Abstract
For many pathogens, the ability to regulate their replication in host cells is a key element in establishing persistency. Here, we identified a single point mutation in the gene for polynucleotide phosphorylase (PNPase) as a factor affecting bacterial invasion and intracellular replication, and which determines the alternation between acute or persistent infection in a mouse model for Salmonella enterica infection. In parallel, with microarray analysis, PNPase was found to affect the mRNA levels of a subset of virulence genes, in particular those contained in Salmonella pathogenicity islands 1 and 2. The results demonstrate a connection between PNPase and Salmonella virulence and show that alterations in PNPase activity could represent a strategy for the establishment of persistency.
The species Salmonella enterica is a classic example of the facultative intracellular pathogen that can cause acute and persistent infections (1–4). Typhoid fever is a serious manifestation of systemic salmonellosis in humans and involves an estimated 16.6 million new cases per year (2) with between 2% and 5% of the cases developing into the carrier state (3, 4).
Much of the knowledge of salmonellosis pathogenesis comes from work on the murine salmonellosis model (1, 5), and this model has been used to define bacterial virulence factors (1, 6, 7). An important virulence trait of Salmonella is its ability to invade and grow within host cells, and many of the genetic determinants essential for these processes have been well characterized (8–11). Invasion of epithelial cells requires the expression of a type III secretion apparatus, which translocates effector proteins from the bacterium to the host cell, inducing the cell to internalize the bacteria by bacterial-mediated endocytosis (8, 11). Proteins involved in this process are encoded by a large cluster of genes collectively termed Salmonella pathogenicity island 1 (SPI 1). The ability to grow intracellularly, once internalized within the host cell, requires the coordinate expression of additional sets of virulence factors (6), such as SPI 2 (1, 10).
Although chronic carriage of Salmonella is recognized as a significant complication that facilitates spread of the disease (2) and predisposes victims to additional clinical conditions (12, 13) the mechanism underlying persistency has remained poorly understood. We discovered the link between polynucleotide phosphorylase (PNPase) and chronic infection while characterizing a mutant of Salmonella typhimurium that was defective in its expression of virulence-associated AgfA fimbriae and known to establish persistent infection in BALB/c mice (14).
Materials and Methods
Bacterial Strains and Manipulations.
S. typhimurium MC1 (SR-11 variant χ3181) and MC2 (χ4665), a spontaneous mutant of MC1, are described in ref. 14. Strain VV 341 (ref. 15; hilA∷kan-334) was used as a source for the hilA-inactivating mutation, and Escherichia coli LM106 (16) was used as a source for pnp∷Tn5, and the resistance elements were transferred by phage transduction. Strains were grown in LB or on Luria agar with antibiotics as appropriate. For plasmid segregation experiments strains were transformed with plasmid pPIR (17) or pHSG422 (18). Transductions were carried out by using bacteriophages P1, P22 HTint 105, and KB1 (19–21). For array RNA extractions and protein labeling, cultures were grown at 37°C until the OD600 reached 1.0–1.2.
Mapping and Sequencing of MC2 Mutation.
A random Tn10camd insertion library from S. typhimurium 14028 (22) was transduced into MC1 by P22 transduction selecting for chloramphenicol-resistant colonies. Transductants were pooled and used to propagate phage KB1, which was then used to transduce MC2. Chloramphenicol-resistant colonies were screened for agf expression on Congo red plates as described (23). A single transductant (MC102) that restored wild-type agf expression in MC2 was identified. Chromosomal DNA flanking the transposon insertion was amplified by arbitrary-primed PCR, and the PCR fragments were sequenced by using the primer GACAAGATGTGTATCCACCTTAAC and the DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia). The sequence flanking the site of insertion was determined by primer walking and deposited in the GenBank database (accession no. AF399929).
Purification of PNPase and Activity Assay.
The whole (codons 1–721) and truncated (codons 1–614) forms of PNPase were amplified from MC1 chromosomal DNA. PCR fragments were amplified by using primer PNP CGGGATCCCGATGCGAGAAGATCGGGTATT with either primer PNP1 CCGCTCGAGCGGCTCGGCCTGTTCGCTCGC or PNP2 CCGCTCGAGCGGGATTTCGATGGTGGTGAAGG to amplify the whole and truncated forms of PNPase, respectively. PCR fragments were digested with BamHI and XhoI and ligated into pET21(c) (Novagen). Proteins were expressed in E. coli BL21 pnp∷Tn5 and purified as described by the manufacturer. PNPase activity of the purified proteins was determined by using a photometric assay (24).
Microarray Analyses.
The microarrays featured 3,169 of the 4,451 (71%) protein coding genes (CDS) identified in S. typhimurium LT2 genome sequence (25). Entire CDS were generated by PCR. In addition to the CDS-specific sequence, all primers (Sigma-Genosys) had a 19-nt 5′ overhang: TCCTAGGAGCTCTTCT for forward primers and TGCCTAGGGCTCTTCG for reverse primers. Taking advantage of the overhangs, all CDS were amplified in two steps. CDS were first amplified in a 25-μl PCR. A 50-fold dilution was then reamplified with universal primers annealing to the overhangs. Both steps were performed under the same conditions. For 100 μl the PCR mix contained 60 pmol of each primer, 1.5 mM MgCl2, 200 μM dNTPs, and 5 units of HotStarTaq polymerase (Qiagen, Chatsworth, CA). As template 10 ng of chromosomal DNA and 5 μl of diluted PCR products were used for the first and second step, respectively. Correct size of the PCR fragments and absence of nonspecific amplification products was confirmed by agarose gel electrophoresis. Validated PCR products were isopropanol-precipitated and resuspended in 50:1 of spotting solution (50% DMSO, 0.3 × SSC). DNA was applied to GAP I-slides (Corning) with an in-house built “Stanford” arrayer (26). Subsequently, slides were treated according to the manufacturer's instructions. cDNA from reference and mutant strains (see below) was labeled with Cy5 and Cy3, respectively. Labeled reference and test cDNA were then combined and hybridized to the microarrays. Each sample was run in quadruplicate and reproducibility was checked with sam software (http://www-stat.stanford.edu/∼tibs/SAM/). For details, see http://www.ifr.bbsrc.ac.uk/Safety/Microarrays/default.html#Protocols.
Northern Blot and RNA Decay Analyses.
Total RNA was prepared by using the FAST RNA-blue kit (Bio 101) according to the manufacturer's instructions and using cultures as above. Samples of 10 μg RNA were analyzed by Northern blotting as described (27). Internal fragments of the genes coding for 16S rRNA (nucleotides 150–651; GenBank accession no. Z49264), sipC (nucleotides 2880–2379; GenBank accession no. X82670), and ompA (nucleotides 408–912; GenBank accession no. X02006) were amplified by PCR, labeled with [α-32P]dCTP (Amersham Pharmacia) with a random prime labeling kit, and used as probes. A radioisotope imaging system (PhosphorImager 445SI; Molecular Dynamics) was used to detect radioactivity.
Protein Analyses.
The purity of proteins was confirmed by SDS/PAGE (20). Labeled S. typhimurium proteins were prepared as described (20). Bacteria were enriched from LB by centrifugation and transferred into methionine-free DMEM (GIBCO) supplemented with glutamine, Hepes, rifampicin, and 35S-Promix (Amersham Pharmacia) at final concentrations of 2 mM, 10 mM, 200 μg/ml, and 50 μCi/ml, respectively. The suspensions were incubated at 37°C for 30 min, the bacteria were separated by centrifugation, and supernatant proteins were precipitated with trichloro-acetic acid, washed with acetone, and dissolved in 0.3% SDS. Isoelectric focusing and second-dimension separations were carried out essentially as described (28). After completed runs, gels were fixed in 5% (vol/vol) methanol and 5% (vol/vol) acetic acid, dried, and exposed to x-ray film. N-terminal sequences were determined from protein bands separated on SDS/PAGE and blotted onto Hybond-P membranes (Amersham Pharmacia) by using a Procise cLC or HT sequences (Applied Biosystems) for Edman degradation (Protein Analysis Center, Karolinska Institute).
Site-Directed Mutagenesis.
A PCR approach was used to introduce a single point mutation (codon 600, GAA to TAA) in pnp on a 1.8-kb fragment amplified from MC1 chromosomal DNA. Two PCR fragments were generated by using the primer pairs 1 ATACCGTTACGCTGTAAACTGGCATGATGG/4 pTGACATGCATGCATGTCGCCTGGATAACAACCTGC and 2 CCATCATGCCAGTTTACAGCGTAAC GGTAT/3 ACG CTCGGTCGACGTCGGTTCTACCGATGTTCAGGT. The PCR products were purified and applied as template by using primers 3 and 4 to amplify the modified PNPase gene. The PCR product was digested with SalI/SphI and ligated into SalI/SphI-digested pCVD442 (29). This plasmid was then used to introduce the amber mutation into MC1 by allelic replacement. The mutation was confirmed by sequencing and immunoblot analysis with anti-PNPase antibodies (30), and the resulting strain was named MC71.
Invasion, Intracellular Replication, and Mouse Experiments.
Bacterial invasion of confluent monolayers of Madin–Darby canine kidney (MDCK) cells has been described (28). Bacterial replication in J774-A.1 mouse macrophage-like cells was assessed (20) by using strains carrying pPir (16). Cells were grown in Hepes-buffered RPMI supplemented with FBS [GIBCO; 10% (vol/vol) final concentration] and 10 μg/ml of gentamicin sulfate. For studying in vivo replication rates, we performed i.p. challenge of 6- to 8-week-old female BALB/c mice at doses of 105 bacteria/mouse (20). For persistency experiments we used oral challenge (14), at doses of 104 for MC1 and 105 for MC2 and MC71.
Results
Mapping a Locus Associated with AgfA Fimbrial Expression and Persistent Murine Salmonellosis.
S. typhimurium strain MC2 is capable of establishing chronic infections in BALB/c mice. Mice chronically infected with MC2 are unable to clear infection from the liver and spleen and secrete the bacteria for long periods, despite mounting an immune response that protects from challenge with virulent MC1 (14). Thus, the carrier state in mice resembles the human carrier state of typhoid fever (2–4).
MC2 was originally isolated as a spontaneous mutant of strain MC1 based on its defective expression of Agf fibers. Agf fibers are fimbrial organelles that bind Congo red and soluble fibronectin (14), contact phase proteins (31), and promote adhesion to the mouse small intestinal epithelial cells (23). As the mutation responsible for the altered agf expression was unknown, we used a transposon mapping approach to identify the locus responsible. This process involved phage transduction of random Tn10camd insertions from MC1 into MC2 (see Materials and Methods). Chloramphenicol-resistant transductants were assayed for AgfA expression on Congo red plates (21), and this screening identified a Tn10camd insertion that restored AgfA expression to MC1 levels in 70% of the transductants, when transduced from MC1 to MC2. In parallel, this AgfA-expressing transductant was shown to cause acute, but not persistent, infection in BALB/c mice (data not shown). Sequencing of the chromosomal DNA flanking the linking transposon positioned it within a gene homologous to yafK of E. coli, indicating that the mutation responsible for altered agf expression was linked to the yafK locus.
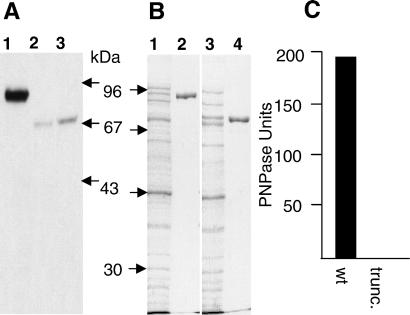
Sequence analysis of an 8-kb region centered on yafK from both MC1 and MC2 revealed a single point mutation in MC2 within a coding sequence showing 97.1% identity to PNPase of E. coli. This mutation altered codon 600 (GAA) to a stop codon (TAA), leading to a truncation of the PNPase protein from 721 aa to 599 aa. Western blot analysis using rabbit antibodies against E. coli PNPase confirmed that MC2 expressed a truncated form of PNPase (Fig. 1A, lanes 1 and 2).
Figure 1.
PNPase of S. typhimurium MC2 is truncated and lacks exonuclease activity. (A) Immunoblot analysis with rabbit antibodies directed against E. coli PNPase (29) was performed on whole-cell lysates of MC1 (lane 1), MC2 (lane 2), and MC71 (lane 3). The migration of marker proteins is indicated on the right with their molecular masses. (B) His-tagged fusion proteins of the whole and truncated form of S. typhimurium PNPase were overexpressed in E. coli (lanes 1 and 3, respectively), and the fusion proteins were purified (lanes 2 and 4). (C) Mean PNPase activities are shown. wt, wild type; trunc., truncated. Purified proteins were assayed for PNPase activity with a photometric assay for PNPase exonuclease activity.
Mutated PNPase Is Defective in 3′ → 5′ Phosphorolytic Activity.
PNPase is a key component in RNA maturation and degradation in E. coli. It acts as a 3′ to 5′ phosphorolytic exonuclease with polyadenylation activity, and as a subunit of the RNA degradosome complex (30, 32, 33). Predictions provided by the rps-blast program (http://www.ncbi.nlm.nih.gov/BLAST/) showed that the truncation in the MC2 pnp ORF would remove the S1 RNA binding domain from the protein. To evaluate whether the truncated PNPase of strain MC2 still retained exonuclease activity, the complete and truncated forms of the Salmonella protein were purified by the overexpression of C-terminal His-tag fusion proteins (Fig. 1B), and the purified proteins were assayed for exonuclease activity. Exonuclease activity was detected only with the whole form of the protein, indicating that MC2 expresses defective PNPase (Fig. 1C).
PNPase Acts as a Global Regulator of Virulence Gene Expression.
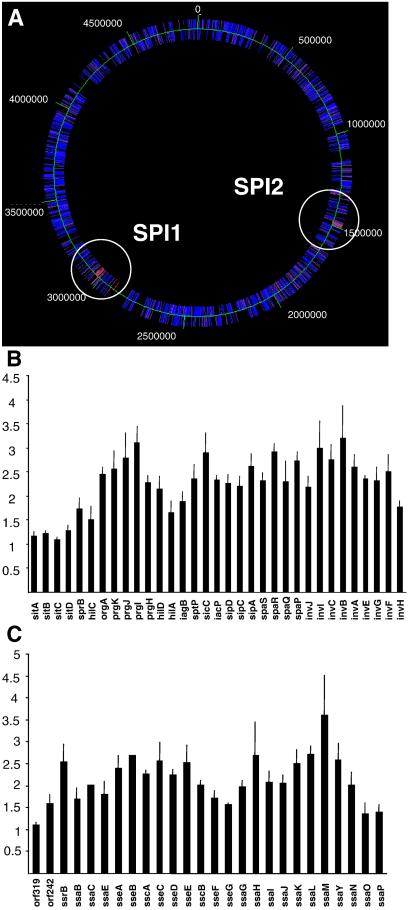
Based on the function of PNPase in E. coli, we anticipated that the mutation in pnp would have pleiotropic effects. We defined the genes controlled by PNPase activity by doing gene expression profiling on MC1 and MC2 (34). In short, bacteria were grown in LB, and RNA was prepared from MC1 and MC2, reverse-transcribed to cDNA, and fluorescently labeled. Labeled cDNA was used to probe 3,169 PCR-amplified ORFs of the S. typhimurium genome by microarray technology. This analysis showed that 87 ORFs had a significantly increased mRNA level in MC2 compared with MC1 (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). The current annotation of the S. typhimurium genome sequence assigns a function to 66 of these genes (25). The majority (50/66) were members of the SPI 1 or SPI 2 (Fig. 2) or represented unlinked genes functionally associated with the SPIs. Conversely, 22 genes showed reduced expression in MC2 (Table 2), indicating that the full expression of certain genes depends on PNPase activity in Salmonella.
Figure 2.
The pnp mutation of MC2 increases the expression of 87 S. typhimurium genes. (A) Color code presentation displaying relative amounts of mRNA originating from 3,169 separate ORFs positioned on the chromosome. Increasing mRNA levels in MC2 are shown on a scale from blue to red. Two clusters encoding increased mRNA are located in SPI 1 and SPI 2. (B and C) The actual increases for individual genes in MC2 are shown, where 1 represents a 1:1 ratio of MC2 to MC1 transcripts. The error bars indicate the standard error of the mean.
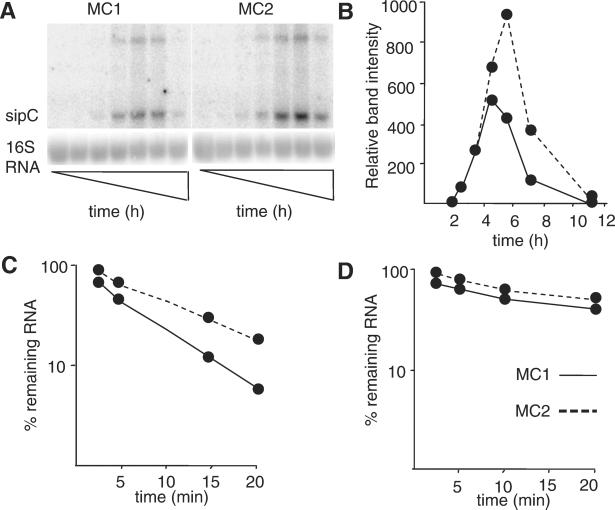
By comparing mRNA stability of the SPI 1 gene sipC and the control gene ompA in the presence of rifampicin, we observed that the half-life of sipC mRNA increased from 2.3 to 4 min in MC2, whereas the half-life of the ompA message was unaffected (Fig. 3).
Figure 3.
Effect of PNPase on the stability of the sipC transcript. (A) Expression of the sipC transcript was followed by Northern blot analysis during growth of MC1 and MC2 in LB at 37°C. Samples were taken after 2, 3, 4, 5, 7, and 11 h. (B) A PhosphorImage quantification of the sipC band intensities is shown. Quantification of sipC (C) or ompA (D) mRNA also was performed on samples taken at intervals after the addition of rifampicin (200 μg/ml). Band intensities of the transcripts were quantified and normalized to the amount of 16S rRNA. The signal strength at time 0 was set as 100%. (C and D) The percentage of remaining transcript at the different times after addition of rifampicin is shown.
The pnp Mutant MC2 Shows Increased Invasion and Intracellular Replication.
It is well established that SPI 1 and SPI 2 effector proteins are involved in invasion and intracellular growth (1, 8). As SPI 1 and SPI 2 expression was increased in MC2, we determined the concomitant effect on bacterial virulence characteristics. To show that any observed alteration in virulence-associated phenotype was caused solely by the mutation in pnp, we introduced the single point mutation into the wild-type MC1 strain, generating strain MC71 by allelic replacement. The expected truncation of PNPase was verified by immunoblotting (Fig. 1A, lane 3), and the mutation designed was confirmed by DNA sequencing. We then tested the abilities of MC1, MC2, and MC71 to express SPI 1 effector proteins, invade MDCK cells, and replicate in BALB/c mice and J774-A.1 cells.
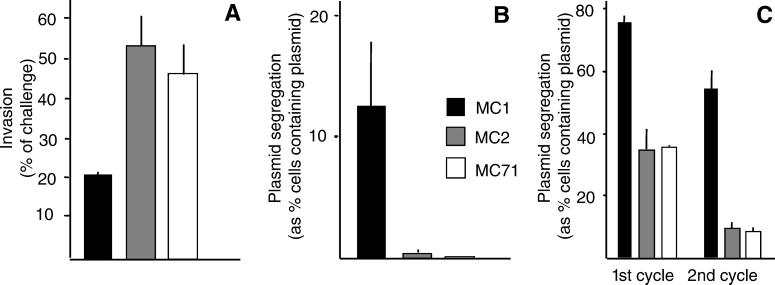
The ability of MC2 and MC71 to invade epithelial MDCK cells was significantly increased (Fig. 4A). This finding was consistent with the observation that selected SPI 1-encoded effector proteins were secreted at higher levels from MC2 and MC71 than MC1 when analyzed by SDS/PAGE (Fig. 5) and two-dimensional gels (data not shown).
Figure 4.
S. typhimurium MC2 and MC71 have enhanced invasion and intracellular growth capability. (A) Invasion of MDCK cells was performed as described (20). Replication in the spleens of BALB/c (B) or macrophage-like cell line J744-A.1 (C) was followed by comparing the segregation rates of plasmid pHSG412 or pPir as a measure of bacterial cell division.
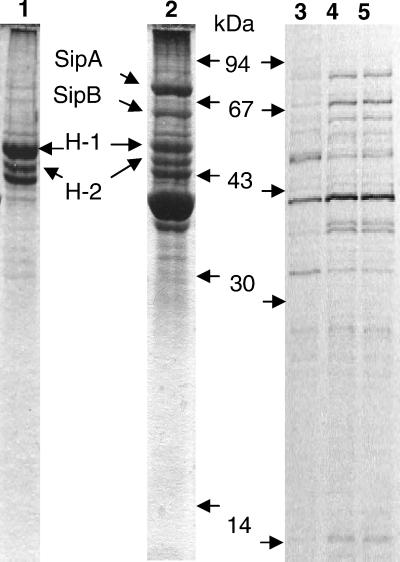
Figure 5.
SDS/PAGE profiles of proteins secreted from S. typhimurium. Lane 1 shows the profile of a MC1 hilA mutant; lane 2 shows wild-type MC1 stained with Coomassie blue. Proteins identified from lane 2 by N-terminal sequencing are indicated between lanes 1 and 2. H1 and H2 show the flagellin proteins of S. typhimurium. Lanes 3–5 show the autoradiograms, respectively, of proteins metabolically labeled and secreted from invasive cultures of MC1, MC2, and MC71. Labeling was performed in the presence of rifampicin (200 μg/ml). The positions of molecular mass markers are indicated between lanes 2 and 3.
Growth rates of the wild type and the bacterial mutants were assessed by measuring the segregation rate of a marker-carrying plasmid with temperature-sensitive replication. During growth above 30°C, each round of replication results in a proportion of bacterial cells that do not inherit the plasmid, which was then used to give a measure of bacterial replication.
PNPase is a defined cold-shock protein in E. coli and Bacillus subtilis, and pnp mutants replicate slower at low temperature (36–38). When grown in LB at 37°C or 39°C, the doubling times of MC1, MC2, and MC71 were the same (approximately 20 min) with no difference in plasmid segregation. At 15°C, however, the generation time of MC1 was 2.4 h, and those of MC2 and MC71 increased to 5.6 h, implying that PNPase also contributes to growth at low temperature in S. typhimurium.
In contrast, when experiments were performed in BALB/c mice after i.p. challenge, plasmid segregation was faster in MC2 and MC71 than in MC1 (Fig. 4B). This phenomenon was also observed during growth during serial passages in J774-A.1 murine macrophage-like cells (Fig. 4C). By measuring the actual growth yields (39), we observed that MDCK cells infected with MC2 or MC71 generated significantly more bacteria than cells infected with MC1 (data not shown). These data show that the pnp mutation increased bacterial growth rates during infection of mice or cultured cells.
The pnp Point Mutation in MC2 Is Sufficient for Establishing Persistency.
To establish the association between the pnp mutation and the establishment of chronic infection, BALB/c mice were challenged orally with MC1, MC2, and MC71, and the appearance of persistently infected mice was determined among the longtime survivors 28–30 days postchallenge (Table 1). At this stage, we could detect neither splenomegaly nor bacteria among the survivors challenged with MC1. In contrast, one-third of mice challenged with MC2 or MC71 showed splenomegaly and splenic bacteria. Mice persistently infected with MC2 and MC71 were also found to contain bacteria in their livers and gall bladders.
Table 1.
Establishment of persistent infections among survivors after challenge with S. typhimurium
| Strain | Proportion persistently infected* | Splenic bacterial load mean (SD) |
|---|---|---|
| MC1 | 0/44 (0%) | — |
| MC2 | 43/144 (30%) | 2.0 × 104 (3.4 × 104) |
| MC71 | 29/79 (37%) | 1.3 × 104 (1.3 × 104) |
BALB/c mice (female, 6–8 weeks of age) were challenged orally (14) with a dose corresponding to LD30. Mice surviving the acute phase of the infection (7–20 days postinfection) were killed 28–30 days postinfection, and their spleens were removed for enumeration of bacteria.
Persistent infection was defined by the presence of splenic bacteria and splenomegaly (mean spleen weight 0.97 ± 0.34 g, as compared to 0.16 ± 0.05 g for sterile spleens recovered from the survivors).
Discussion
Many important bacterial pathogens, such as Brucellae, Mycobacteriae, and Salmonellae, have the ability to establish latent or persistent infections (2, 40–42), yet the bacterial strategies for resisting sterilizing immunity are largely unknown. Model infections with Brucella and Mycobacterium show the process to be complex and including several apparently unconnected functions (40–43). For example, cyclopropanated mycolic acids and isocitrate lyase are involved in peristent Mycobacterium tuberculosis infection in mice (40, 42), whereas glycine dehydrogenase and nitrate reductase have been implicated in persistency of Mycobacterium bovis (44, 45). The recent identification of the response regulator MprA (46) as essential for M. tuberculosis persistency implies that persistency is a multifactorial process that relies both on dedicated effector genes and specific gene regulation. Here we demonstrate that a single point mutation in the gene coding for PNPase affects the level of virulence gene transcripts in S. enterica and enables the establishment of persistent infections in BALB/c mice.
Altering the stability of transcripts is one mechanism by which bacteria alter gene expression and adapt to environmental change (27). For example, in laboratory strains of E. coli one function of PNPase is to degrade cold-shock protein mRNAs, ensuring their transient expression during bacterial cold adaptation (36–38). Conversely, RNase R* of Shigella flexneri has been reported to be required for the expression of the invasion factors IpaB, IpaC, and IpaD (47). Our results with S. enterica show that inactivation of PNPase results in decreased replication efficiency at low temperature, whereas at 37°C or 39°C there is no effect on replication in vitro. An additional pnp-associated phenotype was the inability to bind Congo red at low growth temperature; the wild-type strain MC1 formed rugous dark colonies on Congo red plates, and the mutants MC2 and MC71 formed pale smooth colonies. As binding of this dye correlates with the expression of cold-induced Agf fibers (48), the observation suggests that PNPase activity is required for Agf fiber expression. Indeed, our preliminary observations with agf-lacZ fusions showed a lack of β-galactosidase induction in MC2 and MC71 at low growth temperature.
The fact that a single point mutation in pnp enabled S. enterica to establish persistent infection in mice suggests additional functions for PNPase at physiological temperature. Microarray analyses showed that the genes that were most affected by the pnp mutation at 37°C were contained in two pathogenicity islands, SPI 1 containing genes for invasion and SPI 2 containing genes for intracellular growth. This increase in virulence mRNA also appeared translated into an increase in the corresponding virulence characters. The fact that sipC mRNA of SPI 1 has an increased half-life in the pnp mutant suggests a direct link between PNPase activity and virulence gene expression.
Thus, PNPase seems to repress SPI 1 and SPI 2 expression, but is required for AgfA fiber expression, which leads us to ask why certain mRNAs would be more sensitive to PNPase than others. The exoribonuclease activity of PNPase is known to halt stable stem-loop structures (49), whereas the DNA contained in SPI 1 and SPI 2 are believed to have been acquired through recent horizontal gene transfer events (1) and show significantly higher AT percentage than the S. typhimurium genome in general. Thus, a possible explanation for the PNPase sensitivity of SPI mRNA might originate from infrequent stable stem-loop structures, possibly reflecting the proportion of AT.
Recently, mutants of S. typhimurium have been isolated that show increased replication rates in cultured cells (20, 35). Interestingly, two of the mutations thus identified inactivate the defined virulence gene regulators phoP/phoQ and spvR (35), both of which participate in bacterial intracellular replication (6, 7). In this context it is interesting to note that inactivation of PNPase also resulted in increased intracellular replication. The expression of PNPase is autoregulated, as well as responding to environmental changes in E. coli (36–38). From this finding one may suggest a role for PNPase in degrading mRNAs that adapt the bacteria for host milieu, as does PhoP/PhoQ and SpvR (6, 7), and that a disturbance in such a regulation leads to altered expression of selected virulence functions, which causes altered infection pathogenesis.
Supplementary Material
Acknowledgments
Thanks to Isabelle Hautefort for commenting on the text and to the Oxford laboratory of Chris Higgins for the gift of anti-PNPase antibodies. J.C.D.H., S.L., and A.T. are funded by the Biotechnology and Biological Sciences Research Council. M.R., M.O.C., and J.C.D.H. are grateful for funding from a Training and Mobility of Researchers program of the European Union (Contract Number ERBFMRXCT9) and for Vetenskapsrådet (Medicinska Forskningsrådet) Grant K2000-16X-11593-05A.
Abbreviations
- PNPase
polynucleotide phosphorylase
- SPI
Salmonella pathogenicity island
- CDS
coding genes
- MDCK
Madin–Darby canine kidney
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF399929).
References
- 1.Hansen-Wester I, Hensel M. Microb Infect. 2000;3:549–559. doi: 10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 2.Pang T, Bhutta Z A, Finlay B B, Altwegg M. Trends Microbiol. 1995;3:253–255. doi: 10.1016/s0966-842x(00)88937-4. [DOI] [PubMed] [Google Scholar]
- 3.Buchwald D S, Blaser M J. Rev Infect Dis. 1984;6:345–356. doi: 10.1093/clinids/6.3.345. [DOI] [PubMed] [Google Scholar]
- 4.Edelman R, Levine M. Rev Infect Dis. 1986;8:329–349. doi: 10.1093/clinids/8.3.329. [DOI] [PubMed] [Google Scholar]
- 5.Carter P B, Collins F M. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements M, Eriksson S, Tezcan-Merdol D, Hinton J C D, Rhen M. Ann Med. 2001;33:178–185. doi: 10.3109/07853890109002075. [DOI] [PubMed] [Google Scholar]
- 7.Ohl M E, Miller S I. Annu Rev Med. 2001;52:259–274. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- 8.Collazo C M, Galán J E. Gene. 1997;192:51–59. doi: 10.1016/s0378-1119(96)00825-6. [DOI] [PubMed] [Google Scholar]
- 9.Richter-Dahlfors A, Buchan A M, Finlay B B. J Exp Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hensel M, Shea J E, Waterman S R, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang F C, Holden D W. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 11.Brumell J H, Steele-Mortimer O, Finlay B B. Curr Biol. 1999;9:R277–R280. doi: 10.1016/s0960-9822(99)80178-x. [DOI] [PubMed] [Google Scholar]
- 12.Caygill C P, Braddick M, Hill M J, Knowles R L, Sharp J C. Eur J Cancer Prev. 1995;4:187–193. doi: 10.1097/00008469-199504000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Caygill C P, Hill M J, Braddick M, Sharp J C. Lancet. 1994;343:83–84. doi: 10.1016/s0140-6736(94)90816-8. [DOI] [PubMed] [Google Scholar]
- 14.Sukupolvi S, Edelstein A, Rhen M, Normark S J, Pfeifer J D. Infect Immun. 1997;65:838–842. doi: 10.1128/iai.65.2.838-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hueck C J, Hantman M J, Bajaj V, Johnston C, Lee C A, Miller S I. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 16.McMurry L M, Levy S B. J Bacteriol. 1987;169:1321–1324. doi: 10.1128/jb.169.3.1321-1324.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pósfai G, Koob M D, Kirkpatrick H A, Blattner F R. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulig P A, Doyle T J. Infect Immun. 1993;61:504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 20.Eriksson S, Björkman J, Borg S, Syk A, Pettersson S, Andersson D I, Rhen M. Cell Microbiol. 2000;2:239–250. doi: 10.1046/j.1462-5822.2000.00051.x. [DOI] [PubMed] [Google Scholar]
- 21.McIntire S. J Bacteriol. 1974;117:907–908. doi: 10.1128/jb.117.2.907-908.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott T, Roth J R. Mol Gen Genet. 1988;213:332–338. doi: 10.1007/BF00339599. [DOI] [PubMed] [Google Scholar]
- 23.Sukupolvi S, Lorenz R G, Gordon J I, Bian Z, Pfeifer J D, Normark S J, Rhen M. Infect Immun. 1997;65:5320–5325. doi: 10.1128/iai.65.12.5320-5325.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontanella L, Pozzuolo S, Costanzo A, Favaro R, Deho G, Tortora P. Anal Biochem. 1999;269:353–358. doi: 10.1006/abio.1999.4042. [DOI] [PubMed] [Google Scholar]
- 25.McClelland M, Sanderson K E, Spieth J, Clifton S W, Latrielle P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Nature (London) 2001;423:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 26.Thompson A, Lucchini S, Hinton J C D. Trends Microbiol. 2001;9:154–156. doi: 10.1016/s0966-842x(01)01977-1. [DOI] [PubMed] [Google Scholar]
- 27.Georgellis D, Barlow T, Arvidson S, von Gabain A. Mol Microbiol. 1993;9:375–381. doi: 10.1111/j.1365-2958.1993.tb01698.x. [DOI] [PubMed] [Google Scholar]
- 28.Tezcan-Merdol D, Nyman T, Lindberg U, Haag F, Koch-Nolte F, Rhen M. Mol Microbiol. 2001;39:606–619. doi: 10.1046/j.1365-2958.2001.02258.x. [DOI] [PubMed] [Google Scholar]
- 29.Bloomfield I C, Vaughn V, Rest R F, Eisenstein B I. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 30.Py B, Causton H, Mudd E A, Higgins C F. Mol Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 31.Herwald H, Mörgelin M, Olsén A, Rhen M, Dahlbäck B, Müller-Esterl W, Björck L. Nat Med. 1998;4:298–302. doi: 10.1038/nm0398-298. [DOI] [PubMed] [Google Scholar]
- 32.Liou C G, Jane W N, Cohen S N, Lin N S, Lin-Chao S. Proc Natl Acad Sci USA. 2001;98:63–68. doi: 10.1073/pnas.011535498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coburn G A, Miao X, Briant D J, Mackie G A. Genes Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucchini S, Thompson A, Hinton J C D. Microbiology. 2001;147:1403–1414. doi: 10.1099/00221287-147-6-1403. [DOI] [PubMed] [Google Scholar]
- 35.Cano D A, Martinez-Moya M, Pucciarelli M G, Groisman E A, Casadesús J, García-Del Portillo F. Infect Immun. 2001;69:6463–6474. doi: 10.1128/IAI.69.10.6463-6474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Bechhofer D H. J Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zangrossi S, Briani F, Ghisotti D, Regonesi M E, Tortora P, Deho G. Mol Microbiol. 2000;36:1470–1480. doi: 10.1046/j.1365-2958.2000.01971.x. [DOI] [PubMed] [Google Scholar]
- 38.Robert-Le Meur M, Portier C. EMBO J. 1992;11:2633–2641. doi: 10.1002/j.1460-2075.1992.tb05329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Libby S J, Lesnick M, Hasegawa P, Weidenhammer E, Guiney D G. Cell Microbiol. 2000;2:49–58. doi: 10.1046/j.1462-5822.2000.00030.x. [DOI] [PubMed] [Google Scholar]
- 40.Glickman M S, Cox J S, Jacobs W R., Jr Mol Cell. 2000;5:717–724. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- 41.Hong P C, Tsolis M R, Flicht T A. Infect Immun. 2000;68:4102–4107. doi: 10.1128/iai.68.7.4102-4107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinney J D, Honer zu Bentrup K, Munoz-Elias A, Chen B, Chan W T, Swenson D, Sacchettini J C, Jacobs W R, Jr, Russell D G. Nature (London) 2000;406:683–685. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 43.Ramakrisham L, Fiederspel N A, Falkow S. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 44.Lim A, Eleuterio M, Hutter B, Murugasu-Oei B, Dick T. J Bacteriol. 1999;181:2252–2256. doi: 10.1128/jb.181.7.2252-2256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fritz C, Maass S, Kreft A, Bange F-C. Infect Immun. 2002;70:289–291. doi: 10.1128/IAI.70.1.286-291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zahrt T C, Deretic V. Proc Natl Acad Sci USA. 2001;98:12706–12711. doi: 10.1073/pnas.221272198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng Z-F, Zou Y, Li Z, Rudd K E, Deutscher M P. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 48.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 49.Spickler C, Mackie G A. J Bacteriol. 2000;182:2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.