Abstract
Although both the 5′- and 3′-untranslated regions (5′- and 3′-UTRs) of eukaryotic mRNAs may play a crucial role in posttranscriptional gene regulation, we observe that cis-encoded natural antisense RNAs have a striking preferential complementarity to the 3′-UTRs of their target genes in mammalian (human and mouse) genomes. A null neutral model, evoking differences in the rate of 3′-UTR and 5′-UTR extension, could potentially explain high rates of 3′-to-3′ overlap compared with 5′-to-5′ overlap. However, employing a simulation model we show that this null model probably cannot explain the finding that 3′-to-3′ overlapping pairs have a much higher probability (>5 times) of conservation in both mouse and human genomes with the same overlapping pattern than do 5′-to-5′ overlaps. Furthermore, it certainly cannot explain the finding that overlapping pairs seen in both genomes have a significantly higher probability of having co-expression and inverse expression (i.e. characteristic of sense–antisense regulation) than do overlapping pairs seen in only one of the two species. We infer that the function of many 3′-to-3′ overlaps is indeed antisense regulation. These findings underscore the preference for, and conservation of, 3′-UTR-targeted antisense regulation, and the importance of 3′-UTRs in gene regulation.
INTRODUCTION
Recent estimates suggest that the human and mouse genomes might contain only 20 000 to ∼25 000 protein-coding genes (1,2), similar to other vertebrates, and only slightly more than the simple nematode, Caenorhabditis elegans. It has been suggested that organismal complexity arises from progressively more elaborate regulation of gene expression (3), and that the basis of eukaryotic complexity and phenotypic variation may lie primarily in a control architecture composed of a highly parallel system of trans-acting RNAs that relay state information required for the coordination and modulation of gene expression (4–6). Natural antisense transcripts, as a class of trans-acting RNAs, have been implicated in many levels of eukaryotic gene regulation including translational regulation, genomic imprinting, RNA interference, alternative splicing, X inactivation, RNA editing, gene silencing and methylation [for reviews see (7–9)]. The majority of natural antisense transcripts are cis-encoded and transcribed from the opposite strand of the same genomic loci from their sense counterparts (8). Recent genome-wide analyses suggest that as much as 15 to ∼22% of the mouse and human transcripts (10–16), or even >40% of human transcripts (17), might be involved in (cis-encoded) antisense transcription. Therefore, it is of great interest and importance to study the mechanism of gene regulation mediated by natural antisense RNAs.
It is well known that both the 5′- and 3′-untranslated regions (5′- and 3′-UTRs) of eukaryotic mRNAs play a crucial role in posttranscriptional regulation of gene expression (18–20). However, previous antisense studies show that putative sense–antisense (SA) pairs overlapping at the 3′ ends/3′-UTRs are much more frequent than those overlapping at their 5′ ends/5′-UTRs (10,11,13,21). It is largely unknown why putative SA pairs predominantly overlap at 3′-UTRs rather than at 5′-UTRs. Is such a bias related with antisense-mediated gene regulation (i.e. antisense regulation)? In this study, we employed a robust protocol (14) to identify putative SA pairs in the human and mouse genomes, and found similar phenomena in both genomes. We also observed that putative SA pairs overlapping at the 3′-UTRs have a much higher evolutionary conservation rate between human and mouse genomes than do those overlapping at their 5′-UTRs. Our analyses suggest that a null neutral model, evoking differences in the rate of 3′- and 5′-UTR extension, cannot explain these findings alone. Instead, we infer that the function of many putative SA pairs overlapping at their 3′-UTRs are involved in antisense regulation. Our findings imply that 3′-UTRs might be the preferred binding sites of functional SA pairs that are involved in antisense regulation.
MATERIALS AND METHODS
Identification of transcription clusters in the human and mouse genomes
We employed a robust protocol described in our previous study (14) to identify transcription clusters (i.e. genes) in the human (Homo sapiens; an updated version) and mouse (Mus musculus) genomes based on the recent versions of databases. In brief, transcription clusters were created based on the mRNA and expressed sequence tag (EST) sequences (downloaded from UniGene (22) database; human Build #175; mouse Build #141) alignments to the relevant genome (human Build 35.1; mouse Build 33.1). The transcript sequences and alignments were filtered stringently to ensure the correct orientation: (i) the mRNA and EST sequences had to have an annotated protein-coding region (CDS), and/or both a poly(A) tail and a poly(A) signal (i.e. a polyadenylation site); (ii) all transcript sequences having suspicious splice sites were discarded. The transcript sequences representing highly abundant and tandem duplicate genes such as immunoglobulins and T-cell receptors were excluded. All transcript sequences aligned to the same genomic locus were assembled into one transcription cluster. After assembly, all clusters that contained only one sequence that did not span an intron were excluded.
Classification of bidirectional transcription cluster pairs
As in our previous study (14), the transcript clusters were classified according to the transcribed pattern in the genomes. Clusters containing at least one pair of transcript sequences transcribed from opposite strands of the same genomic locus were called ‘bidirectional clusters’ (BD), while the remaining clusters containing only one-directional transcripts were called ‘non-bidirectional clusters’ (NBD). We further separated each BD cluster into two new clusters (a cluster pair) based on their overlapping patterns: sense (S) and antisense (A) clusters form putative sense–antisense (SA) pairs with exon overlaps (identity ≥94%), while the sense-like (SL) and antisense-like (AL) clusters form non-exon-overlapping bidirectional (NOB) pairs without exon overlaps.
In our previous study (14), we defined the S and A or SL and AL genes in each BD gene pair mainly based on a conventional concept [e.g. (23)] that the S (or SL) gene should exist in more tissues and/or be expressed at a higher level, and thus would have been detected more frequently (i.e. having more transcript sequences deposited in the expressed sequence databases) than its A (or AL) partner. Nevertheless, there is another (even more) common notion that almost all of sense genes are protein-coding genes whereas antisense genes might be coding or ncRNA (7,8,12). The fact that >90% of the defined S (SL) genes in our previous study (14) are protein-coding genes (i.e. with annotated CDS regions) is in accord with this notion. However, in a few pairs, the defined S (or SL) lacks CDS while the corresponding A (or AL) partner has CDS. Thus, in this study, we revised the previous rules as follows: (i) For the SA (or NOB) pairs in which one member has CDS while the other lacks CDS, define the one with CDS as the S (or SL) and the other as the A (or AL); (ii) For the remaining SA (NOB) pairs, the previous rules (14) are employed: (a) define the one containing more transcript sequences as the S or SL cluster, the other as the A or AL cluster; (b) if the sequence numbers were the same, define the one with more mRNA sequences as the S or SL cluster, the other as the A or AL cluster; and (c) if their mRNA sequence numbers were still the same, define the one with intron-spanning sequence(s) as the S or SL cluster while the other one without such intron-spanning sequence(s) would be the A or AL cluster. If none of above conditions were satisfied, define the one mapped to the sense strand of chromosome as the S or SL cluster and the other as the A or AL cluster. After such separation, five categories of unique gene clusters were obtained: S, A, SL, AL and NBD.
Besides putative SA and NOB pairs that are bidirectional gene pairs located on opposite strands of the same genomic loci, there is another type of bidirectional gene pairs that are located on opposite strands of neighboring genomic loci within 1000 bp distance and lack gene overlap (24,25), which we called as neighboring bidirectional (NB) gene pairs. Because putative SA and NOB genes may also form NB gene pairs with their neighboring genes, to avoid NB genes mixed with putative SA or NOB genes, we excluded putative SA and NOB genes from the NB gene pair set for present study.
Classification of subtypes of SA, NOB and NB gene pairs
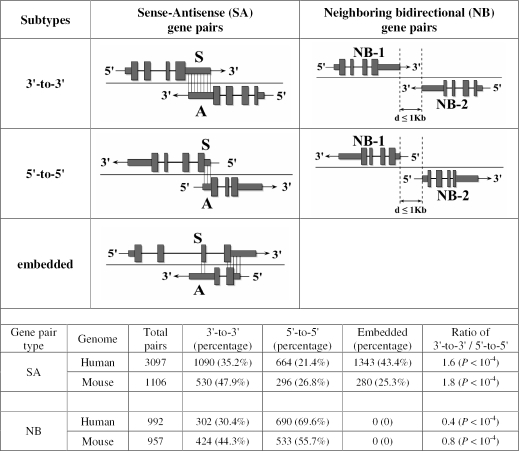
Based on the exonic and genomic overlapping patterns, metazoan putative SA pairs can be divided into three subtypes: 3′-to-3′ (i.e. tail-to-tail exonic overlapping), 5′-to-5′ (i.e. head-to-head exonic overlapping) and embedded (one gene is entirely embedded within the other) putative SA pairs. We observed that >80% of the 3′-to-3′ and >65% of the 5′-to-5′ putative SA pairs are solely or mainly overlapping at the 3′- and 5′-UTRs of the sense genes, respectively (in most cases, also overlapping at the relevant UTRs of the antisense transcripts if they are protein-coding genes). Similar phenomena have been observed by Yelin et al. (13) and Veeramachaneni et al. (21). Thus, 3′-to-3′ and 5′-to-5′ putative SA pairs largely represent 3′- and 5′-UTR-targeted putative SA pairs, respectively. Based on the genomic (non-exonic) overlapping patterns, NOB pairs can be divided into three subtypes as well: 3′-to-3′ (i.e. tail-to-tail genomic overlapping), 5′-to-5′ (i.e. head-to-head genomic overlapping) and embedded (one gene is entirely embedded within the other) NOB pairs. Based on the orientation patterns, NB gene pairs can be divided into two subtypes: 3′-to-3′ (i.e. tail-to-tail orientated) and 5′-to-5′ (i.e. head-to-head orientated) NB pairs. However, because most (>80%) of the NOB pairs belong to embedded NOB pairs and are thus not informative for the present study, we did not perform further analysis on NOB gene pairs. Therefore, only putative SA and NB gene pairs are used in present study (Figure 1).
Figure 1.
Classification and comparison of subtypes of putative SA and NB gene pairs. Based on the overlapping pattern, we divide putative SA pairs into three subtypes: 3′-to-3′ (i.e. tail-to-tail overlap), 5′-to-5′ (i.e. head-to-head overlap) and embedded (one gene is entirely embedded within the other) pairs. Based on the orientation pattern, we divide NB gene pairs into two subtypes: 3′-to-3′ (i.e. tail-to-tail orientated) and 5′-to-5′ (i.e. head-to-head orientated) pairs. Indeed, 3′-to-3′ and 5′-to-5′ putative SA pairs largely represent 3′- and 5′-UTR-targeted putative SA pairs, respectively (Materials and Methods). Coding exons are represented by blocks connected by horizontal lines representing introns. The 5′- and 3′-UTRs are displayed as thinner blocks on the leading and trailing ends of the aligning regions. The distributions and comparison of the subtypes in putative SA and NB pairs are shown in the embedded table. Because many antisense genes have only EST sequences, the proportion of ‘embedded’ putative SA pairs is seriously overestimated here. In fact, a large proportion of the ‘embedded’ putative SA pairs are also mainly overlapping at the 3′-UTRs of the sense genes (data not shown). Regardless of the ‘embedded’ pairs, the 3′-to-3′ putative SA pairs have a significantly higher percentage compared with the 5′-to-5′ putative SA pairs in both genomes. In contrast, a reverse pattern was observed in NB gene pairs.
Analysis of evolutionary conservation of putative SA pairs in the human and mouse genomes
We examined ortholog pairs between mouse and human that were reciprocal best ‘hits’ (matches) between the two genomes. We combined the ortholog pairs from Mouse Genome Informatics Website (ftp://ftp.informatics.jax.org/pub/reports/HMD_HumanSequence.rpt; December 2004) and Ensembl MartView (http://www.ensembl.org/Multi/martview; December 2004). By comparing sequence IDs in our mouse and human gene sets with those in the combined ortholog dataset, we obtained 11 931 one-to-one mouse–human ortholog pairs in our datasets. Of these, 347 putative SA pairs in which at least one member has an ortholog in both the human and mouse genomes are conserved in putative SA form in both genomes, and were called HM-conserved putative SA pairs. Owing to the facts that the number of putative SA pairs in the mouse genome (even in the human genome) are significantly underestimated because of the limitation of qualified transcript sequences (M. Sun, L.D. Hurst, G.G. Carmichael and J. Chen, unpublished data), and that many antisense transcripts are ncRNAs that are not included in the human–mouse ortholog databases, the number of HM-conserved putative SA pairs might be seriously underestimated.
A simulation model
To investigate whether a null neutral model can explain the higher rate of 3′-to-3′ overlapping pairs seen in both mouse and human genomes, we developed a simulation model of the null hypothesis. In this model, we consider the fate of a pair of linked genes A and B, found in the common ancestor of two species. We then consider the evolution of this pair in the two independent lineages and ask how commonly we will find in both lineages that the two genes are overlapping and compare this with the number of occasions on which we would find an overlapping pair in one lineage which is not observed in the second species. The proportion of times that the overlapping pair is found in both species is defined as the conservation ratio. The question we need to ask of the null model is whether this ratio is probable to alter as a function of the relative rates of extension, i.e. do we expect slowly extending 5′-UTRs to have the same ratio as fast evolving 3′ extensions? If we do then the null neutral model is unlikely to account for the above observations of an excess of 3′-to-3′ pairs observed in both species.
In the model we consider the probability per unit time that a pair of genes on opposite strands (either 5′-to-5′ or 3′-to-3′) will extend by 1 U of sequence. This probability is a measure of the extension rate. Both genes are considered separately. We also consider the probability that the two genes might be broken apart by some form of re-arrangement (inversion, translocation and so on). We assume that each gene has a critical length of UTR and that this must be included in the re-arrangement. We additionally assume that following any re-arrangement the gene reverts to using just the necessary UTR parts and not any extensions (which are both neutral and potentially broken by the re-arrangement). When re-arrangement occurs, therefore, we assume that the distance between the genes return on average to what it was prior to the evolution of the neutral extensions to the end. Note that in the null model, overlap of UTRs does not prevent re-arrangement from occuring as the extensions are functionless and hence neutral.
The final important parameter is the distance between the genes. Following re-arrangement this is derived from random sampling from a normal distribution of mean X and standard deviation X/5. We vary X to consider the importance of intergenic distance (IGD). A priori we expect fewer overlaps to evolve when the IGD is big, the extension rate low and the re-arrangement rate high (re-arrangement restarts the growth process). This is confirmed by simulation (data not shown). As 5′-to-5′ and 3′-to-3′ gene pairs need not be equidistant in the common ancestor of our two species, we additionally consider a burn-in period. Here we permit growth and re-arrangement of a linked gene pair. Following re-arrangement we then concentrate on one of the newly linked pairs and so on. From this we can simulate the past history of gene pairs present in the common ancestor that have been linked for some time previously, but not necessarily for all previous time. We find that the distance between the ends of the UTRs of the two genes linearly and negatively co-varies with the extension rate, as might be expected. If the gene pair remained linked in simulation the higher the extension rate the lower the distance between the ends of the genes. If the pair had a past history involving re-arrangement then the distance between the ends of the UTRs will be a function of the time since last re-arrangement and the extension rate. As the time since last re-arrangement is independent of the extension rate, fast extending sequences are physically closer than slow ones.
Note then that in the absence of re-arrangement a high rate of overlapping genes for 3′-to-3′ overlaps might be expected in both lineages, as the common ancestor of the two species may well have had overlapping genes and, if not, the two might independently have grown to overlap. A slowly evolving extension, in contrast, might show the opposite pattern. To understand the dynamics of the model, however, it is necessary to perform the simulation. To this end, after the burn-in we consider the fate of the gene pair. The only distinction now from the burn-in concerns the fate of the re-arranged genes. To have both genes A and B overlapping in both species, we assume that re-arrangement has not occurred. However, to find a gene pair overlapping in one species but not the other there are several possibilities. First, the pair may have remained linked in both lineages but not overlapping in the ancestor with growth-to-overlap occurring in just one of the two lineages (owing to stochasticity). Alternatively, the pair might ancestrally have been linked and overlapping, but the overlap may have been broken by re-arrangement in one of the two lineages. Alternatively, a pair may not have been overlapping in the ancestor but in the lineage with the re-arrangement one of the two genes (A or B) caught in the re-arrangement might by chance be close to the end of its new neighbor and grow to overlap. The number of possibilities are in fact sizeable.
Note that it is possible in the simulations that one or both lineages might be involved in a re-arrangement. Note too that even if both lineages evolve overlap after a re-arrangement this would still be classified as an incidence of an SA pair observed in one lineage but not the other, for no matter which gene pair one examines, if re-arrangement has occurred, an SA pair in one lineage cannot be overlapping in the other. After a re-arrangement we should in principle analyze both genes (A and B) and their new neighbors. However, on the average, for any given UTR end, a new re-arrangement will leave that end on the same strand as the adjoining gene (i.e. it is the 5′ end of the next gene that is nearest the 3′ end) as often as it is on the opposite strand. SA overlap is only possible in the later configuration. Hence by following the fate of one of the two genes from any given pair we will model the rate of overlapping gene formation. We ran the simulation 10 000 times for 100 time units (each time under multiple different parameter settings).
Investigation of co-expression and inverse expression patterns of putative SA pairs in the human and mouse genomes
We evaluated the co-expression and inverse expression of SA pairs at the whole genome level based on their expression profiles obtained from serial analysis of gene expression (SAGE) expression data (26). The procedure is similar to our established procedures (15) with some modifications. We downloaded SAGE expression data (NlaIII SAGE libraries) from the NCBI GEO platform for human (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GPL4; December 1, 2004) and for mouse (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GPL11; December 1, 2004), respectively. For both human and mouse, we constructed 16 tissue-type/cell-type SAGE library combination [for human: blood, brain, breast, colon, lung, ovary, pancreas, prostate, skin, stomach, embryonic stem cells, eye, liver, muscle, placenta and thyroid; for mouse: adipose tissue, brain, bone marrow, cornea, embryonic carcinoma cells, embryonic stem cells, fibroblasts, granulosa cells, heart, hippocampus, kidney, limb (developing), liver, lymph node, T cells and testis] to determine co-expression of gene pairs, and constructed 50 comparison cases to determine inverse expression of gene pairs. Tag counts were converted to counts per million and the expression data were cross-linked to our genes by extracting the 3′-most NlaIII SAGE tag for each transcript in the genes (i.e. transcript clusters). Only tags that matched to a single gene were taken into account. All SAGE tags mapped to the same gene were then combined and the sum of their counts per million in a tissue/cell represented the expression level of that gene in that tissue/cell.
To evaluate the co-expression of an SA pair, we adopted an index of co-expression between two genes a and b (ICEa,b) defined by Lercher et al. (27) that is the number of tissues with common positive expression, weighted by the geometric mean of the two breadths. Note that, unlike the conventional ‘Pearson correlation coefficient (r)’, co-expression in this context refers not to the extent to which levels of transcripts are correlated, but rather to the coupled presence or absence of the transcripts across different tissues or cells (15). ICEa,b ranges from 0 (no co-expression) to 1 (perfect co-expression). We define a pair of genes (e.g. the sense and antisense in an SA pair) to be co-expressed if the ICEa,b ≥ 0.6 in our previous study (15). In this study, we determined ICEa,b values for all gene pairs randomly formed in our gene sets, and found that it is higher than the 99% confidence intervals (i.e. P < 0.01) of the average ICEa,b values of all the possible gene pairs when ICEa,b ≥ 0.6 in humans or ≥0.5 in mice. Thus, we define two genes (a and b; e.g. the sense and antisense in a putative SA pair) to be co-expressed if the ICEa,b ≥ 0.6 in humans or ≥0.5 in mice.
On the basis of our previous study (15), we set up 50 comparison cases for present study, each of which is a pair of two states (two different unique SAGE libraries) at different developmental, differentiation, physiologic or pathological stages/conditions of the same tissue (data not shown). A given gene with positive expression in at least one of the two states of a comparison case would be recognized as being presented in that case. The presence breadth for each gene is the number of cases in which the gene is presented. To measure inverse-expression pattern in a more quantitative way compared with that described previously (15), we defined a new index of inverse expression between two genes a and b (IIEa,b) that is the number of comparison cases (∑fIab,t; t runs over all cases) in which the two partners exhibit an inverse expression pattern between two states (i.e. a member is expressed at a higher level at state 1 but a lower level at state 2 compared with its partner and vice versa) and a significantly greater change of the relative expression ratio of gene a to gene b between two states than expected by chance (i.e. exceeding the 99% confidence interval of the mean changes of all the randomly formed gene pairs), weighted by the geometric mean of the two presence breadths (∑fa,t and ∑fb,t; t runs over all cases):
IIEa,b ranges from 0 (no inverse-expression) to 1 (perfect inverse-expression). Similarly, we define two genes (a and b; e.g. the sense and antisense in a putative SA pair) to be inversely expressed if the IIEa,b is higher than the 99% confidence intervals (i.e. P < 0.01) of the average IIEa,b values of all the randomly formed gene pairs.
The detail list of the 3097 human and 1106 mouse putative SA gene pairs with information of overlapping pattern, evolutionary conservation, co-expression, and inverse expression is in Supplementary Table 1.
RESULTS
Putative SA pairs overlapping at the 3′-UTRs are significantly more frequent than those overlapping at their 5′-UTRs
We employed a robust protocol (14) to identify putative SA pairs in the human and mouse genomes (Materials and Methods). A total of 27 333 human and 19 100 mouse unique genes were identified, each of which represents a single protein- or RNA-coding gene, of which 22.7% (6194) human and 11.6% (2212) mouse unique genes form 3097 and 1106 putative SA pairs, respectively. We further analyze the overlapping patterns of these putative SA pairs. As shown in Figure 1, putative SA pairs can be divided into three subtypes based on their overlapping patterns: 3′-to-3′, 5′-to-5′ and embedded pairs. The 3′-to-3′ and 5′-to-5′ pairs largely represent 3′- and 5′-UTR-targeted putative SA pairs, respectively (Materials and Methods). For each gene, we used the entire gene cluster (i.e. including all of the alternative variants of transcripts of the gene) for the overlapping analysis. Thus, our results would not be affected by alternative splicing. If overlapping is a random or stochastic event, one might expect that the number of 3′-to-3′ overlapping SA pairs is equivalent to that of 5′-to-5′ overlapping SA pairs. However, in both genomes, putative SA pairs overlapping at the 3′ ends are significantly more frequent (35.2% versus 21.4% in humans and 47.9% versus 26.8% in mice; i.e. >1.6 or 1.8 times; P < 10−4) than those overlapping at their 5′ ends (Figure 1). Similar phenomena have been observed in previous SA studies in which different methodologies and data sources were used (10,11,13,21). In contrast, a reverse pattern was observed in NB gene pairs that are located on opposite strands of neighboring genomic loci within 1000 bp distance but which do not overlap (Figure 1), as observed by others (24,25).
Can the ‘null neutral’ model explain the higher conservation rate of 3′-to-3′ overlapping SA pairs compared with 5′-to-5′ overlapping ones?
Why do putative SA pairs predominantly overlap 3′-UTRs rather than 5′-UTRs? While this may reflect a preference (under selection) for 3′-UTR binding in antisense regulation, there is a simpler potential explanation, the ‘null neutral’ model (i.e. the predominant overlaps at 3′-UTRs rather than 5′-UTRs are not under selection related with antisense regulation; instead, it may reflect different freedom of changes in the lengths of 3′- and 5′-UTR sequences). We have observed that the average length of 3′-UTRs has increased significantly from around 300 nt in invertebrates to >800 nt in mice and humans, whereas those of the 5′-UTRs and of the protein-coding (CDS) regions are roughly constant (∼200 and 1600 nt, respectively) in diverse genomes [based on reference sequences; see also ref. (18)], suggesting that 5′-UTR extension is relatively more constrained than 3′-UTR extension. This observation hints that the null neutral model might be upheld. Let us assume two genes start evolving in proximity, but originally do not overlap. From the above evidence we expect evolutionary extension to be predominantly from the 3′ end rather than the 5′ end. Thus, we expect more instances of 3′-to-3′ than 5′-to-5′ overlap, which is observed. Moreover, we expect for NB genes fewer instances of 3′-to-3′ as many of these have evolved into 3′-to-3′ putative SA pairs. This null neutral model hence potentially explains much about the pattern of 3′-to-3′ versus 5′-to-5′ in putative SA and NB pairs (Figure 1).
But is this model adequate? Such a null model might also predict, all else being equal, that 3′-to-3′ and 5′-to-5′ overlapping pairs should have an equal probability of being conserved as a pair over evolutionary time. Examining 3′-to-3′ overlapping pairs in mice, we find the same pair is found in humans ∼50% of the time, while 5′-to-5′ overlapping pairs in mice are found in humans around only 10% of the time (Table 1). Why might this be?
Table 1.
Comparison of the evolutionary conservation rates of different subtypes of putative SA pairs in the human and mouse genomesa
| Genome | Putative SA subtype | Rate of conservation | Rate of same-overlapping-pattern conservation | Ratio of 3′-to-3′/5′-to-5′ in conservation rate (x2-test) |
|---|---|---|---|---|
| Human | 3′-to-3′ | 24.5% (267/1090) | 22.9% (250/1090) | 3.5; 5.4 |
| 5′-to-5′ | 7.1% (47/664) | 4.2% (28/664) | ||
| embedded | 2.4% (33/1343) | 0.6% (8/1343) | (P < 10−4; P < 10−4) | |
| whole | 11.2% (347/3097) | 9.2% (286/3097) | ||
| Mouse | 3′-to-3′ | 53.6% (284/530) | 47.2% (250/530) | 3.5; 5.0 |
| 5′-to-5′ | 15.2% (45/296) | 9.5% (28/296) | ||
| embedded | 6.4% (18/280) | 2.9% (8/280) | (P < 10−4; P < 10−4) | |
| whole | 31.4% (347/1106) | 25.9% (286/1106) |
aNote that, all P-values far <10−4 are also shown as P < 10−4. Rate of conservation refers to the percentage of human or mouse putative SA pairs conserved as putative SA in the mouse or human genome. Rate of same-overlapping-pattern conservation refers to the percentage of human or mouse putative SA pairs conserved as putative SA with the same overlapping pattern (i.e. 3′-to-3′, 5′-to-5′ or embedded) in the mouse or human genome. Owing to the fact that the number of putative SA pairs in the mouse genome (even in the human genome) is significantly underestimated owing to the limitation of qualified transcript sequences (M. Sun, L.D. Hurst, G.G. Carmichael and J. Chen, unpublished data), the rates of conservation and of same-overlapping-pattern conservation of human (probably also mouse) putative SA pairs might be seriously underestimated.
One possible explanation for the higher conservation rate of 3′-to-3′ overlapping pairs is that the prevalence of 5′-to-5′ overlapping pairs, and in turn their conservation rate, might be seriously underestimated, owing to the probability that representation of the 5′ ends of transcripts in the databases is not as complete as that of 3′ ends because of either the difficulty in cloning the full length of cDNAs (28) or 5′-UTRs having a higher possibility of alternative splicing than do 3′-UTRs (29). If so, we would expect that the differences in prevalence and conservation rate between 3′-to-3′ and 5′-to-5′ overlapping pairs should be much smaller in the subsets of gene pairs in which both partners contain roughly full-length, curated reference sequences (RefSeq), compared with those in the whole gene pair sets in which many genes contain only 3′ EST and/or 5′ end-incomplete cDNA/mRNA sequences. However, we observed the opposite phenomenon: (i) 5′-to-5′ overlapping pairs have a similar rate in both types of pair sets, whereas 3′-to-3′ overlapping pairs are much more enriched in the RefSeq gene pair sets, resulting in the observation that the ratios of 3′-to-3′/5′-to-5′ increase from 1.6 to ∼1.8 in the whole gene pair sets to 2.6 to ∼3.3 in the Refseq gene pair sets (Figure 1 and Supplementary Table 2a), and (ii) the ratios of 3′-to-3′/5′-to-5′ in evolutionary conservation rate are similar in both types of gene pair sets (Table 1 and Supplementary Table 2b). Therefore, the higher conservation rate of 3′-to-3′ putative SA pairs probably cannot be explained by the potential incompleteness of 5′ ends.
Thus, the apparently high conservation rates of 3′-to-3′ overlaps might be consistent with different rates of conservation, i.e. where we assume that the putative SA pair was present in the common ancestor of the two species, but the 5′-to-5′ overlapping pairs are less probable to be retained as overlapping pairs compared with the 3′-to-3′ overlapping pairs, possibly because the 3′-to-3′ pairs are more likely to adopt a function. However, this is by no means the only interpretation. For example, the putative SA pair seen in only one of the two species might not have been present in the common ancestor of the two species and, hence, evolved in one of the two lineages rather than being lost in the other. To investigate this further we developed a simulation model of the null hypothesis. On the basis of the simulation model (Materials and Methods), we show that the null model appears to be an unlikely explanation for the observed magnitude differences in conservation rates between 3′-to-3′ and 5′-to-5′ putative SA pairs.
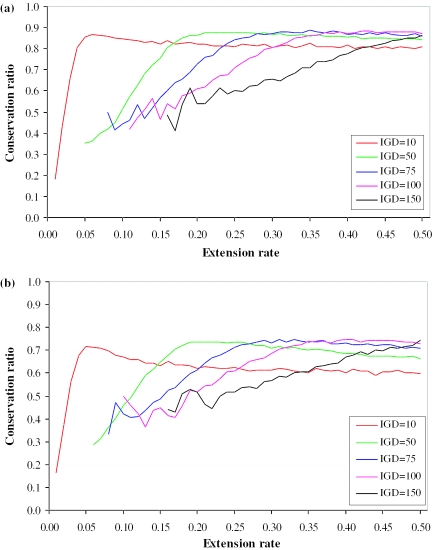
There are three interacting variables (IGD, extension rate and re-arrangement rate), so to consider the relative effects of each we vary each. The results are displayed in Figure 2 and Supplementary Figure 1. For any given value of the IGD, what is notable is that the plot can be split into three phases. In the first phase (very low extension rates) there are no overlapping genes of any variety. These appear blank on the figures as conservation ratio has no meaning (Supplementary Figure 1a–d and Figure 2a and b). In the second phase, a middling extension rate, the conservation rate is increasing with increasing extension rate. In the third phase, the conservation rate hits a plateau. The smaller the IGD the sooner the plateau is reached and the longer the plateau. In this last phase we see that most instances of an overlapping gene being found in one species, but not the other, is largely owing to re-arrangement breaking up overlapping gene pairs.
Figure 2.
The proportion of SA pairs found in both lineages in simulation as a function of the extension rate for different values of IGD. For (a) the re-arrangement rate per unit time is set to 1/1000 in one lineage and 1/5000 in the other. For (b), it is 1/500 in one lineage and 1/1000 in the other. Red lines are for IGD of 10 U, green for 50 U, blue for 75 U, magenta for 100 U and black for 150 U. Only instances in which >10 overlapping pairs out of 10 000 simulants are incorporated in the analysis.
To account for the differences between the apparent conservation ratio difference between 3′-to-3′ pairs and 5′-to-5′ pairs, the slow 5′-to-5′ pairs must have an extension rate that sits in this second phase. Were they in the first phase we would not identify overlapping 5′-to-5′ pairs. Were they in the third plateau phase there would be no difference in the conservation ratio for 5′-to-5′ and 3′-to-3′. From visual inspection of the plots it appears to be unlikely that the null model could account for the observations. When the IGD is low, the second phase tends to be a small domain of parameter space. Moreover, to see a 5-fold difference between 5′-to-5′ and 3′-to-3′, the parameter space in this second phase must also be limited to give such a ratio. When the IGD is on the average high, in contrast, the second phase is more extended and the rise to the plateau more gradual. However, it seems that the lowest conservation ratios possible at these high IGDs are all relatively high. Thus, the difference in conservation ratios that are possible is relatively small, especially if the re-arrangement rate is relatively high. Hence, again, a big difference between gene ends of different extension rates is unlikely to explain large differences in apparent conservation rates. These conclusions appear to be robust to permitting differences in re-arrangement rates between the two lineages (Figure 2a and b), a difference that is probable to be relevant in the mouse–human comparison. Curiously, if one permits large (2-fold) differences in the rate of extension between the two lineages (Supplementary Figure 1d) the conditions under which the null model become valid appear to be broader. However, in the mouse–human comparison any such difference is small as the mean length of mouse and human UTRs is not greatly different: from analysis of the non-redundant UTR database (30), we find that mouse 3′ regions are on average 978 bases ± 6.5 (N = 19 911) while mean human 3′-UTR length is 988 bases ± 5 (N = 37 135); mouse 5′ is 196 bp ± 5 (N = 18 138), human 5′ 277 bp ± 2 (N = 31 663). On the basis of the simulations alone, we cannot definitively reject the null hypothesis, especially if the re-arrangement rate is especially low (Supplementary Figure 1c) or if there is an important difference in the extension rate in the two lineages. However, a priori then, while we cannot be certain about parameter values, the null model appears to be an unlikely explanation for the observed differences in conservation rates between 3′-to-3′ and 5′-to-5′ putative SA pairs.
Many putative SA genes overlapping at 3′-UTRs might indeed function as sense and antisense pairs for gene regulation
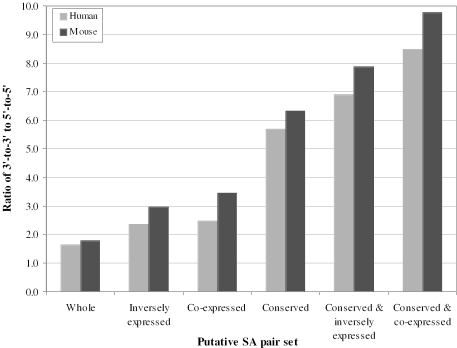
Even were we able to reject the null hypothesis on the above grounds, the evidence presented here, per se, does not demonstrate that 3′-to-3′ overlapping genes are functioning as sense and antisense pairs for gene regulation. One might equally well evoke the idea that the 3′ extensions evolve new functions, perhaps to control the rate of mRNA degradation or to control the location of the transcript. However, is there any compelling reason to hypothesize that the overlaps function in antisense regulation? If the conservation of the putative SA pairs is largely attributed to antisense regulation, we would expect that the two partners in conserved putative SA pairs would have high rates of coordinated expression. For overlapping gene pairs involved in antisense regulation, coordinated expression means both co-expression [i.e. they should be simultaneously expressed (co-expressed) in the same tissue/cell] and inverse expression, whereby a high level of one transcript, relative to the titre of the other, in a given tissue at a given time, is matched by a relative low titre of the same transcript in the same tissue at different time (7,8,15). We evaluated the co-expression and inverse expression of SA pairs at the whole genome level based on their expression profiles obtained from SAGE expression data (26). For both human and mouse, we constructed 16 tissue-type/cell-type SAGE library combination to determine co-expression of gene pairs, and 50 comparison cases to determine inverse expression of gene pairs (for more details see Materials and Methods). Indeed, the rates of co-expression and inverse expression are significantly higher (P < 10−4; almost 2-fold) in the conserved than in the non-conserved pairs, and significantly higher (P < 10−4) in 3′-to-3′ than in 5′-to-5′ and embedded pairs (Table 2). Again, such a finding could not be explained either by the null neutral model or by the potential incompleteness of 5′ ends. This finding together with the higher conservation rate of 3′-to-3′ overlaps suggests that 3′-to-3′ overlapping pairs might be more functionally important in antisense regulation than 5′-to-5′ overlapping pairs. As expected, the bias between 3′- and 5′-UTR-targeted putative SA pairs is much more evident in potentially functional SA pairs in terms of antisense regulation (i.e. inversely expressed, co-expressed and especially conserved pairs): the ratio of 3′-to-3′ to 5′-to-5′ increases significantly from 1.6 in the whole putative SA set to 8.5 in the conserved and co-expressed putative SA pair set in humans, and in mice from 1.8 to 9.8 (Figure 3 and Table 3).
Table 2.
Comparison of the rates of co-expression and inverse expression between different putative SA pair setsa
| Putative SA pair set | Human | Mouse | ||
|---|---|---|---|---|
| Rate of co-expression | Rate of inverse expression | Rate of co-expression | Rate of inverse expression | |
| Conserved | 60.8% (211/347) | 51.9% (180/347) | 56.8% (197/347) | 44.4% (154/347) |
| Non-conserved | 34.9% (961/2750) | 28.5% (784/2750) | 32.4% (246/759) | 28.7% (218/759) |
| x2-test | P < 10−4 | P < 10−4 | P < 10−4 | P < 10−4 |
| 3′-to-3′ | 51.5% (561/1090) | 41.9% (457/1090) | 55.3% (293/530) | 44.2% (234/530) |
| 5′-to-5′ | 34.2% (227/664) | 29.2% (194/664) | 28.7% (85/296) | 26.7% (79/296) |
| embedded | 28.6% (384/1343) | 23.3% (313/1343) | 23.2% (65/280) | 21.1% (59/280) |
| x2-test (3′-to-3′ versus 5′-to-5′; 3′-to-3′ versus embedded) | P < 10−4; P < 10−4 | P < 10−4; P < 10−4 | P < 10−4; P < 10−4 | P < 10−4; P < 10−4 |
aNote that, all P-values far <10−4 are also shown as P < 10−4. The co-expressed putative SA pairs were defined as those in which the two partners are coordinately expressed in the same tissues more often than expected by chance; the inversely expressed putative SA pairs were defined as those with both an inverse expression pattern between two partners and a significantly greater change of the relative expression ratio of sense to antisense between two states of the same tissues than expected by chance; the conserved putative SA pairs are the pairs conserved as putative SA form in both human and mouse genomes. See Materials and Methods for more details.
Figure 3.
Analysis of the ratio of 3′-to-3′ to 5′-to-5′ among different classes of putative SA pair sets. In both genomes, compared with that in the whole putative SA pair set, the percentage of 3′-to-3′ putative SA pairs increases in inversely expressed, co-expressed, and especially conserved putative SA pair sets, while the percentage of 5′-to-5′ putative SA pairs decreases (Table 3). As a result, the ratio of 3′-to-3′ to 5′-to-5′ pair significantly increases from 1.6 and 1.8 in the whole putative SA set, to 8.5 and 9.8 in the conserved and co-expressed putative SA pair set in the human and mouse genome, respectively.
Table 3.
Comparison of subtype proportions in different putative SA pair setsa
| Putative SA pair set | Genome | Total pairs | 3′-to-3′ (percentage) | 5′-to-5′ (percentage) | Embedded (percentage) | Ratio of 3′-to-3′/5′-to-5′ (x2-test) |
|---|---|---|---|---|---|---|
| The whole | Human | 3097 | 1090 (35.2) | 664 (21.4) | 1343 (43.4) | 1.6 (P < 10−4) |
| Mouse | 1106 | 530 (47.9) | 296 (26.8) | 280 (25.3) | 1.8 (P < 10−4) | |
| Inversely expressed | Human | 964 | 457 (47.4) | 194 (20.1) | 313 (32.5) | 2.4 (P < 10−4) |
| Mouse | 372 | 234 (62.9) | 79 (21.2) | 59 (15.9) | 3.0 (P < 10−4) | |
| Co-expressed | Human | 1172 | 561 (47.9) | 227 (19.3) | 384 (32.8) | 2.5 (P < 10−4) |
| Mouse | 443 | 293 (66.1) | 85 (19.2) | 65 (14.7) | 3.5 (P < 10−4) | |
| Conserved | Human | 347 | 267 (77.0) | 47 (13.5) | 33 (9.5) | 5.7 (P < 10−4) |
| Mouse | 347 | 284 (81.8) | 45 (13.0) | 18 (5.2) | 6.3 (P < 10−4) | |
| Conserved & inversely expressed | Human | 180 | 145 (80.6) | 21 (11.6) | 14 (7.8) | 6.9 (P < 10−4) |
| Mouse | 154 | 134 (87.0) | 17 (11.0) | 3 (2.0) | 7.9 (P < 10−4) | |
| Conserved & co-expressed | Human | 211 | 178 (84.4) | 21 (9.9) | 12 (5.7) | 8.5 (P < 10−4) |
| Mouse | 197 | 176 (89.4) | 18 (9.1) | 3 (1.5) | 9.8 (P < 10−4) |
aNote that, all P-values far <10−4 are shown as P < 10−4. The co-expressed putative SA pairs were defined as those in which the two partners are coordinately expressed in the same tissues more often than expected by chance; the inversely expressed putative SA pairs were defined as those with both an inverse expression pattern between two partners and a significantly greater change of the relative expression ratio of sense to antisense between two states of the same tissues than expected by chance; the conserved putative SA pairs are the pairs conserved as putative SA form in both human and mouse genomes. For more details see Materials and Methods.
Although the number of ncRNAs have been rapidly expanded and many of them have been suggested to have regulatory functions, probably an even larger number of ncRNAs have not been identified yet (4–6,31–33). Thus, it might be the case that protein-coding SA pairs may represent only a small fraction of RNA regulation events. In our datasets, among the 3097 human and 1106 mouse putative SA pairs, 1953 (63.1%) and 469 (42.4%) pairs, respectively, have at least one member belonging to ncRNA. In fact, as tissue/time-specific expression data (as well as reliable sequence data) for ncRNAs is limited, the aforementioned percentages might be seriously underestimated, especially for the percentages of such ncRNA SA pairs in the sets of inversely expressed, co-expressed and/or conserved SA pairs (as shown in Supplementary Table 3). Nonetheless, we observed a similar pattern in the ncRNA SA pair sets (Supplementary Table 3) to that in the whole SA pair sets (Table 3), namely that putative SA pairs overlapping at the 3′-UTRs are significantly more frequent than those overlapping at their 5′-UTRs. Thus, there is no intrinsic bias between protein-coding SA pairs and ncRNA SA pairs with regard to the preference of binding at the 3′-UTRs.
DISCUSSION
It has recently been speculated that many important regulatory sequences will differ between species, and are probable to be evolving more rapidly than those encoding analog (protein) components, since their structure–function relationships are less constrained (6). In agreement with this notion, the majority of human putative SA pairs might not be conserved, even among mammalian genomes [Table 1 and see also ref. (21)]. Nonetheless, we demonstrate that many 3′-to-3′ putative SA pairs are probable to have been conserved among mammals (Table 1) and even among vertebrates that have diverged over 300 million years [Supplementary Table 4; see also refs. (23,34,35)]. We have also observed that evolutionarily conserved putative SA genes, especially those in 3′-to-3′ overlapping pairs, are significantly enriched in gene ontology (36) categories that are essential to cell life, including ‘DNA binding’, ‘nucleotide binding’, ‘response to DNA damage stimulus’ and ‘cell growth and/or maintenance’ (M. Sun, L.D. Hurst, G.G. Carmichael and J. Chen, unpublished data), in accord with Duret et al.'s observation (34) in genes containing highly conserved regions. It is reasonable to assume that if ‘essential genes’ are overlapping in a common ancestor, this overlap is expected to be conserved through evolution regardless of RNA-level (e.g. antisense-mediated) regulation as it will be hard to separate these genes by recombination or by gradual changes in transcription start or stop, particularly if their UTRs contain elements important for posttranscriptional regulation. However, many of these conserved overlapping ‘essential genes’ are found to exhibit co-expression and inverse expression (i.e. characteristics of antisense regulation) in the human and mouse genomes (data not shown). Thus, it is possible that a portion of the conserved overlapping of ‘essential genes’ might be related with antisense regulation, i.e. the antisense regulation modes (if any) of the ‘essential genes’ might be under negative (purifying) selection during evolution because such antisense regulation modes might be also essential to cell life.
Therefore, although the null neutral model may generally explain the preference in initial generation of 3′-to-3′ overlaps compared with 5′-to-5′ overlaps, it is unlikely to explain the significantly higher conservation rates of 3′-to-3′ overlaps, especially when coupled with their enrichment in potentially functional SA pair sets (Tables 1–3 and Figure 3). It is possible that the initial generation of gene overlaps is neutral, however, a novel function of antisense regulation might be preferentially added to 3′-to-3′ overlaps, because the 3′-UTR is not under the same rigid structural constraints as the CDS or the 5′-UTRs that need to accommodate the translational machinery (37), and/or because antisense binding at 3′-UTR of the target gene can avoid the mRNA-clearing activity of the ribosome (38). Such a novel regulatory mode might initially occur through very limited events with very weak function, and it is free to evolve owing to the greater degree of freedom of 3′-UTRs. If the rapidly evolving regulatory mode acquires a useful function, positive selection can strengthen its functions and allow it to occur in more events/tissues. Once a functional antisense-regulation mode was established, especially when the target gene is essential to cell life, the mode would be conserved under negative selection.
Intriguingly, a remarkable bias towards 3′-UTR-targeted antisense regulation has also been observed in the study of microRNAs (miRNA). miRNAs are endogenous, ∼22 nt RNAs that can play important regulatory roles in animals and plants by antisense base pairing. These RNAs may be considered as trans-encoded antisense transcripts, as they are transcribed from a different genomic locus than that of the target gene. Notably, in animals, almost all known target sites for miRNAs are in 3′-UTRs [for reviews see refs. (38–41)]. This bias has also been demonstrated in the candidate target genes of human miRNAs predicted by evolutionary conservation analysis (42) or suggested by empirical study (43). Thus, most functional target sites of animal miRNAs reside in the 3′-UTRs, though some may lie in coding regions (42).
In summary, although it is well known that both the 5′- and 3′-UTRs of eukaryotic mRNAs may play a critical role in posttranscriptional gene regulation (18–20), our study together with other studies suggest that both cis- and trans-encoded putative antisense RNAs prefer to bind at the 3′-UTRs of the potential target genes, and such a feature is highly conserved and potentially related to antisense regulation. While miRNA regulation may act on 3′-UTRs in the cytoplasm, 3′-UTR-targeted SA regulation may occur in the nucleus. In that compartment SA interaction could lead to the formation of double-strand RNAs, activation of RNA editing and nuclear retention of transcripts, modification of chromatin, or other yet obscure consequences (44). In agreement with this model, Kiyosawa et al. (45) recently observed that not only is antisense expression widespread, but that a large fraction of natural antisense transcripts are both poly(A) negative and restricted to the nucleus. These results serve to illustrate that much remains to be uncovered with respect to not only the extent of antisense regulation, but also with respect to the mechanisms by which antisense regulation may occur. Owing to the potential dominance and functional importance of 3′-UTR-targeted antisense regulation, it is expected that mutations in 3′-UTRs may lead to perturbations of the regulation and further result in ‘3′-UTR-mediated diseases’. The importance of 3′-UTRs in gene regulation is underscored by the findings that mutations that alter the 3′-UTR can lead to serious pathology (18,37,46).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr Janet D. Rowley for her full support for this study. We also thank Drs David J. Lipman and Sean R. Eddy for their constructive comments and suggestions, and thank Drs W. James Kent and Xiaoqiu Huang for their help in genome Blat analysis and CAP3 assembly, as well as thank two anonymous reviewers for their constructive and helpful comments. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation (J.C.), NIH grant CA84405 (Janet D. Rowley), and the Spastic Paralysis Foundation of the Illinois, Eastern Iowa Branch of Kiwanis International (Janet D. Rowley). L.D.H. was supported by the UK Biotechnology and Biological Sciences Research Council. G.G.C. was supported by NIH grant GM066816. Funding to pay the Open Access publication charges for this article was provided by the G. Harold and Leila Y. Mathers Charitable Foundation (J.C.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.Levinen M., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 4.Mattick J.S. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattick J.S. RNA regulation: a new genetics? Nature Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 6.Mattick J.S. What makes a human? Scientist. 2005;19:32–33. [Google Scholar]
- 7.Kumar M., Carmichael G.G. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 1998;62:1415–1434. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanhee-Brossollet C., Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- 9.Lavorgna G., Dahary D., Lehner B., Sorek R., Sanderson C.M., Casari G. In search of antisense. Trends Biochem Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Lehner B., Williams G., Campbell R.D., Sanderson C.M. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 11.Shendure J., Church G.M. Computational discovery of sense–antisense transcription in the human and mouse genomes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0044. RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiyosawa H., Yamanaka I., Osato N., Kondo S., Hayashizaki Y., RIKEN GER Group; GSL Members Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yelin R., Dahary D., Sorek R., Levanon E.Y., Goldstein O., Shoshan A., Diber A., Biton S., Tamir Y., Khosravi R., et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Sun M., Kent W.J., Huang X., Xie H., Wang W., Zhou G., Shi R.Z., Rowley J.D. Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J., Sun M., Hurst L.D., Carmichael G.G., Rowley J.D. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense–antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Chen J., Sun M., Hurst L.D., Carmichael G.G., Rowley J.D. Human antisense genes have unusually short introns: evidence for selection for rapid transcription. Trends Genet. 2005;21:203–207. doi: 10.1016/j.tig.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 18.Mignone F., Gissi C., Liuni S., Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pesole G., Liuni S., Grillo G., Saccone C. UTRdb: a specialized database of 5′- and 3′-untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 1998;26:192–195. doi: 10.1093/nar/26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkie G.S., Dickson K.S., Gray N.K. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 21.Veeramachaneni V., Makalowski W., Galdzicki M., Sood R., Makalowska I. Mammalian overlapping genes: the comparative perspective. Genome Res. 2004;14:280–286. doi: 10.1101/gr.1590904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuler G.D., Boguski M.S., Stewart E.A., Stein L.D., Gyapay G., Rice K., White R.E., Rodriguez-Tome P., Aggarwal A., Bajorek E., et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 23.Lipman D.J. Making (anti)sense of non-coding sequence conservation. Nucleic Acids Res. 1997;25:3580–3583. doi: 10.1093/nar/25.18.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adachi N., Lieber M.R. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 25.Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 27.Lercher M.J., Urrutia A.O., Hurst L.D. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nature Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki Y., Yamashita R., Nakai K., Sugano S. DBTSS: database of human transcriptional start sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mironov A.A., Fickett J.W., Gelfand M.S. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mignone F., Grillo G., Licciulli F., Iacono M., Liuni S., Kersey P.J., Duarte J., Saccone C., Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy S.R. Non-coding RNA genes and the modern RNA world. Nature Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 32.Eddy S.R. Computational genomics of noncoding RNA genes. Cell. 2002;109:137–140. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 33.Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- 34.Duret L., Dorkeld F., Gautier C. Strong conservation of non-coding sequences during vertebrates evolution: potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res. 1993;21:2315–2322. doi: 10.1093/nar/21.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duret L., Bucher P. Searching for regulatory elements in human noncoding sequences. Curr. Opin. Struct. Biol. 1997;7:399–406. doi: 10.1016/s0959-440x(97)80058-9. [DOI] [PubMed] [Google Scholar]
- 36.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conne B., Stutz A., Vassalli J.D. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nature Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 38.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 39.Bartel D.P., Chen C.Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 40.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 41.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 42.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 44.DeCerbo J., Carmichael G.G. Retention and repression: fates of hyperedited RNAs in the nucleus. Curr. Opin. Cell. Biol. 2005;17:302–308. doi: 10.1016/j.ceb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Kiyosawa H., Mise N., Iwase S., Hayashizaki Y., Abe K. Disclosing hidden transcripts: mouse natural sense–antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 2005;15:463–474. doi: 10.1101/gr.3155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grzybowska E.A., Wilczynska A., Siedlecki J.A. Regulatory functions of 3′UTRs. Biochem. Biophys. Res. Commun. 2001;288:291–295. doi: 10.1006/bbrc.2001.5738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.