Abstract
A theory for the evolution of cellular organization is presented. The model is based on the (data supported) conjecture that the dynamic of horizontal gene transfer (HGT) is primarily determined by the organization of the recipient cell. Aboriginal cell designs are taken to be simple and loosely organized enough that all cellular componentry can be altered and/or displaced through HGT, making HGT the principal driving force in early cellular evolution. Primitive cells did not carry a stable organismal genealogical trace. Primitive cellular evolution is basically communal. The high level of novelty required to evolve cell designs is a product of communal invention, of the universal HGT field, not intralineage variation. It is the community as a whole, the ecosystem, which evolves. The individual cell designs that evolved in this way are nevertheless fundamentally distinct, because the initial conditions in each case are somewhat different. As a cell design becomes more complex and interconnected a critical point is reached where a more integrated cellular organization emerges, and vertically generated novelty can and does assume greater importance. This critical point is called the “Darwinian Threshold” for the reasons given.
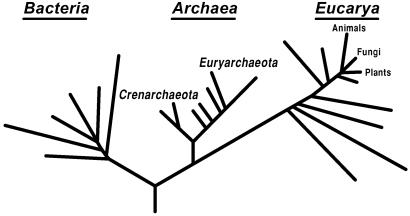
The evolution of modern cells is arguably the most challenging and important problem the field of Biology has ever faced (1, 2). In Darwin's day the problem could hardly be imagined. For much of the 20th century it was intractable. In any case, the problem lay buried in the catch-all rubric “origin of life”—where, because it is a biological not a (bio)chemical problem, it was effectively ignored. Scientific interest in cellular evolution started to pick up once the universal phylogenetic tree, the framework within which the problem had to be addressed, was determined (refs. 3 and 4; Fig. 1). But it was not until microbial genomics arrived on the scene that biologists could actually do much about the problem of cellular evolution.
Figure 1.
The universal phylogenetic as determined by ribosomal RNA sequences analyses. [Reproduced with permission from ref. 15 (Copyright 2000, Proc. Natl. Acad. Sci. USA).]
Initial attempts to frame the issue have typically been in the classical Darwinian mode, and the focus to date has been almost exclusively on modeling the evolution of the eukaryotic cell. The reason, of course, is clear—the appeal of the endosymbiosis concept. Because endosymbiosis has given rise to the chloroplast and mitochondrion, what else could it have done in the more remote past? Biologists have long toyed with an endosymbiotic (or cellular fusion) origin for the eukaryotic nucleus, and even for the entire eukaryotic cell (4–10). These classical explanations have three characteristics: they (i) invoke cells that are basically fully evolved; (ii) evolve the essential eukaryotic cell well after its archaeal and bacterial counterparts (as has always been connoted by the term “prokaryote”); and (iii) focus attention on eukaryotic cellular evolution, which implies that the evolutions of the “prokaryotic” cell types, the archaeal and bacterial, are of a different character—simpler, and, it would seem, less interesting. We cannot expect to explain cellular evolution if we stay locked into the classical Darwinian mode of thinking.
The universal phylogenetic tree in one sense brought classical evolution to culmination. Darwin had said: “The time will come … when we shall have very fairly true genealogical trees of each great kingdom of nature” (11). A century later the universal phylogenetic tree based on molecular (rRNA) sequence comparisons did precisely that and went the further, final step to unify all of the “great kingdoms” into one single “empire” (3). The central question posed by the universal tree is the nature of the entity (or state) represented by its root, the fount of all extant life. Herein lies the door to the murky realm of cellular evolution.
Experience teaches that the complex tends to arise from the simple, and biologists have assumed it so in the case of modern cells. But this assumption is usually accompanied by another not so self-evident one: namely that the “organism” represented by the root of the universal tree was equivalent metabolically and in terms of its information processing to a modern cell, in effect was a modern cell. Such an assumption pushes the real evolution of modern cells back into an earlier era, which makes the problem not directly addressable through genomics. That is not a scientifically acceptable assumption. Unless or until facts dictate otherwise, the possibility must be entertained that some part of cellular evolution could have occurred during the period encompassed by the universal phylogenetic tree.
There is evidence, good evidence, to suggest that the basic organization of the cell had not yet completed its evolution at the stage represented by the root of the universal tree. The best of this evidence comes from the three main cellular information-processing systems. Translation was highly developed by that stage: rRNAs, tRNAs, and the (large) elongation factors were by then all basically in near modern form; hence, their universal distributions. Almost all of the tRNA charging systems were in modern form as well (12). But, whereas the majority of ribosomal proteins are universal in distribution, a minority of them is not. A relatively small cadre is specific to the bacteria, a somewhat larger set common and confined to the archaea and eukaryotes, and a few others are uniquely eukaryotic.
Almost all of the universal translational proteins (as well as those in transcription) show what is called the canonical pattern, i.e., the bacterial and archaeal versions of the protein are remarkably different from one another, so much so that their difference is distinguished as one of “genre” (12). Except for the aminoacyl-tRNA synthetases the corresponding eukaryotic versions are virtually all of the archaeal genre (12). Why canonical pattern exists is a major unanswered question (3). In the overall it would seem that translation, although highly developed at the root of the universal tree, subsequently underwent idiosyncratic modifications in each of the three major cell types.
Transcription seems to have been rather less developed at the root of the universal tree. The two largest (the catalytic) subunits of the DNA-dependent RNA polymerase, β and β′ in bacterial nomenclature, are universal in distribution. But the remaining bacterial subunit (α) is only partially so. Bacterial α exists in two copies in the bacterial polymerase. Its archaeal/eukaryotic counterpart comprises two distinct proteins, each present in single copy in the enzyme and (portions of) each showing homology to (somewhat different) portions of bacterial α and vice versa (13). A structural difference of this magnitude must represent at least some functional distinction. The archaeal transcription apparatus also contains additional (smaller) subunits, none of which are found in bacteria but all of which occur in eukaryotes (13). [As in the case of translation, the (three) eukaryotic mechanism(s) contain additional eukaryote-specific small subunits.] Bacterial transcription initiation does not resemble its archaeal/eukaryotic counterpart (14). Although a universal transcription function seems to have existed by the end of the universal ancestor period, that mechanism seems rudimentary.
A modern type of genome replication mechanism did not exist at the root of the universal tree (14). Virtually no homology (orthology) exists between the bacterial genome replication mechanism and that basically common to the archaea and the eukaryotes (although a number of bacterial and archaeal DNA polymerases, some of which serve repair functions, do show sequence homology). Modern genome replication mechanisms seem to have evolved twice (14).
These fundamental differences in the genetic machinery constitute a prima facie case to the effect that the era of cellular evolution continued well into the evolutionary period encompassed by the universal phylogenetic tree. It would also seem that the order of maturation of the information processing systems was first translation, then transcription, and finally modern genome structure and replication (14).
Horizontal Gene Transfer (HGT)
HGT is one of two keys to understanding cellular evolution. The phenomenon has long been known, but the HGT we thought we knew is not the HGT that genomics reveals. Only a decade ago HGT was generally considered a relatively benign force, which had sporadic and restricted evolutionary impact. However, the HGT that genomics reveals is not of this nature. It would seem to have the capacity to affect the entire genome, and given enough time could, therefore, completely erase an organismal genealogical trace. This is an evolutionary force to be reckoned with, comparable in power and consequence to classical vertical evolutionary mechanisms (15).
How Much Has HGT Eroded the Organismal Trace?
In the late 1990s it became possible to infer universal phylogenetic trees from molecules other than rRNA. Disturbingly, the majority of these yielded universal trees that differ significantly in topology from the rRNA tree (16). The feeling arose that the universal rRNA-based tree was somehow basically flawed (17), or wrongly rooted (18), or that HGT had erased the deep ancestral trace (19), making determination of the universal phylogenetic tree impossible. These hasty conclusions are wrong, as closer examination of the data shows.
The many protein trees that differ in topology from the rRNA tree also differ in topology from one another, the hallmark of HGT. Moreover, some protein-based trees do exhibit topologies in agreement with that of the universal rRNA tree (20). Nearly all of the universal components of translation and transcription do so, as do a small number of other proteins, e.g., HSP-60. The most convincing evidence for a core of genes with a common history that defines an organismal genealogy tracing back to the root of the universal tree comes from the aminoacyl-tRNA synthetases.
The 20-odd aminoacyl-tRNA synthetases have obviously been intensely involved in HGTs (12). Roughly, a third of them yield trees that do not exhibit the above-described canonical pattern (12). The other two-thirds also break with canonical pattern to one extent or another, but that pattern, albeit eroded, is still evident (12). Tellingly, this canonical pattern is seen for synthetases that are specifically related to one another, the valine and isoleucine synthetases, for example. Both of these enzymes demonstrate (highly eroded) canonical pattern but differ completely in how they have violated that pattern (12). The only reasonable explanation here is that the canonical pattern predates the evident HGT that the aminoacyl-tRNA synthetases have undergone, and HGT has failed to erase it completely. Because all have been subject to widespread HGT, and because they are all functionally of a kind, the aminoacyl-tRNA synthetases as a group provide an especially convincing argument that there exists a genetic trace of our descent from some kind of common ancestral condition.
Yet a new realization comes with this finding: although organisms do have a genealogy-defining core of genes whose common history dates back to the root of the universal tree, that core is very small. Our classically motivated notion had been that the genealogy of an organism is reflected in the common history of the majority of its genes. What does it mean, then, to speak of an organismal genealogy when nearly all of the genes in the cell—genes that give it its general character—do not share a common history? This question again goes beyond the classical Darwinian context.
What Shapes HGT?
The most notable characteristic of HGT is the extreme variation in its frequency and phylogenetic range from one gene to another, from one major taxon to another (12). Among those genes important to cellular function and structure, the best example may be the differences in frequency and range of HGT shown by the various componentry of the translation apparatus. On the one hand, the tRNA charging enzymes (as we have seen) are notable for relatively frequent and phylogenetically broad-ranging gene transfers (12). On the other, the ribosomal proteins are notable for the virtual lack thereof. Why do different components of the same mechanism show such different HGT profiles?
A number of factors affects the character of HGT, e.g., proximity of potential donor and recipient organisms (as would occur in microbial consortia, for example) or cellular defenses against alien genetic material. All of these except one are general in nature; they cannot make the fine distinctions among individual genes required to explain the sharp difference between the HGT characteristics of the aminoacyl-tRNA synthetases and other components of the translation mechanism. That one factor is compatibility of an alien gene with the overall organization of the recipient cell (15).
Cellular componentry can be roughly classified according to the degree to which it is connected to the rest of the cell. Loosely connected, or modular, elements define one extreme of the spectrum. Such components tend to be largely self-defining in their structure/function, interacting minimally with other elements in the cell, and are, therefore, obvious candidates for horizontal gene displacement by alien homologs. At the other extreme are the tightly coupled elements, which have extensive, specific, and constraining physical and chemical ties to others of the cellular componentry and, therefore, could seldom, if ever, be sufficiently mimicked by an alien homolog to be displaced by it. The remarkable difference between the HGT profiles of the aminoacyl-tRNA synthetases and others of the translation componentry is thus explained by the loosely coupled, modular nature of the former and the tightly coupled nature of the latter (15, 21).
The Darwinian Threshold.
The degree of connectedness of the componentry of the cell has profound evolutionary implications. If a cell was simple and highly modular in organization, HGT would play a stronger role in its evolution than otherwise (15, 21). Indeed, were that organization simple and modular enough, all of the componentry of a cell could potentially be horizontally displaceable over time. The organismal genealogical record would be ephemeral; no stable record could exist. Suppose that the primitive ancestors of modern cells were of this nature. That would mean that at its beginning, cellular evolution would have been driven in the main by HGT (1, 21).
In its subsequent evolution a primitive cell of this type would become ever more complex, idiosyncratically connected, and thereby increasingly refractory to horizontal gene acquisition, especially the more spectacular forms of it (21). In other words, there would come a stage in the evolution of cellular organization where the organismal genealogical trace (recorded in common histories of the genes of an organism) goes from being completely ephemeral to being increasingly permanent (21). This point in evolution, this transition, is appropriately call the “Darwinian Threshold.” On the far side of that Threshold “species” as we know them cannot exist. Once it is crossed, however, speciation becomes possible (21). The Darwinian Threshold truly represents the Origin of Species, in that it represents the origin of speciation as we know it.
It has a name and criteria by which it can be recognized (21), but what actually is the evolutionary transition called the “Darwinian Threshold?” It could be an otherwise undistinguished mile marker along the road from simple to complex cells. Or it could represent a point in cellular evolution where something drastic occurs. I posit the latter. The cell is a complex dynamic system. As its connectedness increases such a system can reach a critical point, where a phase change occurs, where a new, higher level organization of the whole emerges (22). That, I suggest, is what the Darwinian Threshold represents, a hitherto unrecognized phase change in the organization of the evolving cell.
The Root of the Universal Tree.
The universal tree has no root in the classical sense (1, 21). The root is actually a Darwinian Threshold, the first point at which we can begin to give tree representation to the organismal evolutionary course. A certain “symmetry of descent” is inherent in the classical view that is totally lacking here. In a classical phylogenetic bifurcation both sister lineages and their common ancestor are in essence alike. But, at the root of the universal tree (and in the first branching of the tree) classical presumptions do not hold (21), because the root is not a classical root, the sister lineages resulting from the earliest branchings are in no sense “sisters.” They differ in fundamental ways.
The nonclassical perspective required here takes some getting used to. We need to release all of the classical connotations of “symmetry” in these “bifurcations.” That the cell type on one side of the initial bifurcation has crossed a Darwinian Threshold does not imply that the organisms represented by the other side have done so. Indeed, different cell types would be expected to reach their Darwinian Thresholds more or less independently, at different times (1). The initial bifurcation of the universal tree (Fig. 1) tells us only that the bacterial cell type has crossed its Darwinian Threshold (23). Although the archaea and eukarya are represented by a “common lineage” at that stage, this is deceptive: the two are in effect lumped by forcing tree representation on the situation. Neither has yet to establish a stable genealogical trace. Neither has crossed its Darwinian Threshold. And that is all that their so-called “common ancestral lineage” signifies.
The great similarity between the archaeal and eukaryal information processing systems should be interpreted in this light. This similarity does not result from the implantation of some more or less finished archaeal versions of these systems into a eukaryotic (or other) cell that previously contained nothing of the sort (as is often suggested). Rather, it reflects the fact that at the time of the basal bifurcation of the universal tree, the archaeal and eukaryotic cell types were still being communally forged in the chaos of the universal gene-exchange pool (21), along with other cell types now extinct. Under these evolutionarily fluid conditions, major cellular systems could still be horizontally melded.
Crossing a Darwinian Threshold leads to a more solidified, organized cellular design. In that the bacterial cell design was the first to achieve this condition the bacterial versions of the central (universal) cellular systems represent earlier ancestral versions of these systems than do their archaeal or eukaryotic counterparts.
A question remains as to the order in which the Archaea and Eucarya crossed their Darwinian Thresholds. At this point we have no way of telling which was first. However, I tend to agree with the conjecture of Kandler (23) that it was the archaeal type. My reason is that archaeal versions of the translation, transcription, and genome replication systems (which so strikingly resemble their eukaryotic counterparts) are in all cases simpler in structure, which is consistent, at least, with the archaeal versions being closer to some ancestral form than are the eukaryotic ones.
The Evolution of Translation
The second of the keys to understanding cellular evolution is the nature and evolution of translation. This is a topic dealt with extensively in my earlier writings on cellular evolution (1, 3, 15, 21, 24–29), and, therefore, will merely be summarized here.
Translation increasingly shows itself to be an RNA-defined mechanism (30–33). Its primordial form almost certainly arose in some sort of “RNA-world” context. This simple RNA-based primitive translation process was highly imprecise (in matching anticodon to codon, in maintaining reading frame). Therefore, only small proteins and/or proteins imprecise in sequence (and therefore simple and general in function) could at first evolve (24, 25). The evolution of translation was a boot-strapping process, in which small improvements in translation led to improvement in proteins in general. These second-generation proteins then replaced (most of) their predecessors. Among them, of course, were proteins that further improved translation, and so on—ultimately giving rise to a translation mechanism having modern performance levels (25). Because large proteins of the kind that seem central to modern genome replication mechanisms and the like were initially absent, these information-processing systems were also initially imprecise (25). This fact means that primitive genomes would have been relatively small.
Translation would seem to be the leading edge of a wave of cellular evolution in which proteins evolve to greater and greater levels of complexity and specificity (25). Note that this is in essence the conclusion drawn above from comparative evidence, i.e., the evolution of translation led that of transcription, which in turn led that of modern genome structure and replication.
Cellular Design and Its Evolution
The cell is the essence of biology. At least that is how 20th century molecular biology saw it, and the great goal was to understand how cells were organized and work. This goal, it was assumed, could be accomplished by cataloging (and characterizing) all of the parts of the mechanism, with the tacit assumption that given such a parts list the overall organization of the cell would become apparent. Today, such lists exist for several organisms. Yet an understanding of the whole remains as elusive a goal as ever (34).
The fault here lies with the reductionist perspective of molecular biology. The problem of cellular design cannot be fit into this rigid, procrustean framework. It should be obvious from the foregoing discussion that biological cell design is not a static, temporal, or local problem. Indeed, the problem cannot be understood (as opposed to described) apart from the evolution of that design.
The Doctrine of Common Descent
[P]robably all of the organic beings which have ever lived on this earth have descended from some one primordial form … .
This, Darwin's Doctrine of Common Descent (35), is the primary evolutionary assertion and a cornerstone of modern biology, and it epitomizes classical biological thinking. The Doctrine implies two questions, what was the nature of the primordial form and why was that form unique. Through genomics these can now be approached. However, we are about to find that the two separate questions the Doctrine seems to pose are not separate but part and parcel of one another. The difficulty with the classical Darwinian outlook, as Alfred North Whitehead (36) long ago pointed out, is that it sees evolution as a “procession of forms,” when the focus should instead be on the process that produces them—on the gem, not the reflections from its facets.
The reality of HGT is forcing us to the Whitheadian point of view, making us think more about the process and less about the detailed forms it generates. From this perspective we will see that there was not one particular primordial form, but rather a process that generated many of them, because only in this way can cellular organization evolve. The Doctrine of Common Descent (and classical evolutionary thinking in general) rests on the tacit assumption that the dynamic of the evolutionary process remains unchanged as it gives rise to increasingly complex, specific, etc. cellular forms. Yet the forms in essence are the process. Therefore, fundamental changes in their nature can only mean changes in the underlying evolutionary dynamic. The time has come for Biology to go beyond the Doctrine of Common Descent. Neither it nor any variation of it (invoking, say, several primordial forms) can capture the tenor, the dynamic, the essence of the evolutionary process that spawned cellular organization.
The Dilemma of Cellular Evolution.
Evolving the cell requires evolutionary invention of unprecedented novelty and variety, the likes of which cannot be generated by any familiar evolutionary dynamic. The task can be accomplished only by a collective evolution in which many diverse cell designs evolve simultaneously and share their novelties with one another; which means that (i) HGT (and a genetic lingua franca) is a necessary condition for the evolution of cell designs, and (ii) a cell design cannot evolve in isolation; others will necessarily accompany it.
There is an inherent contradiction in this situation. Although HGT is essential for sharing novelty among the various evolving cell designs, it is at the same time a homogenizing force, working to reduce diversity. Thus, what needs explaining is not why the major cell designs are so similar, but why they are so different. This apparent contradiction can be resolved by assuming that the highly diverse cell designs that exist today are the result of a common evolution in which each of them began under (significantly) different starting conditions. [Initial conditions do not necessarily damp out for complex dynamic processes; indeed, they can lead to vastly different outcomes (22).]
The Course of Cellular Evolution.
From the start the course of cellular evolution is a march toward greater complexity, integration, precision, specificity of cellular design, etc. All of this results in increased idiosyncrasy, which, of course, leads to decreased interactivity with other cell types, decreased capacity to benefit from the universal gene-exchange pool. But in becoming complex, the cell design has created many new directions in which it can evolve on its own; it remains evolutionarily versatile, albeit in a different, more restricted way than when its organization allowed it to partake more freely of the novelty contained in the universal gene-exchange pool.
Cellular evolution is a self-limiting process. As a design evolves to greater complexity, the possible ways in and extent to which that design can be altered continue to diminish. Increased integration and complexity buy specificity, but at the cost of flexibility. An end point is ultimately reached where the organization of the cell can no longer fundamentally change. Only trivial changes are thereafter possible. The evolution of the cell per se is basically complete; the modern cell is born.
When Did the Evolution of Modern Cells Begin?
The origin of proteinaceous cells is the most important single event in evolutionary history and, therefore, had to have occurred at some clearly definable evolutionary stage. I believe that stage is evident. Consider the following: in the evolutionary course there have been a few great junctures, times of major evolutionary advance. Their hallmark is the emergence of vast, qualitatively new fields of evolutionary potential, and symbolic representation tends to underlie such evolutionary eruptions. These “New Worlds” can arise when some existing biological entity (system) gains the capacity to represent itself (what it is and/or does) in some symbolic form (37). The resulting world of symbols then becomes a vast and qualitatively new phase space for evolution to explore and expand. The invention of human language is one such juncture. It has set Homo sapiens entirely apart from its (otherwise very close) primate relatives and is bringing forth a new level of biological organization. The most important of these junctures, however, was the development of translation, whereby nucleic acid sequences became symbolically representable in an amino acid “language,” and an ancient “RNA-world” gave way to one dominated by protein. It is clear that the modern cell could not have evolved except in such a period of great evolutionary expansion. The evolution of modern cells, then, had to begin with the onset of translation.
Transforming an RNA-World.
Everyone seems to have his or her own take on the RNA-world. I see it as an “era of nucleic acid life” (27), a time when (RNA) programmed protein synthesis did not yet exist, and therefore a time when nucleic acid, the most evolutionarily versatile and malleable polymer then extant, defined and dominated the scene. This is not to imply that polypeptides were absent then (29). They existed to the extent that existing (bio)chemical mechanisms could generate them. [Such peptides, however, would be simple and, perhaps more importantly, fundamentally immutable in sequence. In other words, in the era of nucleic acid life, it is the nucleic acids that evolve to accommodate the peptides, not the reverse.] Some source of the monomers and energy to make biopolymers is obviously required, which calls for a “metabolic network,” a network that also produces other compounds, such as cofactors, and, therefore, is biochemically rich and self-sustaining.
Just as cells today are the fundamental units of biological organization, there must have been analogous higher-order architectures designed around nucleic acid componentry, which filled similar niches in the RNA-world. These latter I call “supramolecular aggregates” (or SMAs) to distinguish them from cells of the kinds we know. Within these SMAs, nucleic acids presumably performed roles similar to many of those that proteins perform today. As evolution has populated the biosphere today with a great variety of cellular entities (derived from one of three basic modern cell designs), so the RNAworld would be populated with a collection of diverse SMA architectures.
When translation entered the picture, it produced proteinaceous representations (translations) of preexisting RNA sequences. Initially most such translations must have had no relevance in terms of the existing SMAs, but a small fraction of them likely did—especially any that resembled nontranslationally produced polypeptides already incorporated into the system. Translationally produced peptides would have meaning in either of two contexts: (i) in terms of the SMA of which the translated RNA was already a functional part or (ii) in terms of other SMAs (and other entities) to which the translated RNA per se had no significance. Here then, with the second possibility, was something unique in the (RNA) world, namely, nucleic acids whose primary value lay in their coding capacity. Herein began the need for a commerce in coding nucleic acids.
It is reasonable that in an RNA-world being rapidly transfigured into a proteinaceous one, some of the basic SMA types served as starting platforms for proteinaceous cell designs. Here then is the reason why extant cellular designs are so different. If they start under different initial conditions, each from a different SMA platform, they will end up as different designs, despite the homogenizing influence of HGT.
Because of their origin in SMA architectures, cell designs would initially use nucleic acids in a variety of function/structure roles, many of them catalytic. With the translation apparatus as the prime example, it is clear that nucleic acids still play important functional roles in cells today—something that becomes ever clearer as interest in small cellular RNAs increases (38). One can picture (translated) proteins first serving to facilitate nucleic acid-defined functions, and later those proteins evolving to assume those functions outright. Also, some of the RNA components of early cells could subsequently have been recruited into new roles (6). Translation would seem to be an intermediate case, where functionality defined by RNA clearly still exists, but it seems facilitated if not enabled by proteins (32).
The present model strives to release the fetters classical Darwinian thinking imposes on the concept of cellular evolution. In evolving the basic cellular fabric, endosymbiosis (interactions between mature cells) has now been relegated to a secondary role, if not disallowed completely. The core componentry of the cell was forged in the high evolutionary temperatures (21) of the communal ancestral gene pools that the various cell designs shared. Having to rationalize those features unique to each of the three major cell types (10, 39) on the assumption they evolved late in the game (in the period covered by the universal tree) is clearly problematic. Now, these and other features of the cell have time to develop, to spawn gene families, and so on, in the vast expanses of the prehistoric past—in the chaotic era of modular cell designs, cosmopolitan proteins, and the like—long before any Darwinian Threshold had been reached.
Recapitulation
To begin to understand cellular evolution, one has to go beyond the classical concepts of cells, ancestry, and genealogy. The theory presented here is intended to provide a starting framework for conceptualizing cellular evolution in general, to move beyond endosymbiosis models and the like, to view the evolutions of the three major modern cell designs in comparable terms, and to give the picture of cellular evolution a continuity it heretofore lacked. The following points summarize the theory.
- 1.
Modern cells necessarily begin to evolve with the origin of the translation process, in an RNA-world-like setting.
- 2.
The RNA-world was populated by a variety of SMAs, the counterparts of cells today. Different ones of these SMAs would serve as the starting architectures around which various aboriginal proteinaceous cell designs were initially fashioned. These differing initial conditions are what underlie the differences in organization among the three primary cell types today.
- 3.
The type and extent of novelty required to evolve a (proteinaceous) cell is impossible to generate in the context of the highly limited kind of gene pool (and type of cellular organization) that is associated with an individual lineage today. Only global invention arising in a diverse collection of primitive entities is capable of providing the requisite novelty.
- 4.
Therefore, cellular evolution must begin in a collective mode. (i) The (early) evolution of a cell design can occur only in a context wherein a variety of other cell designs are simultaneously evolving. (ii) Mechanisms must exist whereby novelty can be globally disseminated (horizontally exchanged), which include a universal genetic code (lingua franca) carried by a standard, generic translation apparatus, one readily exchanged among the various cell types. (iii) The componentry of primitive cells needs to be cosmopolitan in nature, for only by passing through a number of diverse cellular environments can it be significantly altered and refined. (iv) Early cellular organization was necessarily modular and malleable.
- 5.
Because of their loose construction and the cosmopolitan nature of their componentry, primitive cells initially did not have stable genealogical records.
- 6.
Individual lineages, species as we know them, emerged from this common ancestral chaos only when cellular organization achieved a certain degree of complexity and connectedness. This transition is marked by certain key functions, such as translation (themselves complex in nature), becoming fixed, “custom-fitted” into the emerging cellular fabric, which made them hard to impossible to displace through horizontal gene exchange. I have called the stage at which this new, more complex, integrated organization arises the Darwinian Threshold, the first occurrence of which corresponds to the emergence of a modern type of cellular organization and is conventionally perceived as the root of the universal tree.
- 7.
Extant life on Earth is descended not from one, but from three distinctly different cell types. However, the designs of the three have developed and matured, in a communal fashion, along with those of many other designs that along the way became extinct.
Acknowledgments
Grateful thanks to Norman Pace, Claudia Reich, Jürgen Brosius, Frank Harold, Charles Kurland, and Harry Noller for their various helpful comments and suggestions. My work is supported by the National Aeronautics and Space Administration and the Department of Energy.
Abbreviations
- HGT
horizontal gene transfer
- SMA
supramolecular aggregate
References
- 1.Woese C R. Zentralbl Bakteriol Hyg I Abt Orig C. 1982;3:1–17. [Google Scholar]
- 2.Brenner S. Science. 1998;282:1411–1412. [Google Scholar]
- 3.Woese C R. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zillig W, Klenk H P, Palm P, Leffers H, Puhler G, Grupp F, Garrett R. Endocytol Cell Res. 1989;6:1–25. [Google Scholar]
- 5.Margulis L. Origin of Eucaryotic Cells. New Haven, CT: Yale Univ. Press; 1970. [Google Scholar]
- 6.Hartman H. Spec Sci Technol. 1984;7:77–81. [PubMed] [Google Scholar]
- 7.Sogin M L. Curr Opin Genet Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- 8.Moreira D, Lopez-Garcia P. J Mol Evol. 1998;47:517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- 9.Martin W, Mueller M. Nature (London) 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 10.Hartman H, Fedorov A. Proc Natl Acad Sci USA. 2001;99:1420–1425. doi: 10.1073/pnas.032658599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkhardt F, Smith S. The Correspondence of Charles Darwin. Vol. 6. Cambridge, U.K.: Cambridge Univ. Press; 1990. pp. 1856–1857. [Google Scholar]
- 12.Woese C R, Olsen G J, Ibba M, Soll D. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langer D, Hain J, Thuriaux P, Zillig W. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen G J, Woese C R. Trends Genet. 1996;12:377–379. doi: 10.1016/0168-9525(96)30092-9. [DOI] [PubMed] [Google Scholar]
- 15.Woese C R. Proc Natl Acad Sci USA. 2000;97:8392–8396. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown J R, Doolittle W F. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennisi E. Science. 1998;280:672–674. doi: 10.1126/science.280.5364.672. [DOI] [PubMed] [Google Scholar]
- 18.Pennisi E. Science. 1999;284:1305–1307. doi: 10.1126/science.284.5418.1305. [DOI] [PubMed] [Google Scholar]
- 19.Nesbo C L, Boucher Y, Doolittle W F. J Mol Evol. 2001;53:340–350. doi: 10.1007/s002390010224. [DOI] [PubMed] [Google Scholar]
- 20.Brown J R, Douady C J, Italia M J, Marshall W E, Stanhope M J. Nat Genet. 2001;28:281–285. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- 21.Woese C. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodwin B. How the Leopard Changed its Spots: The Evolution of Complexity. New York: Simon & Schuster; 1996. [Google Scholar]
- 23.Kandler O. J Biol Phys. 1994;20:165–169. [Google Scholar]
- 24.Woese C R. Proc Natl Acad Sci USA. 1965;54:1546–1552. doi: 10.1073/pnas.54.6.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woese C R, Fox G E. J Mol Evol. 1977;10:1–6. doi: 10.1007/BF01796132. [DOI] [PubMed] [Google Scholar]
- 26.Woese C R. Organization and Control in Prokaryotic and Eukaryotic Cells. In: Charles H P, Knight B D J G, editors. Soc. Gen. Microbiol. Symp. 20. Cambridge, U.K.: Cambridge Univ. Press; 1970. pp. 39–54. [Google Scholar]
- 27.Woese C R. In: Exobiology. Ponnamperumapp C, editor. Amsterdam: North–Holland; 1972. pp. 301–341. [Google Scholar]
- 28.Woese C R. In: Evolution from Molecules to Men. Bendall D S, editor. Cambridge, U.K.: Cambridge Univ. Press; 1983. pp. 209–233. [Google Scholar]
- 29.Woese C R. The Genetic Code: The Molecular Basis of Genetic Expression. New York: Harper & Row; 1967. [Google Scholar]
- 30.Cech T. Science. 2000;289:878. doi: 10.1126/science.289.5481.878. [DOI] [PubMed] [Google Scholar]
- 31.Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 32.Woese C R. RNA. 2001;7:1055–1067. doi: 10.1017/s1355838201010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brosius J. Trends Biochem Sci. 2001;26:653–656. doi: 10.1016/s0968-0004(01)01972-7. [DOI] [PubMed] [Google Scholar]
- 34.Harold F M. Microbiology. 1995;141:2765–2778. doi: 10.1099/13500872-141-11-2765. [DOI] [PubMed] [Google Scholar]
- 35.Darwin C. On the Origin of Species. MA, and London: Harvard Univ. Press Cambridge; 1859. p. 484. [Google Scholar]
- 36.Whitehead A N. Process and Reality. Toronto: Collier–Macmillan Canada; 1929. [Google Scholar]
- 37.Barbieri M. The Organic Codes. Ancona, Italy: Casa Editrice Pequod; 2001. [Google Scholar]
- 38.Hüttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie J-P, Brosius J. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham D E, Overbeek R, Olsen G J, Woese C R. Proc Natl Acad Sci USA. 2000;97:3304–3308. doi: 10.1073/pnas.050564797. [DOI] [PMC free article] [PubMed] [Google Scholar]