Abstract
Transcriptional interference between genes and the regulatory elements of simple eukaryotes such as Saccharomyces cerevisiae is an unavoidable consequence of their compressed genetic arrangement. We have shown previously that with the tandem arranged genes GAL10 and GAL7, inefficient transcriptional termination of the upstream gene inhibits initiation of transcription on the downstream gene. We now show that transcriptional interference can occur also with S. cerevisiae RNA polymerase II genes arranged convergently. We demonstrate that when the GAL10 and GAL7 genes are rearranged in a convergent orientation, transcriptional initiation occurs at full levels. However, as soon as the two transcripts begin to overlap, elongation is restricted, resulting in a severe reduction in steady-state mRNA accumulation. This effect is observed only in cis arrangement, arguing against RNA-interference effects acting on the potential generation of antisense transcripts. These data reinforce the necessity of separating adjacent RNA polymerase II transcription units by efficient termination signals.
The genomic sequence of Saccharomyces cerevisiae is highly compact, with over 70% represented by ORFs (1). This high gene density is largely a result of the short intergenic regions that average 618 bp apart for diverging promoters and 517 bp for tandem (head-to-tail) genes using promoter/terminator combinations (2). Convergent genes average only 326 bp apart (2), with several examples of less than 300 bp (3–6). Such a limited intergenic sequence would allow only ≈150 bp per gene for proper termination outside of the neighboring ORF, which is less sequence than the few termination regions defined thus far in S. cerevisiae genes (7–9). Analysis of adjacent gene sets in S. cerevisiae determined that less than 6% of coexpressed, adjacent genes are in a convergent orientation, with a bias to divergent transcription (10). These statistics suggest that there may be evolutionary pressure to select against convergently arranged, cotranscribed genes (2).
Transcription of RNA polymerase II (PolII) in S. cerevisiae has been studied extensively, with both the mechanism of initiation and elongation recently described in atomic detail (11, 12). The mechanism for termination and coupled mRNA 3′-end processing is understood less completely. However, mutations in cleavage/polyadenylation factors prevent properly regulated termination, highlighting the interdependence of these functions (13). The polyadenylation signals in budding yeast genes are less sequence-specific than in mammalian systems. In mammals, an almost invariant AAUAAA hexonucleotide is located 15–20 nt upstream of the cleavage site, which works in conjunction with a less well defined GU- or U-rich element downstream to promote proper 3′-end formation. In S. cerevisiae, a more degenerate poly(A) signal is assisted by an upstream efficiency element (consensus UAUAUA) to promote 3′-end formation. Additionally, at 15–30 nt downstream of the cleavage site a positioning element (consensus AAAAAA or AAUAAA) appears to direct cleavage at the preferred site Y(A)n (see ref. 14 for review).
With such a densely packed genome it is not surprising that interference has been detected between transcribing genes (15–17). Transcriptional interference (TI) may occur when an elongating polymerase fails to terminate properly and thus reads into a cotranscribed downstream transcription unit, reducing promoter activity. TI also has been documented to affect the proper use of both ARS and CEN elements and in so doing inhibits either replication or chromosomal segregation in mitosis (18–20). Sequence elements that mediate enhanced meiotic gene conversion also are susceptible to TI (21). At the nascent level, the Schizosaccharomyces pombe genes nmt1 and nmt2 were shown to transcribe into two poorly defined convergent downstream ORFs (22) with little detectable interference. These nmt genes both are induced to transcribe at high levels, whereas the convergent genes avn2 and gut2 are ubiquitously expressed at much lower levels. The possibility that these convergent genes are expressed separately remains to be determined. In contrast, steady-state analysis in S. cerevisiae has shown that the elimination or constitutive activation of one promoter in a convergent pair of genes often affects the steady-state levels of mRNA generated from the opposing gene (4, 15).
In the tightly regulated metabolic pathway of galactose fermentation, the tandem GAL10 and GAL7 genes are affected by TI (16). A fraction of polymerases elongate beyond GAL10 and have an inhibitory effect on the initiation of the downstream gene, GAL7. Furthermore mutations of the GAL10 poly(A) signal prevent all PolII termination and thus have a more drastic effect on the levels of GAL7 gene expression. Interestingly, GAL7 expression is recoverable by overexpression of Gal4p (23). Presumably TI blocks the access of the Gal4p transcription factor to the GAL7 promoter and in so doing prevents transcriptional activation. The coinduction of these genes by galactose makes an ideal system to study TI in different gene orientations.
In these studies we have manipulated the orientation of the GAL10 and GAL7 genes to model the effect of convergent, cotranscribed genes. By eliminating the intergenic region of these convergent genes, we have investigated the effects of improper termination on convergent transcription. Our results graphically illustrate the need for efficient PolII termination. In its absence, inhibition of both genes occurs by a mechanism we refer to as transcriptional collision.
Materials and Methods
S. cerevisiae Strain.
All experiments use ΔGAL S. cerevisiae strain 6-1/13 (MATα his3Δ200 met15Δ0 trpΔ63 ura3Δ0 gal10-7∷URA3) (16). The strain was grown on synthetic complete (SC) medium lacking uracil and supplemented with 2% raffinose.
Plasmids.
pYC10-7 (wild type, WT) was based on Ycplac22 (16). pYC10-7Con (Con) was created by XbaI digestion of pYC10-7 and partial digestion with EcoRV. The 7,658-bp product (lacking GAL7) was isolated. A GAL7 fragment was generated by PCR with a unique XhoI site 103 bp after the poly(A) site (see Primers).
pYC10-7Fus (Fus) was created by ligating long-range PCR products generated from Con by using Pfu DNA polymerase (Stratagene) and kinased primers Fuse7 and Fuse10 located directly inside the ORFs for both genes (see Primers). The 209-bp GAL7 poly(A) site fragment (GAL7, 1,118–1,327) in the forward orientation for GAL7 was inserted into the SacI site of the vector backbone upstream of the GAL10 promoter (−442).
pYC10-7FusΔTATAGAL7 (Fus-Δ7P) had a 9-bp deletion of the TATA box, 5′-ATATAAAA- 3′ (GAL7, −179 to −188), in the Fus construct generated by long-range PCR with primer ΔTATAa and ΔTATAb (see Primers).
pYC10-7FusΔUASGAL10 (Fus-Δ10P) had the EcoRI fragment of Fus replaced with the EcoRI fragment from the pΔ-UASG10 plasmid (16), which created Fus with a deletion that eliminates −136 to −414 of GAL10, removing all UAS sites.
pYC10-7FusΔTATAGAL7/ΔUASGAL10 (Fus-Δ7P/Δ10P) was generated by replacing the EcoRI fragment from the Fus-Δ7P plasmid with the EcoRI fragment from Fus-Δ10P.
pYC10-7FusΔTATAGAL7&ΔUASGAL10 (Fus-Δ7P&Δ10P) was a cotransformation of Fus-Δ7P and Fus-Δ10P. See Growth and Transformation of S. cerevisiae Strains below.
Primers.
The primer used were: Con GAL7 insert, GAL7+100 (CTCGAGACTTACAAGCTGCATTGTATTC) and UpGAL7 (GGAGTTCATTTCGTTACTTTTG); Fus construct, Fuse7 (CCACTTTCTTTTTACAGTCTTTGTAGATAATG), and Fuse10 (GCTGGCAAATCAGGAAAATCTGTAGAC); Δ7P construct, Δ7TATAa (CTTTGCTAGCCAAACTAACTGAACATAG), and Δ7TATAb (GCAGGTCGGAAATATTTATGGGCATTA).
Growth and Transformation of S. cerevisiae Strains.
Strain 6-1/13 was grown on SC medium lacking uracil with 2% raffinose (Sigma). Strains transformed with the plasmids were plated onto SC plates lacking uracil and tryptophan (24). Fus-Δ7P&Δ10P was created by transforming 6-1/13 carrying the Fus-Δ7P plasmid with Fus-Δ10P plasmid and selecting on URA−, TRP− SC plates with 2% galactose (Sigma) and 20 μg/ml ethidium bromide. All growth was with URA−, TRP− SC medium, and 2% galactose with this strain.
Before transcriptional run-on (TRO) analysis, strains (except WT and Fus-Δ7P&-Δ10P) were grown on URA−, TRP− SC medium and 2% raffinose until early log phase and then induced with 2% galactose for 30–60 min. WT was grown in URA− SC medium and 2% raffinose. Fus-Δ7P&-Δ10P were grown on URA−, TRP− SC medium, and 2% galactose.
M13 Probe Constructs.
Single-stranded DNA probes were made (see Table 1) by cloning PCR-generated fragments (using Pfu DNA polymerase, Stratagene) from Con into the double-digested XbaI and EcoRI sites of M13mp18 and M13mp19, which generated probes for the sense strands of both GAL7 and GAL10. The M13 control probe (m) contained no insert. The positive control actin probe (a) had a 567-nt fragment containing 277–844 nt 3′ of the ACT1 start codon inserted into the HincII site of M13mp18.
Table 1.
TRO M13 probes
| Probe | Location | Size, nt |
|---|---|---|
| A | GAL7: −401/−212 | 189 |
| B | GAL7: 73/252 | 179 |
| C | GAL7: 638/793 | 155 |
| D | GAL7: 800/944 | 144 |
| E | GAL7: 938/1,119 | 181 |
| F | GAL7: 1,110/1,294 | 184 |
| G | GAL7: 1,184/1,354 | 170 |
| H | GAL7: 1,266/1,362 | 177 |
| GAL10: 2,406/2,325 | ||
| I | GAL10: 2,245/2,055 | 190 |
| J | GAL10: 2,070/1,884 | 186 |
| K | GAL10: 1,885/1,727 | 158 |
| L | GAL10: 1,720/1,572 | 148 |
| M | GAL10: 1,537/1,379 | 158 |
| N | GAL10: 330/152 | 178 |
| O | GAL10: −187/−371 | 184 |
TRO Analysis.
TRO analysis was performed as described (25) except that 100-ml cultures grown to an OD600 of ≈0.1 were induced with 2% galactose. Additionally, the TRO reaction was allowed to proceed for 5 min and then washed briefly with AE buffer (50 mM NaAc/10 mM EDTA, pH 5.0). All TRO analyses were reproduced in at least three independent experiments.
Northern Blot Analysis.
Total RNA was analyzed (16). The Northern probe was a 1.5-kb fragment generated from the WT plasmid by BglII and SalI digestion, which contains 427 bp of 5′ of GAL7 and 403 bp of 3′ of GAL10. The ACT1 probe was a 567-bp fragment (277–844 bp 3′ of the ACT1 start codon) from the ACT1 gene of S. cerevisiae.
Results
Transcription of the GAL10 and GAL7 Genes Arranged in Convergent Orientation.
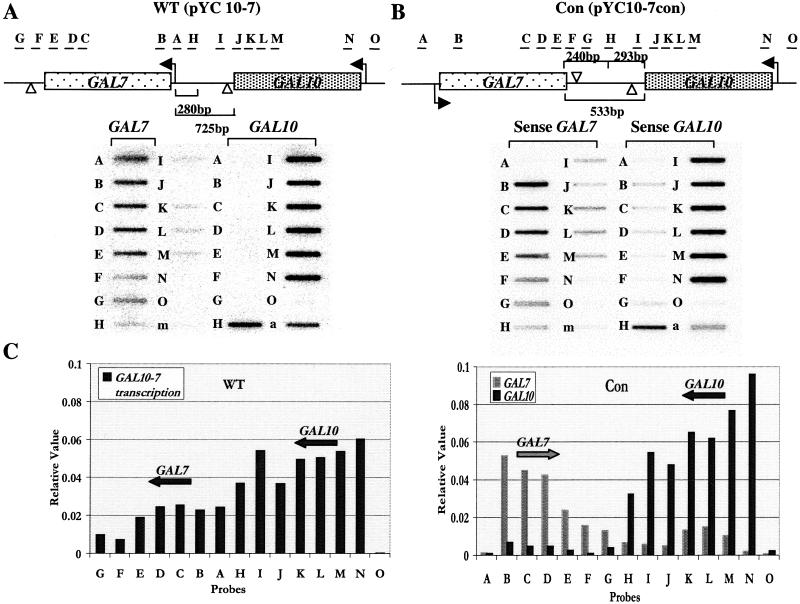
The GAL10 and GAL7 genes are organized naturally in a tandem arrangement on chromosome II. A previous study determined the transcription profile for nascent transcription in these genes for the WT conformation. These results were obtained by TRO analysis of a centromeric plasmid containing the GAL10-GAL7 genes (pYC10-7) transformed into the ΔGAL strain (16). This TRO analysis is repeated in Fig. 1A. As indicated, higher nascent transcription is seen over the GAL10 than GAL7 gene. Also, most transcription fails to terminate beyond the GAL10 poly(A) signals and reads into and accumulates over the GAL7 promoter. In the previous study (16) analysis of a plasmid containing the GAL10-GAL7 genes with the GAL7 promoter inactivated revealed that most GAL10 transcription terminates upstream of the GAL7 transcription start site. Consequently, run-on signals over the GAL7 gene shown in Fig. 1A are likely to derive from transcription initiation on the GAL7 promoter.
Figure 1.
TRO analysis of transformed GAL10-7 plasmids. (A) TRO analysis of pYC10-7 (WT) transformed cells. The diagram shows the tandem arrangement of the GAL10 and GAL7 genes, with the direction of transcription indicated with an arrow. The position of the single-stranded M13 probes used are shown and are as described in Table 1. Probe m is an M13 probe without an insert serving as a negative hybridization control. Probe a is an M13 probe with a fragment of the S. cerevisiae actin gene serving as a positive transcription control. The distances between the ORFs of GAL10 and GAL7 in the WT orientation are labeled. Open arrowheads indicate polyadenylation sites. The GAL7 promoter region is indicated also. (B) TRO analysis of pYC10-7Con (Con)-transformed cells. The diagram shows convergent orientation of GAL10 and GAL7, with the intergenic region's distance labeled. This construct includes 240 bp downstream of GAL7 and 293 bp downstream of GAL10 to make a hybrid intergenic region. Sense and antisense single-stranded M13 DNA probes were selected as appropriate. (C) Quantitation of the data shown in A and B. Signals were quantified in a PhosphorImager (Molecular Dynamics). The values were corrected for background hybridization and U content in each probe. Signals were normalized to the actin positive transcription probe a on each filter to allow direct comparison of transcription levels.
To investigate the effect of concurrent, convergent transcription, we inverted the GAL7 gene to generate pYC10-7Con (Con, Fig. 1B). The natural polyadenylation sites and a portion of the GAL10-7 intergenic region were maintained. The intergenic region between GAL10 and GAL7 (from the 3′ end of the GAL10 ORF to the 5′ end of the GAL7 ORF) is 725 bp including the 280-bp promoter region of GAL7. In Con, the intergenic region is 533 bp from the 3′ end of the GAL10 ORF to the 3′ end of the GAL7 ORF, which includes 240 bp beyond the 3′ end of the GAL7 ORF and 293 bp downstream of the 3′ end of the GAL10 ORF.
TRO analysis of the ΔGAL yeast strain transformed with Con yields a nascent termination profile similar to the WT situation with a few notable exceptions. Transcription in the GAL10 and GAL7 genes appeared to terminate over the same probes regardless of orientation. However, the level of read-through transcription from GAL10 decreased in Con (compare the GAL10 probe G of Con with the GAL7 probe A of WT). GAL7 transcription terminated shortly after the WT poly(A) site in both orientations. Additionally, the levels of transcription appeared higher for both GAL10 and GAL7 genes in Con. Compare the levels of GAL10 and GAL7 nascent transcripts to the endogenous actin probe (a) for WT and Con plasmids. In particular for GAL7, there was a 2-fold increase in the level of nascent transcription from GAL7, which may be the result of alleviation of promoter occlusion occurring from read-through transcription beyond the GAL10 poly(A) site into the GAL7 promoter. The relative increase in GAL7 gene expression is reflected by the steady-state mRNA analysis shown in Fig. 2 (lanes 1 and 2), where GAL7 mRNA levels increase 2.3-fold with the Con construct. However, we also note that GAL10 mRNA levels increase 1.7-fold in the Con construct as compared with WT. In this case TI is unlikely to be important, because no TRO signal is detectable over the GAL10 promoter (probe O) in WT. It therefore is possible that other factors such as supercoiling effects may be involved.
Figure 2.
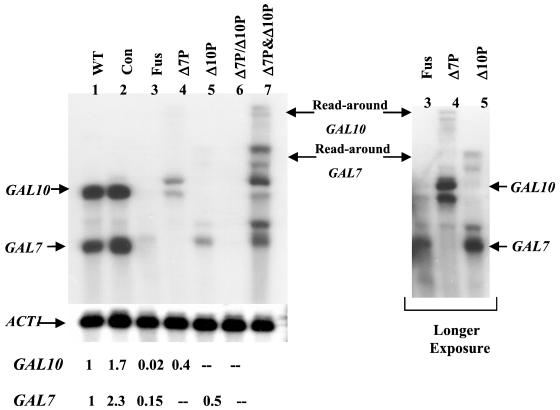
Steady-state analysis of GAL constructs. Northern blot with total RNA from pYC10-7 (WT, lane 1), pYC10-7Con (Con, lane 2), pYC10-7Fus (Fus, lane 3), Fus-Δ7P (lane 4), Fus-Δ10P (lane 5), Fus-Δ7P/Δ10P (lane 6), and Fus-Δ7P&Δ10P (lane 7). Longer exposure shows the read-around bands for Fus-Δ7P (lane 4) and Fus-Δ10P (lane 5). The filter was striped and reprobed with ACT1 as a loading control. Quantitated levels of steady-state GAL10 and GAL7 mRNA are corrected for actin and compared by using the steady-state level of GAL10 and GAL7 mRNA from WT as 1.
Removal of Termination Signals Promotes Transcriptional Collision in Convergent Transcription.
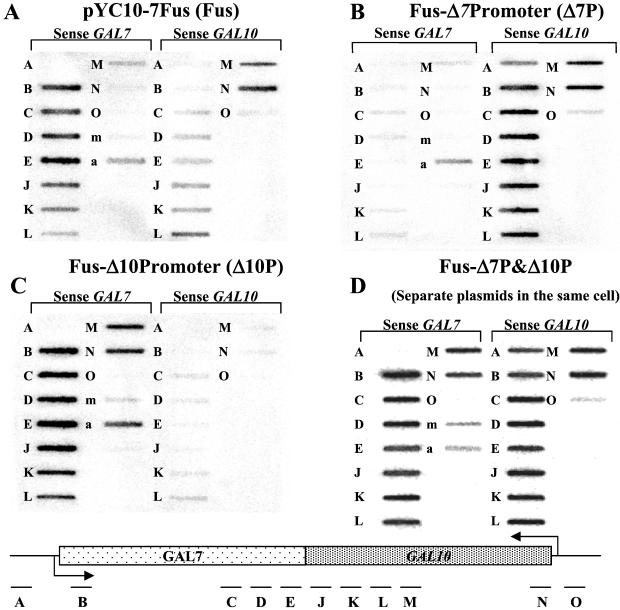
To test the effect of TI in the convergent orientation, the entire intergenic region was deleted from Con to make the plasmid pYC10-7Fus (Fus), effectively fusing together the GAL7 and GAL10 genes. The deleted GAL7 poly(A) site was reinserted into the vector backbone upstream of GAL10 in the forward orientation for GAL7, because this poly(A) signal is known to function bidirectionally. We anticipated that it would act to polyadenylate both the GAL10 and GAL7 transcripts in Fus (Fig. 3A). This construct left the intact ORFs of each gene but eliminated any termination or processing signals beyond the 3′ end of either ORF. In so doing, sequences complimentary to the probes F, G, H, and I were deleted.
Figure 3.
TRO analysis of transformed plasmids as indicated (A–D). The positions of single-stranded M13 probes used are shown in the diagram at the bottom of the figure and are as described in Table 1.
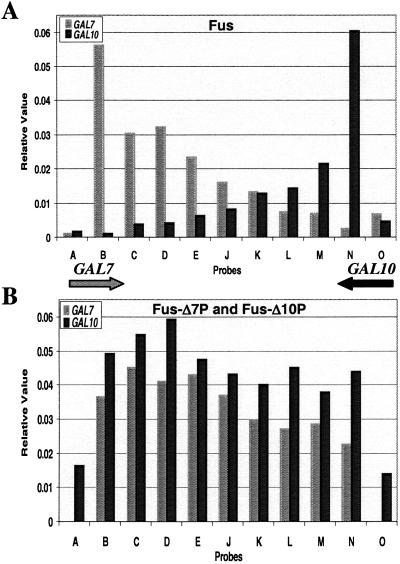
TRO analysis of ΔGAL yeast transformed with Fus showed dramatic TI when these genes were transcribed simultaneously (Figs. 3A and 4A). For GAL10, transcription signals in Fus showed full levels at initiation but then dropped abruptly with probe M and more gradually to background levels from probe L to B. The initiation level of GAL7 transcription was similar to Con, but as with GAL10 signals it abruptly dropped at probe C. However, they then continued at a significant level, leading to a more gradual decline through the GAL7 ORF, tailing off beyond the region at which it normally would contain a poly(A) site in the WT construct. Thus TI of convergent transcripts seems to have no effect on initiation but acts during transcriptional elongation. We refer to this process as transcriptional collision. Steady-state analysis of GAL10 and GAL7 mRNA produced by the Fus construct is entirely consistent with the TRO analysis. Thus GAL10 mRNA levels were reduced 20-fold and GAL7 levels 4-fold as compared with the single transcription unit constructs Fus-Δ7P and Fus-Δ10P, respectively (Fig. 2, compare lane 3 with lanes 4 and 5). It is clear that transcriptional collision of convergent transcripts results in a drastic reduction in gene expression.
Figure 4.
Quantitation of Fus and Fus-Δ7P and Fus-Δ10P transcription profiles. (A) TRO analysis of Fus transformed plasmid. The signals were corrected as described for Fig. 1. Arrows indicate the directions of transcribing polymerases. (B) TRO analysis of Fus-Δ7P and Fus-Δ10P. In the Fus-Δ10P plasmid (giving GAL7 transcription), the O probes have been deleted with the deletion of the GAL10 promoter. In the GAL10 transcription profile, decrease in GAL10 transcription over probe A is likely the result of cryptic polyadenylation that is seen at the steady-state level (Fig. 2, lane 4). Also, the above background levels of transcription over probe O (before the GAL10 promoter) are most likely to be the result of read-around transcription seen at steady state (Fig. 2, lane 4). The single-stranded M13 probes used are described in Table 1.
Transcriptional Collision Requires Convergent Transcription in cis.
Because convergent transcription will generate separate antisense nascent transcripts, it is possible that the effects we observe are caused by an RNA-interference (RNAi) effect in vivo or more trivially by prehybridization of the antisense TRO RNA before filter hybridization during the experimental analysis. We therefore sought to demonstrate that transcriptional collision depends on transcription from the same template by creating control constructs that separated the transcription of the genes in the Fus plasmid. Elimination of the TATA box from the GAL7 promoter inactivated GAL7 gene transcription (Fus-Δ7P, Fig. 3B). The nascent transcription profile in this construct shows very low polymerase density over GAL7 and higher levels of transcription on GAL10 over the length of the template up to probe A. At this point nascent transcription begins to terminate (Fig. 3B). Steady-state analysis shows that GAL10 uses cryptic poly(A) sites to generate a doublet of detectable mRNA bands (Fig. 2, lane 4). The lower level of these steady-state GAL10 transcripts presumably reflects the inefficiency of this cryptic mRNA processing.
Similar analysis was done with the deletion of the UAS of the GAL10 gene (Fus-Δ10P) showing an up-regulation of transcription across the GAL7 gene and the elimination of transcripts from the GAL10 promoter (Fig. 3C). Polymerase density appears relatively consistent across the length of the probes (probe O was deleted with the deletion of the GAL10 promoter region). Steady-state analysis shows that Fus-Δ10P generates low levels of steady-state GAL7 mRNA again by using inefficient cryptic poly(A) signals (Fig. 2, lane 5). Read-around bands can be seen also in both this construct and Fus-Δ7P (Fig. 2, longer exposure lanes 4 and 5), which reflect transcripts that have used the same cryptic poly(A) sites the second time around the plasmid. Finally, the Fus-Δ7P/Δ10P construct demonstrated that the deletion of both promoters in the same construct resulted in little transcription from either promoter at nascent (data not shown) or steady-state (Fig. 2, lane 6) levels.
With the above single GAL7 and GAL10 transcribing plasmids defined, we finally carried out the critical experiment of cotransforming both of these plasmids into the ΔGAL deletion strain followed by TRO analysis. At both the nascent (Fig. 3D) and steady-state (Fig. 2, lane 7) levels we observe profiles similar to the sum of both Fus-Δ7P and Fus-Δ10P plasmids. These data demonstrate that the transcriptional collision effect we have observed require the GAL10 and GAL7 genes to be present in a cis arrangement. Trivial explanations for the observed inhibition of GAL10 and GAL7 transcription through antisense effects are effectively excluded.
Discussion
The aim of these studies was to investigate the effect that convergent gene transcription has on PolII transcriptional activity in S. cerevisiae. In the presence of intact mRNA 3′-end-processing signals the convergent GAL7 and GAL10 genes behaved similarly to the tandem-arranged genes as found in the WT situation. However, once the intergenic region, including the termination signals, was deleted there is a drastic effect on the level of transcriptional processivity in both genes, a phenomenon that we term transcriptional collision.
Both GAL7 and GAL10 transcriptional initiation appear unperturbed. Instead, the impact of interference is seen in the elongation stage of the transcription. The precise biological cause for this collision effect remains to be identified. One possibility is that as transcription proceeds, the polymerases collide and cause one or both to release from the template. Alternatively, a polymerase complex, when confronted with one polymerase on the opposite strand, stalls and is unable to advance further. This collision effect could be a direct physical impediment to the transcription machinery or an indirect effect caused by supercoiling changes to the DNA template during transcription (26–29). In the later scenario, a transcription bubble progresses along a template, creating positive supercoils. If these positive supercoils were generated from both strands simultaneously, it would create a region of hyper-supercoiling that would be predicted to prevent further advancement in either direction on the template. Such a mechanism explains the steady-state and nascent drop in detectable transcription.
The transcriptional collision seen in this investigation occurred in cis and was alleviated when the opposing polymerase was prevented from transcribing simultaneously. Separate expression of GAL10 (Fus-Δ7P) and GAL7 (Fus-Δ10P) in the same cell (Fus-Δ7P&Δ10P; see Fig. 3D) showed no interference even though it created antisense transcripts. These data demonstrate that simultaneous transcription of these genes is occurring and also argues against any RNAi effects in this S. cerevisiae strain. Previous research investigating RNAi in S. cerevisiae has detected RNAi in cis but not in trans. It seems plausible that these so-called cis RNAi effects actually may be caused by transcriptional collision on the same template rather than true RNAi (ref. 30 and references therein).
This study also provides further information about the capacity of GAL10 and GAL7 to interact in their WT arrangement. In the construct Con, elimination of transcribing polymerases over the promoter for GAL7 (Fig. 1B, probe A) presumably permits the unimpeded binding of Gal4p transcription factor (23) and thus increases transcription of GAL7. The level of transcription before the GAL7 promoter (GAL7 probe A) in WT clearly illustrates the extent of read-through transcription into the GAL7 promoter region, which is in contrast to the same region in Con, which showed no detectable signal before the promoter of GAL7. The quantified 2-fold increase in GAL7 nascent transcripts from WT to Con could underrepresent the true increase in transcripts that initiated at the GAL7 promoter, because the WT level of transcription detected in the GAL7 region is a combination of transcripts that initiated at GAL7 and the read-through from the GAL10 promoter. However, this observed 2-fold increase is consistent with the increase at steady-state levels (Fig. 2, lane 2). Thus we observe an increase of 2.3-fold in GAL7 mRNA for Con as compared with WT. These data further support the existence of TI in the WT orientation of GAL10 and GAL7.
In this paper we have defined a new type of TI termed transcriptional collision that can be alleviated when transcription on the opposing strand is suppressed. The relevance of this phenomenon to the endogenous genes of S. cerevisiae remains to be established. However, it is abundantly clear that coexpressed adjacent genes must guard against TI when arranged in tandem or transcriptional collision when convergent. The role of efficient PolII-termination signals is critical to allow the full expression of such genes. It seems plausible that restricted or regulated gene expression could be effected by the regulation of PolII termination.
Acknowledgments
We are grateful to members of the Proudfoot lab for advice throughout these studies and to Nick Kent for reading the manuscript. E.M.P. was supported by the HLP Educational Trust. This research was supported by Wellcome Trust Program Grant 062329 (to N.J.P.).
Abbreviations
- PolII
RNA polymerase II
- TI
transcriptional interference
- WT
wild type
- TRO
transcriptional run-on
- RNAi
RNA interference
References
- 1.Feldmann H, Aigle M, Aljinovic G, Andre B, Baclet M C, Barthe C, Baur A, Becam A M, Biteau N, Boles E, et al. EMBO J. 1994;13:5795–5809. doi: 10.1002/j.1460-2075.1994.tb06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dujon B. Trends Genet. 1996;12:263–270. doi: 10.1016/0168-9525(96)10027-5. [DOI] [PubMed] [Google Scholar]
- 3.Malavasic M J, Elder R T. Mol Cell Biol. 1990;10:2809–2819. doi: 10.1128/mcb.10.6.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson J A, Myers A M. Nucleic Acids Res. 1993;21:5500–5508. doi: 10.1093/nar/21.23.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGrath J P, Varshavsky A. Nature (London) 1989;340:400–404. doi: 10.1038/340400a0. [DOI] [PubMed] [Google Scholar]
- 6.Barker D G, White J H M, Johnston L H. Eur J Biochem. 1987;162:659–667. doi: 10.1111/j.1432-1033.1987.tb10688.x. [DOI] [PubMed] [Google Scholar]
- 7.Aranda A, Perez-Ortin J E, Moore C, del Olmo M. Nucleic Acids Res. 1998;26:4588–4596. doi: 10.1093/nar/26.20.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aranda A, Perez-Ortin J E, Moore C, del Olmo M. RNA. 1998;4:303–318. [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne B I, Guarente L. Proc Natl Acad Sci USA. 1989;86:4097–4101. doi: 10.1073/pnas.86.11.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruglyak S, Tang H X. Trends Genet. 2000;16:109–111. doi: 10.1016/s0168-9525(99)01941-1. [DOI] [PubMed] [Google Scholar]
- 11.Fu J H, Gnatt A L, Bushnell D A, Jensen G J, Thompson N E, Burgess R R, David P R, Kornberg R D. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 12.Poglitsch C L, Meredith G D, Gnatt A L, Jensen G J, Chang W H, Fu J H, Kornberg R D. Cell. 1999;98:791–798. doi: 10.1016/s0092-8674(00)81513-5. [DOI] [PubMed] [Google Scholar]
- 13.Birse C E, Minvielle-Sebastia L, Lee B A, Keller W, Proudfoot N J. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Hyman L, Moore C. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puig S, Perez-Ortin J E, Matallana E. Curr Microbiol. 1999;39:369–373. doi: 10.1007/s002849900474. [DOI] [PubMed] [Google Scholar]
- 16.Greger I H, Proudfoot N J. EMBO J. 1998;17:4771–4779. doi: 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Springer C, Valerius O, Strittmatter A, Braus G H. J Biol Chem. 1997;272:26318–26324. doi: 10.1074/jbc.272.42.26318. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka S, Halter D, Livingstonezatchej M, Reszel B, Thoma F. Nucleic Acids Res. 1994;22:3904–3910. doi: 10.1093/nar/22.19.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder M, Sapolsky R J, Davis R W. Mol Cell Biol. 1988;8:2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill A, Bloom K. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocco V, Demassy B, Nicolas A. Proc Natl Acad Sci USA. 1992;89:12068–12072. doi: 10.1073/pnas.89.24.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen K, Birse C E, Proudfoot N J. EMBO J. 1998;17:3066–3077. doi: 10.1093/emboj/17.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greger I H, Aranda A, Proudfoot N. Proc Natl Acad Sci USA. 2000;97:8415–8420. doi: 10.1073/pnas.140217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz R D, Schiestl R H. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 25.Birse C E, Lee B A, Hansen K, Proudfoot N J. EMBO J. 1997;16:3633–3643. doi: 10.1093/emboj/16.12.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunaway M, Ostrander E A. Nature (London) 1993;361:746–748. doi: 10.1038/361746a0. [DOI] [PubMed] [Google Scholar]
- 27.Tsao Y P, Wu H Y, Liu L F. Cell. 1989;56:111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- 28.Giaever G N, Wang J C. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 29.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atkins D, Arndt G M, Izant J G. Biol Chem Hoppe-Seyler. 1994;375:721–729. doi: 10.1515/bchm3.1994.375.11.721. [DOI] [PubMed] [Google Scholar]