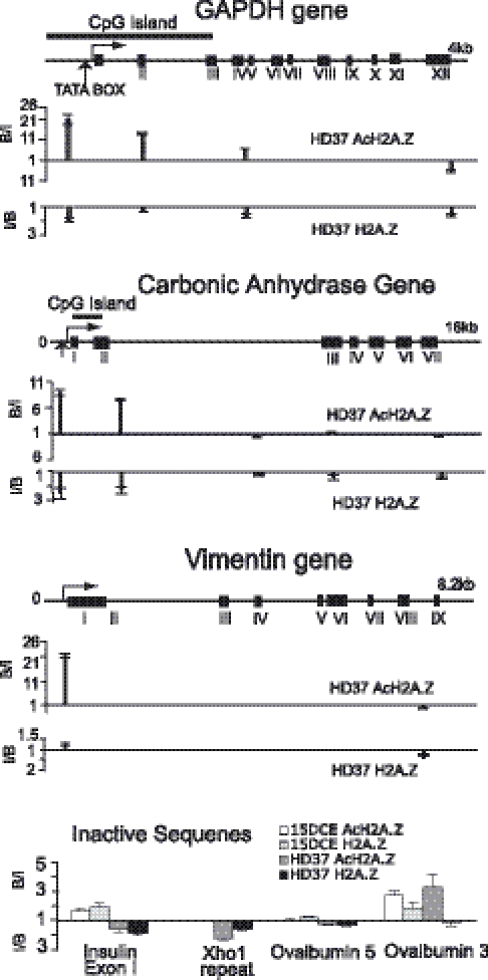
Figure 2 shows the distribution of H2A.Z at the tissue-specific carbonic anhydrase (CA) gene and at the housekeeping GAPDH and vimentin genes in HD37 cells. A consistent overall distribution is seen in which AcH2A.Z is abundant at the promoter proximal and 5′ ends of the genes, falling off substantially in a 3′ direction to very low levels. In contrast, for unacetylated H2A.Z, only depletions are observed with the sole exception of a very small enrichment (1.3-fold) at the vimentin promoter. In the case of CA, AcH2A.Z is maximal ∼650 bp upstream of the transcriptional start site and still substantial within IntronII, ∼850 bp beyond the end of the CpG island. At the shorter GAPDH gene, a significant fall off is seen ∼2 kb into the gene at ExonIV, just beyond the end of the CpG island. Thus, although the location of AcH2A.Z approximately coincides with that of the CpG islands, there is no absolute link. Similar results were obtained for these genes in ChIPs using 15DCE input nucleosomes (data not shown). AcH2A.Z thus appears to be a feature of active genes with a concentration at the 5′ end, rather like that found at these genes for acetylated H3 and H4 (10–12) and di- or tri-methyl K4/H3 (12,13). There is no evidence for the presence of unacetylated H2A.Z at these active housekeeping genes.
Figure 2.
Distributions of H2A.Z and AcH2A.Z at two housekeeping genes, GAPDH and vimentin and the tissue-specific gene carbonic anhydrase in HD37 erythroblasts. The bottom panel shows four amplicons at inactive sequences in HD37 cells and 15DCE. B/I values (the ratio of the Bound to the Input signal for an amplicon) above the unity lines represent ‘fold enrichment’ achieved by the antibodies. I/B values are plotted below the unity lines when the Bound signal is less than that of the Input: this ratio represents ‘fold depletion’.
A recent publication (14) described the distribution of H2A.Z at the human GAPDH and cMyc genes using xChIP with an antibody recognizing an epitope in the C-terminal region of H2A.Z, i.e. blind to its state of acetylation. For both these genes, substantial levels of H2A.Z were found immediately upstream of the start site, extending into the transcribed sequences but falling off before the transcription termination points. Since this distribution is similar to the present distributions of AcH2A.Z at housekeeping genes, it is reasonable to suppose that Farris et al. (14) monitored the acetylated forms of H2A.Z.
The presence of H2A.Z was also monitored at inactive genes in both HD37 cells and 15DCE (Figure 2): at the insulin gene and at both ends of the ovalbumin transcribed region, very small depletions/enrichments were found with both antibodies. We conclude that inactive genes carry neither unacetylated nor acetylated H2A.Z. Thus, although AcH2A.Z is a characteristic of active genes, it is not, for example, always present at genes in an unmodified form for subsequent acetylation on gene activation. Since H2A.Z is enriched in the pericentric heterochromatin of mouse embryos (8), the XhoI repeat region of the chicken W-chromosome (15) was tested. Using HD37 nucleosomes, small depletions were observed (Figure 2), i.e. the XhoI repeat does not contain H2A.Z or AcH2A.Z in these cells.
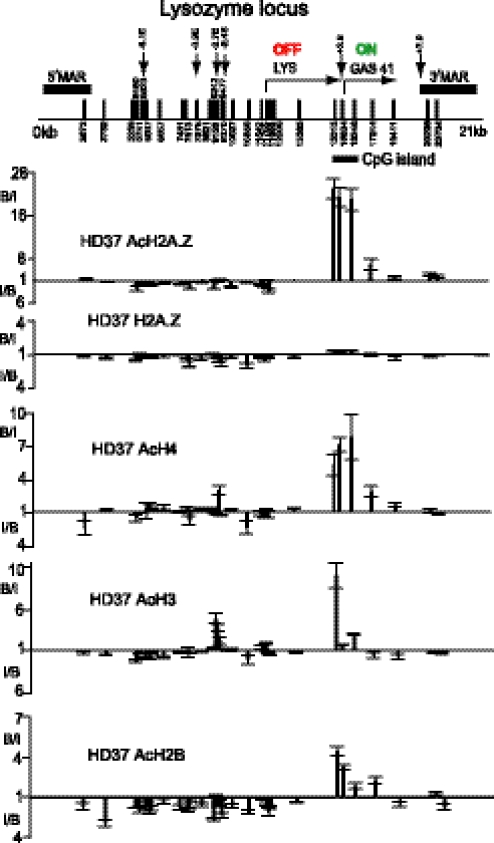
Figure 3 shows the distribution of H2A.Z at the lysozyme locus in HD37 cells. This short locus includes the Gas41 housekeeping gene, located only 200 bp downstream of the tissue-specific cLYS gene which is active in macrophages and oviduct but not in HD37 cells (16). The DNase I hypersensitive sites (DHSs) at −6.1, −3.9 and −2.7 kb linked to cLYS activity are absent in these cells, while the silencer DHS at −2.4 kb is present (17). The distribution of AcH2A.Z at the active Gas41 gene parallels that at other housekeeping genes: substantial enrichments at the promoter proximal region (15 315), at the CpG island close to the +3.9 constitutive DHS between the cLYS and Gas41 genes (15 634) and in ExonIII (16 246), but by ExonVI (17 244, ∼2 kb beyond the promoter) the level has dropped. In contrast, at the promoter of the inactive cLYS gene (11 755), on the gene itself and at the several upstream cis elements, depletions were found with both antibodies. The lack of H2A.Z at the cLYS gene and its upstream elements accords with the observations at the insulin and ovalbumin genes that inactive genes do not carry H2A.Z of either form.
Figure 3.
Distributions of H2A.Z, AcH2A.Z, AcH4, AcH3 and AcH2B at the lysozyme locus in HD37 erythroblasts. Vertical arrows = DNase I hypersensitive sites and vertical bars with numbers indicate amplicon positions. The two genes, their transcriptional status, the 5′ and 3′ matrix attachment regions (MARs) and the CpG island are indicated. B/I values (the ratio of the Bound to the Input signal for an amplicon) above the unity lines represent ‘fold enrichment’ achieved by the antibodies. I/B values are plotted below the unity lines when the Bound signal is less than that of the Input: this ratio represents ‘fold depletion’.
The distributions of the hyperacetylated forms of the other three core histones were determined at the lysozyme locus (lower panels of Figure 3) using antibodies generated against hyperacetylated H4, H3 and H2B which recognize the most highly acetylated forms of these core histones and were previously used to map these modifications in 15DCE (11,18). It is seen that at the lysozyme locus in HD37 cells they all have a similar distribution to that of AcH2A.Z: substantial amounts at the 5′ end of the active Gas41 gene, particularly for AcH4, and mostly depletions at the cLYS gene, at its promoter and at its upstream regulatory elements. An exception is the small concentration of hyperacetylated H3 in the region of the −2.7 kb enhancer and a small peak of H4 acetylation at the −2.4 kb silencer. There is no evidence of any acetylation of the core histones at the flanking matrix attachments sites. The mapping of acetylation of all four core histones in Figure 3 demonstrates a clear division of the chicken lysozyme locus in HD37 cells into the active Gas41 region and the inactive upstream lysozyme region.
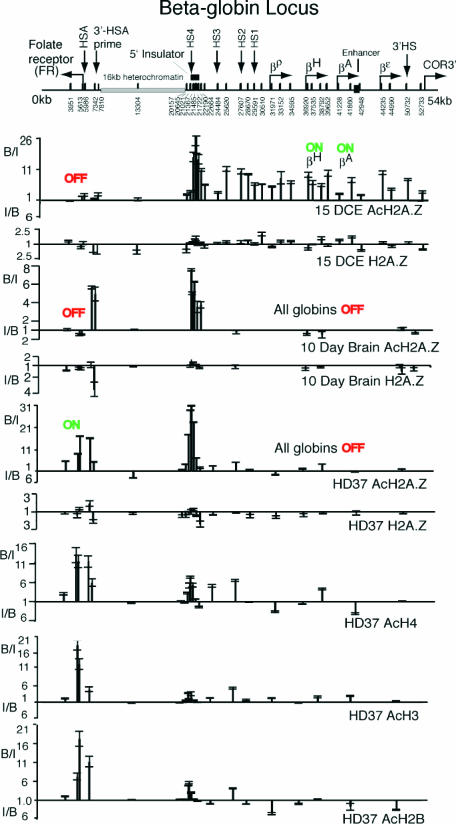
Figure 4 shows distributions of H2A.Z at the β-globin locus in 15DCE, cells in which the adult (βA) and hatching (βH) genes are active and the folate receptor (FR) gene is inactive. In 15DCE, there is a substantial peak of AcH2A.Z at the 5′ insulator (HS4) and significant levels throughout the β-globin locus, with no obvious preference for the active genes as compared with inactive or intergenic regions. In contrast, within the adjacent 16 kb of heterochromatin and at the FR gene, essentially no enrichments are seen. This distribution of AcH2A.Z is broadly similar to that of acetylated forms of the other three core histones at the β-globin locus in 15DCE (10,11,18,19). A significant number of nucleosomes in this active locus therefore contain hyperacetylated forms of all four core histones. Levels of unacetylated H2A.Z are low throughout the β-globin locus, the 16 kb heterochromatin and at the FR gene in 15DCE cells.
Figure 4.
Distributions of H2A.Z, AcH2A.Z, AcH4, AcH3 and AcH2B at the β-globin locus/folate receptor gene in 15-day chicken embryo erythrocytes (15DCE), 10-day chicken brain tissue (10DCB) and HD37 chicken erythroblasts. Nomenclature as in Figure 3. B/I values (the ratio of the Bound to the Input signal for an amplicon) above the unity lines represent ‘fold enrichment’ achieved by the antibodies. I/B values are plotted below the unity lines when the Bound signal is less than that of the Input: this ratio represents ‘fold depletion’.
In HD37 cells, where there is no globin expression but the FR gene is active, there are again substantial amounts of AcH2A.Z at the 5′ insulator (HS4) but very little across the remainder of the β-globin locus. The FR gene itself carries high levels of AcH2A.Z, tailing off towards its 3′ end, becoming low by Intron4, a distribution similar to that at the Gas41, CA, GAPDH and vimentin genes in HD37 cells. Unacetylated H2A.Z is absent from the β-globin locus, the 5′ insulator region and the FR gene. In 10DCB, where both the β-globin and the FR genes are inactive, again there is a concentration of AcH2A.Z at the 5′ insulator but none within the β-globin locus, nor at the constitutive 3′HS at its 3′ end (50 732). Strikingly, however, AcH2A.Z is present upstream of the FR gene, at 7342 and 7810 the 3′-HSA prime element. In contrast, there is no AcH2A.Z neither at the FR promoter (5986/5613), nor on the gene itself (3951), the same situation as found at other silent genes. In HD37 cells, the hyperacetylated forms of the other three core histones, see lower panels of Figure 4, are substantially enriched at the active FR gene, similarly to AcH2A.Z, but at the 5′ insulator there are no strong peaks of H4/H3/H2B acetylation comparable with that observed for AcH2A.Z.
So what is the role of AcH2A.Z at the 5′ insulator and 3′-HSA prime (locations tightly flanking the 16 kb heterochromatin) in HD37 and 10DCB cells? In HD37 cells the AcH2A.Z at 3′-HSA prime might be related to FR activity but that is not the case in 10DCB cells where both the globin and FR genes are silent. The 5′ insulator is known to harbour at least two activities: enhancer blocking and barrier activity (20). Since there is no AcH2A.Z at 3′HS, the constitutive DHS at the 3′ end of the locus that binds CTCF to generate enhancer blocking activity (21), AcH2A.Z cannot be a feature of this activity. AcH2A.Z could, however, be associated with a barrier activity of the 5′ insulator and 3′-HSA prime, i.e. to stop spreading of the 16 kb heterochromatin in both directions. If this is a function of AcH2A.Z, the absence of AcH2A.Z from 3′HS is expected since there is an immediately adjacent olfactory receptor gene [COR3′ (22)] and no heterochromatin blocking is required. It is worth noting that at 3′-HSA prime in 10DCB, there is neither H3 acetylation, nor H3/K4 methylation (activating modifications) nor H3/K9 methylation (a repressive modification) (23), so the presence of AcH2A.Z is exceptional for this location. If the presence of AcH2A.Z marks a barrier activity, then its absence from 3′-HSA prime in 15DCE is unexpected; however, it has been postulated (24) on the basis of increased DNA methylation in the region of 3′-HSA prime that the heterochromatin spreads across the FR gene in erythrocytes, i.e. no barrier activity is expressed in these cells. A search for co-factors of human CTCF (25), a protein linked to enhancer blocking activity, identified H2A.Z (among others); although this might correspond to the presence of AcH2A.Z at the 5′ insulator, the CTCF site at 3′HS (50 732) is clearly not associated with AcH2A.Z, nor are the potential CTCF sites just upstream of the FR gene, (located at 5986, at least 1 kb from the points at which AcH2A.Z is found in 10DCB). An obligatory association of H2A.Z with CTCF does not therefore appear to be the case. A concentration of AcH2A.Z at sites immediately flanking the 16 kb heterochromatin in HD37 and 10DCB might be a correlate of the finding in yeast that H2A.Z is involved in preventing the spread of telomeric heterochromatin (3) and in mammalian cells is enriched at pericentric heterochromatin (8).
Figure 4 also shows distributions at the β-globin locus of the hyperacetylated forms of the other three core histones in HD37's, a cell more developed than HD24's but at an earlier developmental stage than 6C2 cells (19). Correspondingly, the FR gene shows high levels of core histone acetylation at its promoter, HSA and at 3′-HSA prime, falling to a low level by IntronIV (3951), distributions similar to that of AcH2A.Z. No enrichments are seen within the 16 kb heterochromatin. Within the inactive β-globin locus itself, only for AcH4 are modest enrichments seen: at HS4, HS3, HS2 and at the βA-promoter. By comparing with the FR gene it is clear that at the 5′ insulator the acetylation of H2A.Z dominates that of the other three core histones.
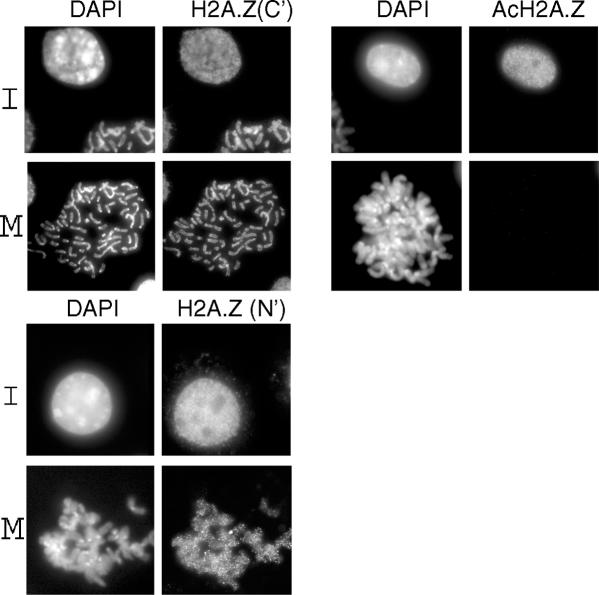
Since the mapping experiments suggest that active genes carry AcH2A.Z at their 5′ ends, immuno-staining at interphase should display intensity throughout nuclei. Figure 5 shows this to be the case in mouse L929 cells when three different antibodies are used: the two used here for the ChIP experiments, H2A.Z(N′) and AcH2A.Z, both raised to the N-terminal tail, and an antibody raised to a C-terminal epitope, H2A.Z(C′), that is independent of the state of acetylation (8). At metaphase (M), condensed chromosomes are visualized with both the H2A.Z(N′) and H2A.Z(C′) antibodies, with significant depletions at centromeric heterochromatin, demonstrating that substantial amounts of H2A.Z remain at metaphase. In contrast, when the AcH2A.Z antibodies are used for staining at M, the condensed chromosomes are invisible, i.e. the acetylation is completely erased.
Figure 5.
Mouse L929 cell nuclei stained with DAPI and three different antibodies: H2A.Z(N′) and AcH2A.Z raised to the N-terminal tail of H2A.Z (the present antibodies) and an antibody previously raised (8) to a C-terminal epitope, H2A.Z(C′).
CONCLUSIONS
The data show that in chickens, H2A.Z is present at active genes, moreover in a hyperacetylated form but absent from inactive genes. The link between gene activity and the presence of H2A.Z found in S.cerevisiae (2) is now established for acetylated H2A.Z in a vertebrate. Similar data recently obtained at two housekeeping genes in human HL60 cells (14) using antibodies insensitive to the state of H2A.Z acetylation suggest that these conclusions may also apply to mammalian cells.
The presence of AcH2A.Z at sites flanking the 16 kb of heterochromatin at the β-globin locus suggests a link to barrier activity. The acetylated form of H2A.Z could occur in nucleosomes either because a HAT acetylates pre-exchanged H2A.Z or exchange occurs using already acetylated H2A.Z. The human equivalent of the S.cerevisiae NuA4 complex (26,27) contains the expected yeast homologues but also subunits homologous to those in the S.cerevisiae SWR1 complex known to incorporate H2A.Z into nucleosomes (5,28,29). The larger human complex thus appears to combine the H4/H2A HAT activities of NuA4 with the remodelling and H2A.Z exchange capabilities of SWR1 (30), so that H2A.Z could be deposited into nucleosomes in an acetylated form (31); this would be consistent with the widespread lack of unacetylated H2A.Z.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The Portsmouth and Leeds laboratories acknowledge the BBSRC of Great Britain for research funding and also for a Studentship for K.B. The authors acknowledge the technical help of Flavie Faist. I.G. is supported by an Australian NHMRC grant. P.L. is supported by the Wellcome Trust. Funding to pay the Open Access publication charges for this article was provided by the University of Portsmouth.
Conflict of interest statement. None declared.
REFERENCES
- 1.Redon C., Pilch D., Rogakou E., Sedelnikova O., Newrock K., Bonner W. Histone H2A variants H2AX and H2A.Z. Curr. Opin. Genet. Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 2.Santisteban M.S., Kalashnikova T., Smith M.M. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 3.Meneghini M.D., Wu M., Madhani H.D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 4.Dhillon N., Kamakaka R.T. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell. 2000;6:769–780. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 5.Krogan N.J., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 6.Allis C.D., Glover C.V., Bowen J.K., Gorovsky M.A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eukaryote, Tetrahymena thermophila. Cell. 1980;20:609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- 7.Faast R., Thonglairoam V., Schulz T.C., Beall J., Wells J.R., Taylor H., Matthaei K., Rathjen P.D., Tremethick D.J., Lyons I. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 8.Rangasamy D., Berven L., Ridgway P., Tremethick D.J. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003;22:1599–1607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rangasamy D., Greaves I., Tremethick D.J. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nature Struct. Mol. Biol. 2004;11:650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- 10.Hebbes T.R., Clayton A.L., Thorne A.W., Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers F.A., Chong W., Evans D.R., Thorne A.W., Crane-Robinson C. Acetylation of histone H2B mirrors that of H4 and H3 at the chicken beta-globin locus but not at housekeeping genes. J. Biol. Chem. 2003;278:36315–36322. doi: 10.1074/jbc.M305822200. [DOI] [PubMed] [Google Scholar]
- 12.Kouskouti A., Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell. Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 14.Farris S.D., Rubio E.D., Moon J.J., Gombert W.M., Nelson B.H., Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J. Biol. Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- 15.Kodama H., Saitoh H., Tone M., Kuhara S., Sakaki Y., Mizuno S. Nucleotide sequences and unusual electrophoretic behavior of the W chromosome-specific repeating DNA units of the domestic fowl, Gallus gallus domesticus. Chromosoma. 1987;96:18–25. doi: 10.1007/BF00285878. [DOI] [PubMed] [Google Scholar]
- 16.Chong S., Riggs A.D., Bonifer C. The chicken lysozyme chromatin domain contains a second, widely expressed gene. Nucleic Acids Res. 2002;30:463–467. doi: 10.1093/nar/30.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonifer C., Jagle U., Huber M.C. The chicken lysozyme locus as a paradigm for the complex developmental regulation of eukaryotic gene loci. J. Biol. Chem. 1997;272:26075–26078. doi: 10.1074/jbc.272.42.26075. [DOI] [PubMed] [Google Scholar]
- 18.Myers F.A., Evans D.R., Clayton A.L., Thorne A.W., Crane-Robinson C. Targeted and extended acetylation of histones H4 and H3 at active and inactive genes in chicken embryo erythrocytes. J. Biol. Chem. 2001;276:20197–20205. doi: 10.1074/jbc.M009472200. [DOI] [PubMed] [Google Scholar]
- 19.Litt M.D., Simpson M., Recillas-Targa F., Prioleau M.N., Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung J.H., Whiteley M., Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 21.Saitoh N., Bell A.C., Recillas-Targa F., West A.G., Simpson M., Pikaart M., Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulger M., van Doorninck J.H., Saitoh N., Telling A., Farrell C., Bender M.A., Felsenfeld G., Axel R., Groudine M. Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes. Proc. Natl Acad. Sci. USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litt M.D., Simpson M., Gaszner M., Allis C.D., Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 24.Prioleau M.N., Nony P., Simpson M., Felsenfeld G. An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yusufzai T.M., Tagami H., Nakatani Y., Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 26.Cai Y., Jin J., Tomomori-Sato C., Sato S., Sorokina I., Parmely T.J., Conaway R.C., Conaway J.W. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 27.Doyon Y., Selleck W., Lane W.S., Tan S., Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. ATP-driven exchange of histone H2A.Z variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H., Richardson D.O., Roberts D.N., Utley R., Erdjument-Bromage H., Tempst P., Cote J., Cairns B.R. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell. Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doyon Y., Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:0587–0599. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.