Abstract
Gene amplification is a common form of genomic instability in a wide variety of organisms and is often associated with tumor progression in mammals. One striking feature of many amplified genes is their organization as large inverted duplications (palindromes). Here, we describe a molecular mechanism for palindrome formation in mammalian cells that is also conserved in protists. We introduced a short (79 or 229 bp) inverted repeat into the genome of Chinese hamster ovary cells and showed that it promoted the formation of a large DNA palindrome after an adjacent DNA double-strand break. This finding suggests that short inverted repeats in the mammalian genome can have a critical role in the initiation of gene amplification. This specific mechanism may provide a novel target for cancer therapies.
Many tumors amplify regions of their genome to vastly increase the copy number of specific oncogenes that drive tumor progression (1, 2). In many cases characterized, these amplified regions contain large “head-to-head” duplications (palindromes) of sequences that can be tens to hundreds of kb in size (3–8). Cytogenetic studies have further revealed that large inverted duplications of a chromosomal region are formed at early stages of gene amplification (9–11). Thus, the formation of a DNA palindrome has been suggested as a crucial step that leads to further DNA amplification (7, 9–11). However, little is known regarding the exact molecular mechanism for the initial palindrome formation in mammalian cells. This problem is a difficult one to study partly because, in most systems, only a small fraction of cells undergoes the relevant initial events. Lack of an appropriate model system has limited our ability to identify factors involved in this process and develop methods for preventing it in tumor progression.
Amplification of palindromic DNA has also been observed in several unicellular eukaryotes including budding and fission yeast, Leishmania, Physarum, and Tetrahymena (12–16). Studies of ribosomal RNA gene amplification in the ciliated protozoan Tetrahymena have identified critical steps necessary for palindrome formation: a DNA double-stranded break (DSB) occurring near a short inverted-repeat (IR) sequence of 29 bp or longer facilitates an intramolecular recombination between the repeats, which generates a large palindrome after one round of replication (Fig. 1A; refs. 17–19). This mechanism seems to be shared by other unicellular eukaryotes, as short IR sequences also facilitate palindrome formation in yeast, either following a DSB (20) or when MRE11 is mutated (21). Short IRs also are found at the genomic sites that form the junctions of palindromes in other organisms, including Leishmania and the fission yeast (15, 16). In mammalian cells, the importance of DSB in gene amplification has been suggested in recent studies (22–27), although little is known about its direct role. If palindrome formation in mammalian cells also depends on a similar short IR-mediated mechanism, the susceptibility of a gene to undergo gene amplification could be influenced by its proximity to short IR sequences, which are abundant in mammalian genomes (28). Also, it would suggest that mammalian gene amplification occurs through a conserved process that utilizes an intrachromosomal homologous recombination mechanism, which may provide novel targets for cancer therapies.
Figure 1.
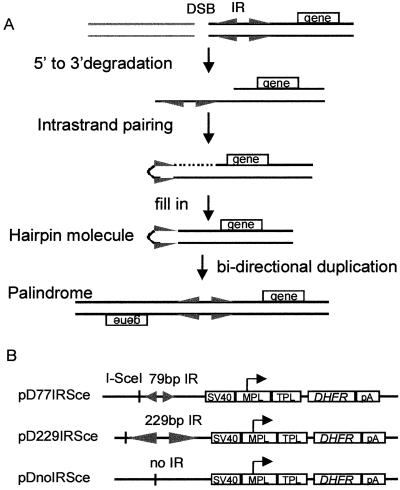
(A) Intramolecular recombination model of palindrome formation. For simplicity, one possible mode of recombination, single-strand annealing version, is shown (19, 20). After a DSB, one strand of a short IR is exposed by 5′ to 3′ degradation, followed by intrastrand pairing, to generate a hairpin molecule, which is converted to a large palindrome by bi-directional replication. Lines represent DNA sequences, and a triangle represents one repeat of a short IR. (B) pDhfrSce constructs. Each construct has an I-SceI recognition sequence and either a 79-bp IR (pD79IRSce), 229-bp IR (pD229IRSce), or noIR (pDnoIRSce). Each short IR has a 29-bp nonpalindromic center. Simian virus 40 (SV40), SV40 enhancer element and origin of replication; MPL, adenovirus major late promoter; TPL, tripartite leader from adenovirus late mRNA and small intervening sequence; pA, polyadenylation signal from SV40 early region.
In this study, we directly test whether a short IR, with an adjacent DSB, can mediate the formation of a large DNA palindrome in mammalian cells. We used the homing endonuclease I-SceI to induce a DSB (29) and a mouse DHFR gene as an amplification marker in DHFR-deficient Chinese hamster ovary (CHO) cells (30). Here, we show that a 79- or 229-base pair IR in the genome promotes the formation of a large DNA palindrome following an adjacent DNA double-stranded break. This result suggests a common pathway for palindrome formation among eukaryotes and an important role for short IR sequences in genome instability.
Materials and Methods
Plasmid Constructs.
pD79IRSce, pD229IRSce, and pDnoIRSce were derived from the DHFR expression vector pED (gift from R. J. Kaufman, University of Michigan Medical School, Ann Arbor, MI; ref. 31). The internal ribosome expression site of pED was deleted by digestion with XbaI and XhoI, followed by Klenow treatment to generate pED (-EMC). A synthetic I-SceI site with BamHI overhangs was created by annealing the two oligonucleotides 5′-GATCCGCTAGGGATAACAGGGTAATATA-3′ and 5′-GATCTATATTACCCTGTTATCCCTAGCG-3′ together, which was inserted into the BamHI site of pUC18 to produce pUC18Sce. The EcoRI and SacI fragment of pPCB08 (20), which includes the 42-bp IR of Tetrahymena with a 28-bp spacer, was cloned into the EcoRI and SacI site of pUC18Sce to generate pUC18/229IRSce. The EcoO109I fragment of pCB08 was inserted into the AccI site of pUC18Sce after Klenow treatment to generate pUC18/79IRSce. The EcoRI/HincII fragment of pUC18/229IRSce and the EcoRI/HindIII fragment of pUC79IRSce were treated with Klenow and inserted into Klenow-treated NdeI site of pED (-ECM), located 216 bp upstream of the SV40 enhancer element, to generate pD229IRSce and pD79IRSce. The clone pDnoIRSce was derived by a slightly different route. The BstBI fragment of pD5H8 (18), which contains 38 bp of the Tetrahymena inverted repeats, was inserted into HincII site of pUC18 after Klenow treatment to generate pUC18/38IR. The synthetic oligonucleotide containing I-SceI site was inserted into the BamHI site of pUC18/38IR to produce pUC18/38IRSce. The EcoRI/HindIII fragment of this plasmid was treated with Klenow and ligated into Klenow-treated NarI site of pED (-ECM), which is 164 bp upstream of the SV40 enhancer element, to generate pD38IRSce. pDnoIRSce was obtained by deleting the 38-bp inverted repeats by digestion with XbaI and PstI, which flank the inverted repeats. Most of the cloning was done by using SURE supercompetent cells (Stratagene).
Cell Transfections and Southern Analysis.
DHFR-deficient CHO cells (CHOdhfr−; ref. 30) were grown in DMEM supplemented with 10% (vol/vol) FBS, HT supplement, and MEM Amino Acid Solution (nonselective media, GIBCO/BRL). To obtain single-copy transformants, 50 ng of each of the three plasmids (pD79IRSce, pD229IRSce, and pDnoIRSce) was linealized by digestion with AhdI and electroporated into CHOdhfr− cells. Transformed cells were selected for 12 to 14 days in DMEM with 10% (vol/vol) dialyzed FBS (GIBCO/BRL) and analyzed by Southern blotting to identify single-copy transformants. Each transformant was cultured in media with 0.05 μM, 0.2 μM, 0.4 μM, and 0.8 μM methotrexate (MTX) to determine the lowest concentration necessary for selecting MTX-resistant colonies after DSB. I-SceI expression vector pCMV3xnls-I-SceI (5 μg; gift from Maria Jasin, The Sloan–Kettering Institute, New York; NY; ref. 29) or the vector without I-SceI coding sequence (pCMVnoSce) was transfected into 3 × 105 cells by using Superfect (Qiagen, Chatsworth, CA). Forty-eight hours after transfection, 1 × 105 cells were plated in media with MTX. Resistant colonies were scored after 10 days and picked for DNA extraction. To extract DNA, cells were incubated in the lysis buffer including 100 mM NaCl/10 mM Tris⋅HCl, pH 8.0/25 mM EDTA/0.5% SDS/Proteinase K, followed by phenol/chloroform extraction and ethanol precipitation.
Bisulfite Modification of Genomic DNA and PCR.
Bisulfite treatment of genomic DNA was done following published procedures (32). The primers used in the nested PCR amplifications were as follows: Dpab418F, 5′-GTTGTGATTGGGGAAAATTGTGG-3′; Dpab917R, 5′-AATCACCCTTCCCAACAATTACACAACCTA-3′; Dpab1123R, 5′-ACAAAAACCAAAACCACCTCAACCTCTAAACT-3′; Dpab898F, 5′-TTGTTATTTAGGTTGTGT-3′; and Dpab1052R, 5′-CACTAACCATCATTTTACAACATCATA-5′.
In Situ Hybridization.
Fluorescence in situ hybridization (FISH) was performed as described (27). The XbaI-AhdI fragment of pD79IRSce and 5 kb of flanking genomic sequence of the transgene in the 79IR-8 transformant were used together as a probe. The flanking sequence was cloned as two separate fragments by arbitrarily primed PCR (33) with the following modifications. To clone longer genomic fragments, 6-bp recognition sequences of restriction enzymes including BamHI, KpnI, EcoRI and XbaI, instead of a 5-bp specific sequence, were added to the 3′end of the primary arbitrary primers, and the Expand Long Template PCR system (Roche Applied Science) was used for PCR.
Results
Increased MTX-Resistant Subclones After Chromosomal DSB.
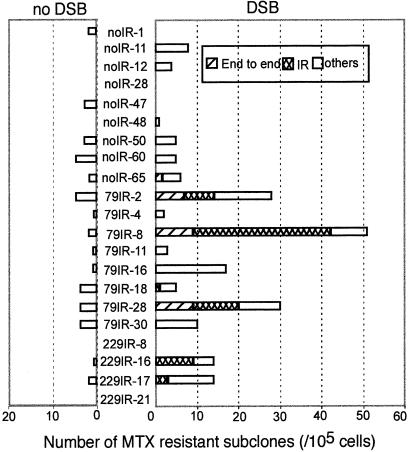
To determine whether an IR sequence near a DSB facilitates palindrome formation and gene amplification in mammalian cells, we studied the amplification of a DHFR transgene in DHFR-deficient CHO cells (CHOdhfr−; ref. 30). We constructed a DHFR expression vector with an I-SceI restriction site and a short IR of different lengths (0, 79, or 229 bp) with a 29-bp spacer placed near the 5′ end of the cassette (Fig. 1B). These constructs were introduced into CHOdhfr− cells, and several transformants with a chromosomally integrated single-copy DHFR transgene were obtained. They were transiently transfected with an I-SceI expression vector (pCMV3xnls-I-SceI; ref. 29) to induce a chromosomal DSB, or with a control vector that does not express I-SceI (pCMVnoSce), and were grown in 0.2–0.8 μM MTX to select for subclones with elevated DHFR activity. In the absence of an I-SceI-induced DSB, very few MTX-resistant subclones were formed (Fig. 2). In contrast, an I-SceI-induced DSB resulted in significantly higher numbers of MTX-resistant subclones from the parental transformants containing a short IR (79IR+DSB vs. 79IR-DSB, P = 0.02, Wilcoxon rank-sum test), but not from transformants without a short IR (noIR-DSB vs. noIR+DSB, P = 0.27). For example, the induction of a DSB in transformant 79IR-8 increased the frequency of MTX-resistant subclones by more than 20-fold. Because the estimated efficiency of transient transfection with pCMV3xnls-I-SceI was approximately 10–20%, the actual increase in MTX-resistant subclones could be more than 100-fold. Thus, a DSB adjacent to an IR significantly facilitates the generation of subclones with increased MTX resistance.
Figure 2.
Number of MTX-resistant subclones with each type of palindrome analyzed in all transformants. Nine, eight, and four transformants of noIR, 79IR, and 229IR respectively were subjected to DSB induction and MTX selection. The number of MTX-resistant subclones from cells with DSB (Right) and without DSB (Left) are indicated by the lengths of the bars. The presence of various types of palindromes in the MTX-resistant subclones of every transformant was determined by Southern hybridization analysis, and their fractions are indicated by using different shadings. Many subclones from transformants 79IR-2, -8, and -28 contained short IR-mediated palindromes. 229IR-16 and -17 contained only IR-mediated palindromes. Transformants of noIR produced a lower number of MTX-resistant subclones than 79IR transformants (79IR+DSB vs. noIR+DSB, P = 0.02). The “others” category includes subclones with unrearranged DNA, other types of rearranged DNA, or mixtures of DNA types that might include palindromes.
Palindrome Formation After DSB.
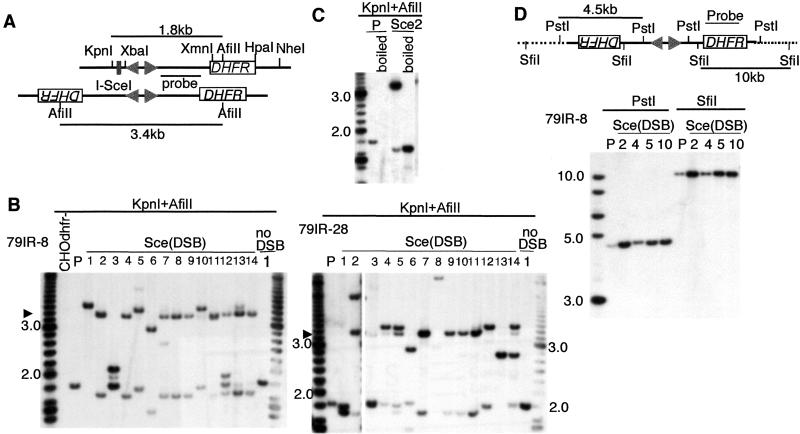
We analyzed several MTX-resistant subclones from each 79IR transformant and found two types of palindromes, in addition to other types of rearrangements. The representative data are shown in Fig. 3. If a palindrome is formed by using the short IR as the center, a 3.4-kb fragment is produced after double digestion with KpnI and AfiII, in contrast to the 1.8-kb fragment of the original single-copy transformant (Fig. 3A). Analysis of subclones from parental transformants 79IR-8 and 79IR-28 (both contain a 79-bp IR) demonstrated that most of the subclones (23/23 from 79IR-8 and 12/14 from 79IR-28) had indeed lost the 1.8-kb fragment and gained a 3.4-kb fragment following DSB (Fig. 3B). This fragment remained the same size when using XbaI instead of KpnI for the digestion (data not shown), which is consistent with our model. The presence of palindromic DNA was further verified by using additional restriction enzymes, including XmnI, HpaI and NheI (data not shown). A 1.7-kb fragment of lower intensity was present in addition to the 3.4-kb AfiII fragment and represents the “snap-back” form of the palindromic DNA that was probably generated during genomic DNA isolation or restriction digestion. In support of this point, the 3.4-kb fragment was converted to the 1.7-kb fragment through heating and rapidly cooling the sample before electrophoresis (Fig. 3C), a condition that favors intramolecular hybridization. Instead of the 3.4-kb AfiII fragment, a few clones contained a 3.6-kb band with a 1.8-kb snap-back form (e.g., subclones 5 and 10 from 79IR-8; Fig. 3B). Additional restriction enzyme analysis confirmed that they were also palindromes (data not shown). Based on SfiI digestion, the palindromic structure appeared to extend into the flanking genomic sequence for at least 10 kb to each side (Fig. 3D). Copy numbers of the DHFR gene in these subclones were estimated by using a PhosphorImager (Molecular Dynamics) with the p53 gene as a control. Most of the subclones from the 79IR-8 transformant contained only two to four copies of DHFR sequence (i.e., two to three copies in subclone Sce2 and Sce5, four in Sce4; data not shown), indicating that they are at the very early stage of gene amplification.
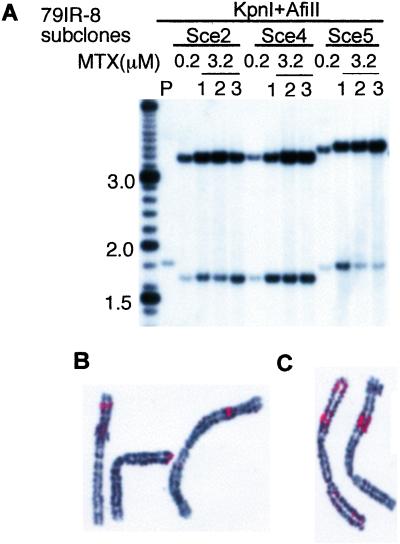
Figure 3.
Palindrome formation after DSB induction in 79IRSce transformants. (A) Restriction maps of the expected structures. In the original construct, the AfiII-KpnI fragment detectable by the probe indicated is 1.8 kb. If palindrome formation occurs after cleavage at the I-SceI site, this KpnI site will be lost, and the fragment will be replaced by a 3.4-kb AfiII fragment. Solid lines represent sequences of the transgene. (B) Southern blotting analysis of DNA from MTX-resistant subclones. Subclones from transformants 79IR-8 and 79IR-28 are shown as examples. After DSB, many subclones produced the 3.4-kb AfiII fragment indicative of the palindrome predicted. In some subclones (e.g., subclones 5 and 10 from 79IR-8), a slightly larger fragment is seen that represents a variant palindrome. In other subclones (e.g., subclones 12 and 13 of 79IR-8 and subclones 2 and 5 of 79IR-28), multiple fragments, including the 3.4-kb fragment, were detected. They could contain a mixture of rearrangements or be a mixture of cells with different rearrangements and were not counted as palindromes in Fig. 2. Size markers (200-bp ladder) are shown on both sides of the gel. P, parental transformant. (C) Snap-back analysis of the palindromic DNA. The 3.4-kb fragment is always accompanied by a l.7-kb fragment of lower intensity, which could be a snap-back form of the palindrome. After heat denaturation and rapid cooling (boiled), the 3.4-kb fragment disappears and the 1.7-kb fragment becomes dominant. (D) Extension of palindromic structure outside of the transgene. DNA from the parental transformant 79IR-8 (P), subclones with a short IR-mediated palindrome (2, 4), and end-to-end joining palindrome (5, 10) were digested with SfiI or PstI and probed with the DHFR gene. In all cases, only one fragment was detected, suggesting that both arms of the palindrome are identical to that of the parental transformant and extend beyond the transgene. Solid lines represent sequences of the transgene, and dotted lines represent the flanking genomic sequence. A 1-kb ladder is used as size marker.
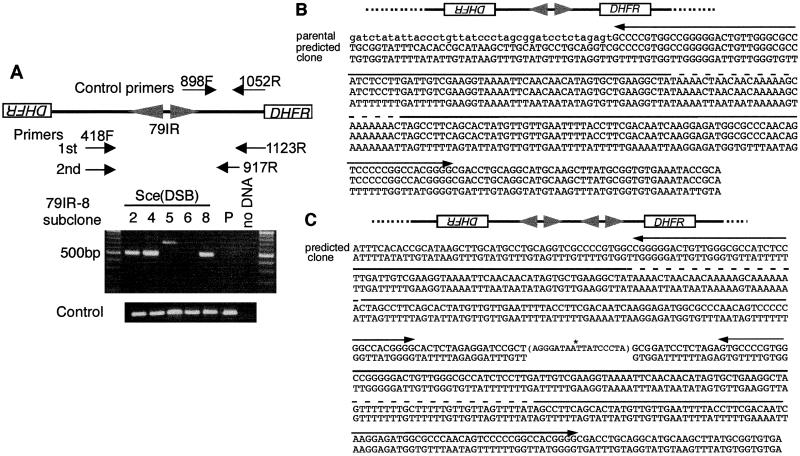
We amplified the sequences around the junction of both types of palindromes by PCR. Palindromes are known to be technically challenging for PCR, largely because they fold back rapidly through base pairing between the arms (34). Bisulfite modification of DNA converts each cytosine to uracil, rendering the repeats imperfect and allowing PCR to occur (Fig. 4A). The products were sequenced and compared with the predicted sequence of the palindromic junction. In MTX-resistant subclones with a 3.4-kb AfiII fragment (subclones 2, 4, and 8 of the transformant 79IR-8), two copies of the transgene were joined at the 79-bp IR (Fig. 4B), creating a palindrome exactly like those found in Tetrahymena, a short IR-mediated palindrome. In contrast, the 3.6-kb variant palindrome (subclones 5) contained junctions at or near the I-SceI recognition site with small deletions on both arms (Fig. 4C), possibly mediated by nonhomologous end-to-end joining (NHEJ) between sister chromatids (end-to-end joining palindrome). To assess the possibility that the short IR-mediated palindrome is derived from the end-to-end joining palindrome through further rearrangements, we cultured subclones 5 and 10 of the 79IR-8 (Fig. 3B) for an additional 48 cell doublings and could not detect any sign of such conversion (data not shown). Therefore, we conclude that these different palindromes arise independently, probably by distinct mechanisms.
Figure 4.
PCR amplification and sequencing of the bisulfite-modified DNA from the palindromic center. (A) PCR amplification. Nested PCR was used to amplify the junctions of the rearranged DNA. The locations of the primers used, including those for the positive controls, are indicated. DNA from 79IR-8 parental cells (P) and MTX-resistant subclones 2, 4, 5, 6, and 8 were treated with bisulfite and amplified. Subclones 2, 4, and 8 produced an ≈500-bp PCR fragment with the palindromic structure diagrammed. Subclone 5 produced an ≈700-bp fragment. There is no amplification product from subclone 6 or a parental transformant. (B) Sequence of the short IR-mediated palindrome from subclone 2. PCR fragments described above were cloned and sequenced (bottom line, bisulfite modified) and compared with the predicted palindromic sequence (middle line). Bisulfite treatment converts C in the middle lines to T in the bottom line. The parental nonpalindromic sequence in this region is also shown (top line). The sequence upstream of the 79-bp IR is shown in lowercase. The two divergent arrows indicate the locations of the 79-bp IR, and the dashed line indicates the nonpalindromic center. A schematic drawing is shown at the top. (C) Sequence of the end-to-end joining palindrome from subclone 5. The sequence after bisulfite modification (bottom line) was shown along with the predicted sequence (top line). The two palindromic arms are joined head to head at the I-SceI cleaved site (asterisk) with 9 bp and 8 bp of deletions from the two sides (end-to-end joining palindrome), which is shown in the parenthesis. Note that two pairs of the short IR (arrows), including the nonpalindromic center (dashed line), are present in this sequence, compared with only one in the sequence in B.
We also analyzed the transformants without a short IR (noIR) and those with a 229-bp IR (229IR). Of the nine noIR transformants studied, only one produced a few MTX-resistant subclones with end-to-end joining palindromes, whereas two of the four 229IR transformants analyzed produced palindromes; all of them are IR-mediated (Fig. 2). An analysis of data generated from all 21 transformants studied (Fig. 2) reveals that both a DSB and a short IR are necessary for the formation of short IR-mediated palindromes. The data also suggest that a longer IR (229-bp) favors the formation of IR-mediated palindromes over the end-to-end joining palindromes. This point is in agreement with previous studies in Tetrahymena and yeast, which suggested that IR-mediated palindrome formation depends on homologous recombination (18, 20).
Intrachromosomal Amplification of the DHFR Gene.
To determine if further DHFR amplification can occur from these initial palindromes, we grew subclones 79IR-8 Sce2, Sce4, and Sce5 at a higher MTX concentration (3.2 μM). Resistant subclones were obtained at higher rates (about 10−2) than from the parental transformant without DSB (about 10−5); both kinds of palindromes (short IR-mediated palindrome in subclones Sce2 and Sce4, end-to-end joining palindrome in Sce5) were stable during further amplification (Fig. 5A).
Figure 5.
Intrachromosomal amplification of DHFR. (A) Subclones with short IR-mediated palindrome (subclones 2 and 4) and end-to-end joining palindrome (clone 5) isolated in 0.2 μM MTX from transformant 79IR-8 were further selected with 3.2 μM MTX, and resistant subclones were isolated after 10 days of growth. The genomic DNA was digested with KpnI and AfiII and analyzed by Southern blotting. (B) Intrachromosomal palindrome formation of DHFR transgene in subclone 2. Chromosomes shown are from three different cells. FISH was performed by using DNA fragment from pD77IRSce and 5 kb of the flanking genomic sequence as a probe. Each signal (red) probably represents one palindrome of the DHFR transgene. (C) In the highly amplified Sce2–1 subclone, large blocks of signals were seen in different cells on the same chromosome, which, in some cases, showed fused sister chromatids (Left).
We also cultured these subclones in nonselective media, in which cells without DHFR can grow, and determined the presence of both types of palindromes after 72 cell doublings by Southern blotting (data not shown). We found that these palindromes were stable without selection during this period, suggesting that they were integrated in a chromosome. We then carried out FISH to confirm the intrachromosomal location of these amplified genes. In subclone 77IR-8 Sce2, hybridization signals were seen on the same chromosome arm in all (71) metaphases examined (Fig. 5B). Signals also were detected at the same location in another subclone 77IR-8 Sce4, which was derived independently from the same parental transformant. The identical location in these two subclones strongly suggests that the formation of the initial palindrome and its further amplification took place intrachromosomally at this locus. However, we could not detect the single DHFR transgene in the parental transformant by FISH presumably because of the small size of the probe available (<10 kb). The varied lengths of the material between the DHFR signals and the chromosomal end in different cells suggests that these chromosomes are undergoing recurrent rearrangement, probably through breakage-fusion-bridge (BFB) cycles, as observed in other studies (11). Additional FISH signals were detected in the more highly amplified Sce2–1 clone, which is estimated by PhosphorImager analysis to have more than 20 DHFR copies (Fig. 5C). One cell exhibited two large blocks of DHFR signal and fused sister chromatids (left), both suggestive of a history of BFB cycles.
Discussion
In this study, we show that a short IR adjacent to a DSB promotes large DNA palindrome formation in DHFR-deficient CHO cells. This finding provides one mechanism for the initiation of gene amplification in mammalian cells. Gene amplification has been observed in a wide variety of organisms, but studies of the initial molecular process have been limited by the difficulty of detecting these rare events in complex organisms. Studies in protists, including Tetrahymena and yeast, have revealed an interesting mechanism that initiates the process through a DSB adjacent to a short IR sequence (18–20). Despite the difference in genome complexity and utilization of DNA-repair pathways, it seems that the same or a similar mechanism can operate in CHO cells. The presence of a short IR can increase the frequency of MTX-resistant subclones by more than 20-fold, with at least 50% accompanied by the formation of a palindrome. This process depends strictly on a nearby DSB. Moreover, the majority of the palindromes generated have the structure predicted from our model: a perfect palindrome separated in the center by the original spacer of the short IR. We envision a mechanism similar to that described in protists: the repair of the DSB involves an intramolecular recombination event at the short IR to generate a hairpin structure that is resolved into a giant palindrome after one round of DNA replication (19, 20).
DSB is a common form of DNA damage, which can result from radiation, hypoxia, chemicals, endogenous DNA enzymes, and replication errors (35). Several recent studies have suggested the involvement of chromosome breakage in gene amplification (22–27). Our study presents a possible mechanism through which a DSB can initiate palindrome formation and gene amplification. Significantly, a relatively short IR (79 bp) is sufficient to promote palindrome formation and gene amplification. Random DSBs could occur adjacent to a short IR and generate a palindrome through the mechanism described here. Therefore, the locations of short IR sequences in the human genome can be a critical factor in the susceptibility of a region to undergo amplification in human cancers.
By using bisulfite-modified DNA, we were able to analyze the sequence of the junction. We found two types of structure at the center of the palindrome: the type predicted from the model (short IR-mediated palindrome) and another that is probably derived from NHEJ between sister chromatids. In our experimental system, I-SceI cleaves DNA at the identical location in both sister chromatids and allows end-to-end joining palindromes to occur, which would likely be infrequent in the absence of a targeted endonuclease in nature. In contrast, short IR-mediated palindrome formation requires only one single DSB adjacent to a short IR. For this reason, we think the short IR-mediated mechanism is more relevant to palindrome formation observed in nature. This view agrees with the observation that mice with impaired NHEJ activities still develop tumors that show oncogene amplification (36–38).
Gene amplification is generally considered to be associated with tumor progression and provides a predictive factor of clinical outcome and a target for therapeutics (1, 2). We have shown that a short IR can have a critical role for the initial events that lead to palindrome formation and low-level gene amplification. This process occurs within the chromosome in the one case studied, which, according to our model, likely produces a dicentric chromosome and leads to further rearrangements and gene amplification through subsequent BFB cycles. A future challenge is to identify the genes and factors that regulate this process. Agents that block the early events of palindrome formation should prevent subsequent gene amplification and the associated tumor progression.
Acknowledgments
We thank Hillary Massa for FISH analysis, Drs. Maria Jasin, Randal J. Kaufman, and Dusty Miller for providing plasmids and a cell line, Dr. Ted Gooley for statistical analysis, and Drs. Mark Groudine and Daniel E. Gottschling for critical reading of the manuscript. This work was supported by National Institutes of Health Grants R01GM26210 (to M.-C.Y.), and R01AR45113 and R01AR45203 (to S.J.T.). H.T. is a recipient of Interdisciplinary Dual Mentor Fellowship of Fred Hutchinson Cancer Research Center (1999 and 2001).
Abbreviations
- DSB
double-stranded break
- IR
inverted repeat
- MTX
methotrexate
- FISH
fluorescence in situ hybridization
- SV40
simian virus 40
References
- 1.Tlsty T D. Curr Top Microbiol Immunol. 1997;221:37–46. doi: 10.1007/978-3-642-60505-5_4. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 3.Ford M, Fried M. Cell. 1986;45:425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- 4.Saito I, Stark G R. Proc Natl Acad Sci USA. 1986;83:8664–8668. doi: 10.1073/pnas.83.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Looney J E, Hamlin J L. Mol Cell Biol. 1987;7:569–577. doi: 10.1128/mcb.7.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Passananti C, Davies B, Ford M, Fried M. EMBO J. 1987;6:1697–1703. doi: 10.1002/j.1460-2075.1987.tb02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz J C, Wahl G M. Mol Cell Biol. 1988;8:4302–4313. doi: 10.1128/mcb.8.10.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyrien O, Debatisse M, Buttin G, de Saint Vincent B R. EMBO J. 1988;7:407–417. doi: 10.1002/j.1460-2075.1988.tb02828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith K A, Stark M B, Gorman P A, Stark G R. Proc Natl Acad Sci USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo F, Le Roscouet D, Buttin G, Debatisse M. EMBO J. 1992;11:2665–2673. doi: 10.1002/j.1460-2075.1992.tb05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C, Martin S, Trask B, Hamlin J L. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 12.Vogt V M, Braun R. J Mol Biol. 1976;106:567–587. doi: 10.1016/0022-2836(76)90252-7. [DOI] [PubMed] [Google Scholar]
- 13.Karrer K M, Gall J G. J Mol Biol. 1976;104:421–453. doi: 10.1016/0022-2836(76)90280-1. [DOI] [PubMed] [Google Scholar]
- 14.Walton J D, Paquin C E, Kaneko K, Williamson V M. Cell. 1986;46:857–863. doi: 10.1016/0092-8674(86)90067-x. [DOI] [PubMed] [Google Scholar]
- 15.Ouellette M, Hettema E, Wust D, Fase-Fowler F, Borst P. EMBO J. 1991;10:1009–1016. doi: 10.1002/j.1460-2075.1991.tb08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albrecht E B, Hunyady A B, Stark G R, Patterson T E. Mol Biol Cell. 2000;11:873–886. doi: 10.1091/mbc.11.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao M C, Yao C H, Monks B. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- 18.Yasuda L F, Yao M C. Cell. 1991;67:505–516. doi: 10.1016/0092-8674(91)90525-4. [DOI] [PubMed] [Google Scholar]
- 19.Butler D K, Yasuda L E, Yao M C. Mol Cell Biol. 1995;15:7117–7126. doi: 10.1128/mcb.15.12.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler D K, Yasuda L E, Yao M C. Cell. 1996;87:1115–1122. doi: 10.1016/s0092-8674(00)81805-x. [DOI] [PubMed] [Google Scholar]
- 21.Lobachev K S, Gordenin D A, Resnick M A. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 22.Windle B, Draper B W, Yin Y X, O'Gorman S, Wahl G M. Genes Dev. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- 23.Poupon M F, Smith K A, Chernova O B, Gilbert C, Stark G R. Mol Biol Cell. 1996;7:345–354. doi: 10.1091/mbc.7.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 25.Paulson T G, Almasan A, Brody L L, Wahl G M. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pipiras E, Coquelle A, Bieth A, Debatisse M. EMBO J. 1998;17:325–333. doi: 10.1093/emboj/17.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singer M J, Mesner L D, Friedman C L, Trask B J, Hamlin J L. Proc Natl Acad Sci USA. 2000;97:7921–7926. doi: 10.1073/pnas.130194897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenger J E, Lobachev K S, Gordenin D, Darden T A, Jurka J, Resnick M A. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- 29.Liang F, Han M, Romanienko P J, Jasin M. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urlaub G, Chasin L A. Proc Natl Acad Sci USA. 1980;77:4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufman R J, Davies M V, Wasley L C, Michnick D. Nucleic Acids Res. 1991;19:4485–4490. doi: 10.1093/nar/19.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulin R, Grigg G W, Davey M W, Piper A A. Nucleic Acids Res. 1998;26:5009–5010. doi: 10.1093/nar/26.21.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, Umezu K, Kolodner R D. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 34.Devine S E, Chissoe S L, Eby Y, Wilson R K, Boeke J D. Genome Res. 1997;7:551–563. doi: 10.1101/gr.7.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieber M R. Am J Pathol. 1998;153:1323–1332. doi: 10.1016/s0002-9440(10)65716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y, Ferguson D O, Xie W, Manis J P, Sekiguchi J, Frank K M, Chaudhuri J, Horner J, DePinho R A, Alt F W. Nature (London) 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 37.Frank K M, Sharpless N E, Gao Y, Sekiguchi J M, Ferguson D O, Zhu C, Manis J P, Horner J, DePinho R A, Alt F W. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 38.Sharpless N E, Ferguson D O, O'Hagan R C, Castrillon D H, Lee C, Farazi P A, Alson S, Fleming J, Morton C C, Frank K, et al. Mol Cell. 2001;8:1187–1196. doi: 10.1016/s1097-2765(01)00425-7. [DOI] [PubMed] [Google Scholar]