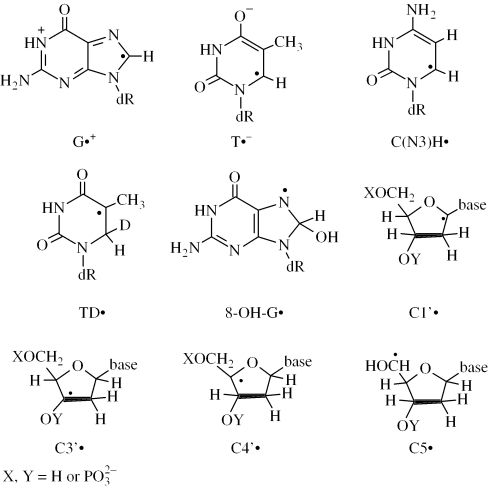
Abstract
This work presents evidence that photo-excitation of guanine radical cations results in high yields of deoxyribose sugar radicals in DNA, guanine deoxyribonucleosides and deoxyribonucleotides. In dsDNA at low temperatures, formation of C1′• is observed from photo-excitation of G•+ in the 310–480 nm range with no C1′• formation observed ≥520 nm. Illumination of guanine radical cations in 2′dG, 3′-dGMP and 5′-dGMP in aqueous LiCl glasses at 143 K is found to result in remarkably high yields (∼85–95%) of sugar radicals, namely C1′•, C3′• and C5′•. The amount of each of the sugar radicals formed varies dramatically with compound structure and temperature of illumination. Radical assignments were confirmed using selective deuteration at C5′ or C3′ in 2′-dG and at C8 in all the guanine nucleosides/tides. Studies of the effect of temperature, pH, and wavelength of excitation provide important information about the mechanism of formation of these sugar radicals. Time-dependent density functional theory calculations verify that specific excited states in G•+ show considerable hole delocalization into the sugar structure, in accord with our proposed mechanism of action, namely deprotonation from the sugar moiety of the excited molecular radical cation.
INTRODUCTION
Free radicals on the deoxyribose moiety of DNA are precursors to DNA strand breaks, with the double strand break known to be the most biologically significant radiation-produced lesion (1–4). Elucidation of the production and nature of DNA sugar radicals is, therefore, critical to understanding their damaging effects in DNA. The deoxyribose radical formed by hydrogen atom loss at the C1′ position (C1′•) is known to result in an alkali labile strand break, whereas the C3′•, C4′• and C5′• sugar radicals can lead to frank strand breaks (5–7). Although some progress has been made in understanding the sugar radicals that are present in DNA irradiated at low temperatures, using both low and high LET radiation (8–13), there is, as yet, no clear picture of the radiation yield of the individual sugar radicals formed and stabilized at low temperatures in irradiated DNA. Thus, there is a substantial ongoing effort to identify and characterize the radicals formed on the deoxyribose-phosphate backbone in irradiated DNA and on the sugar moiety in DNA nucleoside/tide model compounds (8–15). Two mechanisms for sugar radical formation have been recently described. In one, photo-excitation of a base radical cation results in charge and unpaired spin transfer to the sugar followed by deprotonation from the sugar (12,13); a second invokes low energy electron induced C–O or P–O bond dissociation resulting in radicals from phosphate loss (16,17).
Radiation studies have been employed in previous work to produce deoxyribose radicals in DNA and in model compounds in a variety of systems, such as single crystals, aqueous glasses and frozen aqueous solutions; this literature has been reviewed [(8–17) and references therein]. At low temperature, in irradiated hydrated-DNA, both C1′• and C3′• have been identified (8–19). However, to the best of our knowledge, there have been no reliable identifications of C2′•, C4′• or C5′• stabilized in hydrated DNA at low temperatures (12,13).
Characterizing sugar radicals in γ-irradiated DNA at low temperatures using ESR spectroscopy has been fraught with difficulty (8–19) because they comprise only a small percentage (10–15%) of all radicals formed, and because of the variety of sugar radical conformations defined by the pseudo-rotation cycle (20). Since ESR spectral parameters for any sugar radical will strongly depend on the sugar conformation, the latter complicates the identification and characterization of sugar radicals even apart from their low yields (21–26).
This work follows our recent work, which showed that photo-excitation of guanine radical cation (G•+) in DNA and in 2′-deoxynucleosides/tides at 77 K results in the formation of deoxyribose radicals (15). In γ-irradiated DNA, the guanine radical cation (G•+) is the dominant electron loss center (8–19) consistent with the fact that among the DNA bases, guanine has the lowest oxidation potential at pH 7 (7,27–31). In this work, we report experimental and theoretical results that clarify the mechanism of formation of sugar radicals in DNA- and guanine-based model systems. Specifically, we report the wavelength dependence for the formation of C1′• from photo-excited G•+ in dsDNA and, in doing so, correct an earlier report (15). Furthermore, using 2′-dG, 5′-dGMP and 3′-dGMP, we identify and further characterize C1′•, C3′• and C5′• formed via photo-excitation of guanine radical cations; selective deuteration at C3′, C5′ and C8 is used to confirm radical assignments. Most importantly, by performing the photo-excitation at elevated temperatures (∼143 K) and controlling conditions, such as pH, we have been able to achieve near complete conversion of guanine radical cations to specific sugar radicals. We note that this work shows that guanine radical cations in DNA and model systems readily convert to deoxyribose radicals via photo-excitation, and that this observation has implications for many studies [(32) and references therein] in which holes (guanine radical cations, G•+) in DNA are subjected to visible illumination.
MATERIALS AND METHODS
DNA sample preparation
Salmon testes DNA (sodium salt, 57.3% AT and 42.7% GC), was obtained from Sigma Chemical Company (St Louis, MO). Deuterium oxide (99.9 atom% D) was obtained from Aldrich Chemical Company Inc. (Milwaukee, WI). These were used without any further purification.
Ice-like samples of DNA, with or without Tl3+, were prepared using procedures described previously [(15) and references therein]. The Tl3+ loading used (1/10 bp) is sufficient to thoroughly suppress the reductive-damage pathway (33) and allows observation of the formation of sugar radicals from photo-excited G•+ (15). All samples were stored at 77 K.
Model compound sample preparation
2′-Deoxyguanosine (2′-dG), 2′-deoxyguanosine 3′-monophosphate (3′-dGMP), 2′-deoxyguanosine 5′-monophosphate (5′-dGMP) and lithium chloride (99% anhydrous, SigmaUltra) were obtained from Sigma Chemical Company (St Louis). Potassium persulfate (crystal) was purchased from Mallinckrodt, Inc. (Paris, KY). 3′-D-2′-deoxyguanosine (3′-D-2′-dG) and 5′,5′-D,D-2′-deoxyguanosine (5′-D,D-2′-dG) were obtained from Omicron Biochemicals, Inc. (South Bend, IN). All chemicals were used without further purification. Deuteration at C-8 in the guanine moiety of 2′-dG, 5′-D,D-2′-dG, 3′-dGMP and 5′-dGMP was performed according to Huang et al. (34) using triethylamine (TEA) from Fischer Scientific, NJ; the degree of deuteration (≥96%) was determined by 1D NMR signal integration using a Bruker 200 MHz NMR.
To prepare glassy samples, ∼3 mg of nucleoside or nucleotide were dissolved in 1 ml of 7 M LiCl in D2O in the presence of 5 mg K2S2O8. If required, the pH was adjusted by adding appropriate amounts of 1 M NaOH in D2O or concentrated HCl under ice-cooled conditions. All pHs were measured using pH paper; because of this and because of the high ionic strength of the glasses, the pHs reported here are approximations. The solutions were then thoroughly bubbled with nitrogen. Using these solutions, the glassy samples were then prepared by cooling to 77 K as reported earlier (15). All samples are stored at 77 K.
γ-Irradiation
DNA and DNA-Tl3+ ice samples were γ-irradiated (60Co) with an absorbed dose of 15.4 kGy. Glassy samples of nucleosides and nucleotides were γ-irradiated with an absorbed dose of 2.5 kGy. All irradiations were performed at 77 K.
Annealing and illumination of samples
γ-Irradiated DNA and DNA-Tl3+ ice samples were annealed to 130 K to remove the ESR signal of •OH (35,36); since the •OH is in the separate ice phase, annealing does not result in additional DNA radicals (35,36). These samples were then illuminated with light at two different temperatures. At 77 K, either of the two sources was used: (i) a 200 W high pressure Xe lamp (Oriel Corporation), with cut-off filters which cut off light ≤310 nm, ≤480 nm, or ≤540 nm, or band-pass filters (340–370 nm and 380–480 nm), (ii) a Nd-yag laser at 521 nm, with 40 J/pulse and a 10 Hz repetition rate. At 143 K, a 250 W tungsten lamp was used with and without a variety of cut-off filters.
Glassy guanine model compound samples were annealed to 150 K for 10–12 min (see Results) which resulted in the loss of (light yellow) Cl2•− and the concomitant formation of G•+ as evidenced by the ESR spectrum and color development in the sample: red–violet at pH ≤ 9, and blue at pH ≥ 11. The samples of deoxyribonucleosides/tides were illuminated at a variety of temperatures using a 250 W tungsten lamp with and without cut-off or band-pass filters. The phrase ‘visible light illumination’ used throughout this paper refers to illumination with this lamp with wavelengths ≤310 nm cut-off; because of the small size of the sample and small solid angle it subtends, only a small fraction of the 250 W impinges on the sample.
Electron spin resonance
After γ-irradiation, annealing and light illumination, the ice or glassy sample was immediately immersed in liquid nitrogen, and an ESR spectrum recorded at 77 K and 40 dB (20 µW) [(11,15,33) and the references therein]. Fremy's salt (with g = 2.0056, AN = 13.09 G) was used for field calibration.
Analysis of ESR spectra
ESR spectra presented in this paper have been computer analyzed for component radicals by use of benchmark spectra. The fractional compositions of radicals observed in experimental spectra were determined using least-square fittings of benchmark spectra with the help of programs (ESRPLAY and ESRADSUB) developed in our laboratory [(11,15,33) and the references therein]. A sharp singlet spike from irradiated quartz at g = 2.0006 was subtracted from all spectra before analysis.
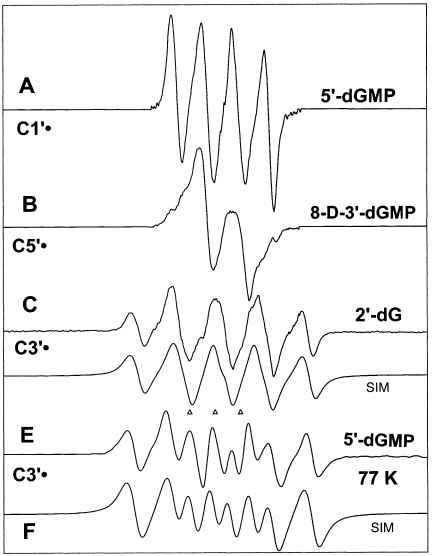
The origin of the benchmark spectra used is described in the Appendix. The benchmark spectra for nucleosides/tides shown in Figure 1 are those for C1′• (Figure 1A), C5′• (Figure 1B) and C3′• (Figure 1C and E). Two benchmark spectra for C3′• are used (Figure 1C and E) because the three major hyperfine coupling constants vary slightly with compound and temperature of illumination (Table 2). For analysis of the spectra obtained from DNA, the benchmark spectra used include the guanine one-electron oxidized radical cation (G•+), the one-electron reduced species, namely T−• and C(N3H)•, and a composite spectrum of neutral radicals found in low temperature irradiated DNA [(15,33) and references therein].
Figure 1.
Benchmark spectra used for computer analysis. (A) C1′•, produced from G•+ in 5′-dGMP. (B) C5′•, produced from G•+ in 8-D-3′-dGMP (C) C3′•, produced from G•+ in 2′-dG. (D) Computer simulated spectra of C3′•, to match experimental spectrum of C. (E) C3′•, found after photo-excitation of G•+ at 77 K in 5′-dGMP. (F) Computer simulated spectra of C3′•, to match experimental spectrum of E. (See Table 1 and Appendix for details).
Table 2.
Wavelength dependence of radical formation from G•+ in 2′-dGa
| Wavelength of illumination (nm) | C1′• | C3′• | C5′• |
|---|---|---|---|
| 380–480 | 7% | 35% | 58% |
| >540 | 3% | 40% | 57% |
aHundred minutes of illumination at 143 K. Percentages to ±5%.
Time-dependent density functional theory (TD-DFT) calculations
TD-DFT calculations were performed to determine the energy and the nature of the molecular orbitals in geometry optimized 2′-dG•+. Optimization was performed with DFT using B3LYP functionals with the 6–31G(d) basis set provided in the Gaussian 03 program package (37–39). TD-DFT calculations were performed using the 6–31G(d) and 6–311++G(d,p) (see Supplementary Data) basis sets for a total of 16 electronic transitions. Calculations for 24 transitions using the 6–31G(d) basis set gave identical results for the first 12 states which are considered in this work.
RESULTS
UV-VIS absorption spectra of G•+
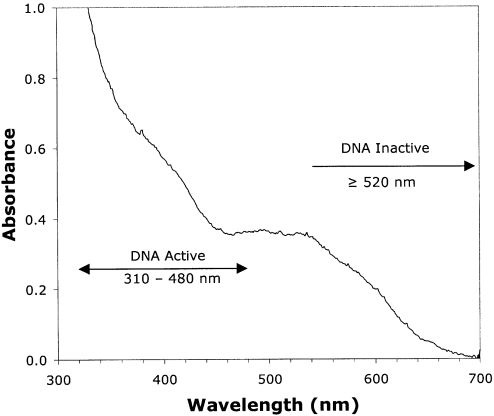
Figure 2 shows the UV-visible absorption spectrum of G•+ in 2′-dG, taken at 77 K in a 7 M LiCl glass. This spectrum is very similar to that reported by others for G•+ in TpdG, dCpdG and ApG in aqueous solution at room temperature (40) as well as in aqueous solutions of double-stranded oligonucleotides containing a GGG sequence (31).
Figure 2.
The UV-visible absorption spectrum of G•+ produced by Cl2•− oxidation of 2′-dG, as described in Materials and Methods, at 77 K in 7 M LiCl/D2O.
For DNA, the conversion of photo-activated G•+ to sugar radicals occurs with light of wavelength 310–480 nm, but not with wavelengths ≥520 nm. In contrast, the deoxynucleosides/tides investigated show no wavelength dependence for the conversion of G•+ to sugar radicals at wavelengths from 310 nm and above (vide infra).
Frozen solutions of DNA
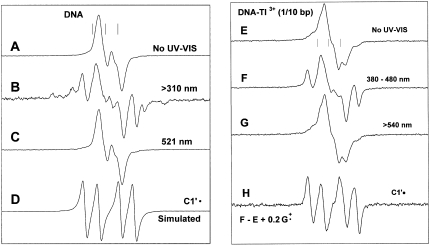
Figure 3A shows the ESR spectrum obtained from γ-irradiated samples of frozen aqueous solutions of DNA. The spectrum is identical to that obtained in previous investigations and has been previously assigned to 85–90% base radicals [36% G•+, 36% C(N3)H•, 15% T•−] and 10–15% neutral (largely deoxyribose) radicals (13–15). The spectrum in Figure 3B was obtained after illumination of the sample at 77 K for 1 h (200 W xenon lamp) using a filter which cuts off wavelengths ≤310 nm. This spectrum has prominent line components associated with the quartet ESR spectrum of C1′• and small line components from the thymine deuterium atom adduct, TD• (15). This observation is the same as that reported earlier (15), but with an important amplification and clarification of the wavelength of light that causes the photolytic conversion. In this regard, Figure 3C shows the spectrum of an identically prepared sample that initially gave the ESR spectrum in Figure 3A, but that was illuminated with 521 nm light from an Nd-YAG laser for 30 min. This spectrum is identical to that shown in Figure 3A and indicates that 521 nm light does not cause the photolytic conversion observed in Figure 3B. Figure 3D shows a simulated ESR spectrum of C1′• in DNA using the couplings in Table 1 and a line width of 5 G (15). The line components of the simulated spectrum nicely match those in Figure 3B.
Figure 3.
(A) ESR spectrum of DNA ice samples (50 mg/ml D2O) γ-irradiated to 15.4 kGy dose, annealed to 130 K to eliminate •OH. (B) After illumination at 77 K for 1 h with light >310 nm. (C) After illumination of an identically prepared sample as in (A), at 521 nm for 30 min. (D) Simulated spectrum of C1′• using the parameters shown in Table 1. This spectrum matches line components in spectrum B. (E) ESR spectrum of DNA (50 mg/ml) with 1 Tl3+/10 base pairs γ-irradiated to 15.4 kGy dose, annealed to 130 K to eliminate •OH. (F) After illumination, at 77 K, of the sample in E with 380–480 nm light for 110 min; (G) After illumination, at 77 K, of an identically prepared sample as in E, using light with wavelength >540 nm, for 30 min. (H) Subtraction of E from F, plus addition of 0.2G•+ showing the growth of the spectrum from C1′•; addition of the G•+ spectrum added compensates for its loss in F on photolysis.
Table 1.
| Compoundc | Hyperfine coupling constants (G) | g-value |
|---|---|---|
| C1′• | ||
| 2′-dG, 5′-D,D-2′-dG, 5′-dGMP | 16 (1βH), 32 (1βH) | 2.0029 |
| 3′-dGMP | 15.5 (1βH), 35(1βH) | 2.0029 |
| DNA (77 K illumination) | 15 (1βH), 37 (1βH) | 2.0029 |
| C5•′ | ||
| 2′-dG, 5′-D,D-2′-dG, 3′-dGMP | ∼19 (1αH) | 2.0025 |
| C3′• | ||
| 2′-dG, 5′-D,D-2′-dG | ∼21 (1βH), 25 (1βH), 42 (1βH) | 2.0032 |
| 5′-dGMPd | ∼20 (1βH), 30 (1βH), 41 (1βH) | 2.0032 |
aValues are for radicals at 77 K, formed by illumination at 143 K and at native pH (∼5), unless otherwise stated.
bDNA in frozen aqueous solutions (ice). Other samples in aqueous glasses (7 M LiCl/D2O).
cDeuteration at C8 in 2′-dG, its 3′- and 5′-nucleotides and in 5′-D,D-2′-dG has no effect on the values reported here.
d77 K illumination.
In spectra 3F to 3H, we present ESR spectra that result from illumination of G•+ in DNA-Tl3+ ice samples (77 K, 200 W Xe lamp), at different wavelengths. Figure 3E shows the spectrum of a sample before illumination. In agreement with previous results, analyses using the benchmark spectra shown in Figure 1 and those in ref. (15) indicate that this spectrum is a composite originating with ∼50% G•+, a small amount of T•− and C(N3)H• and the remainder sugar radicals (15). Spectrum F was obtained by photo-excitation of the sample for 100 min using a 380–480 nm band-pass filter. This spectrum shows prominent line components from C1′•. Both spectra 3E and 3F have approximately the same total integrated radical intensities, and thus it is evident that most of C1′• originates with a photolytic conversion. The isolated spectrum of C1′• shown in Figure 3H results from the subtraction of spectrum 3E (100%) from 3F with an addition of G•+ (20%) to compensate for the loss of G•+ by photolysis.
Photo-excitation (200 W Xe lamp) of DNA-Tl3+ ice samples with light in the 340–370 nm band yields results identical to those from the 380 to 480 nm band. However, at wavelengths >540 nm, C1′• is not produced in these samples at 77 K (Figure 3G). As observed in DNA-ice samples lacking Tl3+, illumination of DNA-Tl3+ samples with light <310 nm does not contribute significantly to the formation of C1′•.
In summary, these experiments show that, in DNA, illumination with light in the 310–480 nm range causes conversion of G•+ to C1′•, and light under 310 nm or ≥520 nm does not cause the conversion (Schemes 1 and 2).
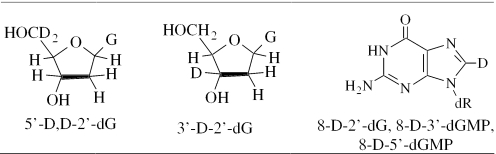
Scheme 1.
Isotopically substituted compounds used.
Scheme 2.
Radicals described.
MODEL COMPOUNDS
Formation of base radical cations in aqueous glass
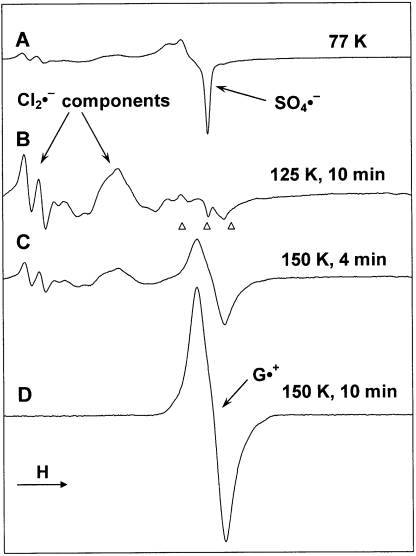
Figure 4 illustrates the method used to prepare G•+ in LiCl glasses. Figure 4A shows the ESR spectrum (77 K) of a sample of a nucleoside in 7 M LiCl containing K2S2O8, γ-irradiated to a dose of 2.6 kGy; in this instance the nucleotide is 5′-D,D-2′-dG, but the identity of the nucleoside/tide is unimportant in the process described. The spectrum shows the central portion of the 700 G wide multiplet from Cl2•− and a sharp singlet from SO4•−, produced, respectively, from radiation-produced holes and electrons (reactions 1 and 2). Annealing at 125 K for 10 min results in the oxidation of Cl− by SO4•− to yield Cl2•− (reaction 3) and its spectrum (Figure 4B). Further annealing at 150 K for 10 min permits Cl2•− to diffuse to and react with the guanine moiety of the nucleoside (reaction 4). The resulting spectrum (Figure 4D) originates, in this case, from G•+, without observable spectra from any other radical and is the same as that observed earlier for G•+ (8–19). In fact, the ESR spectra from G•+ in all of the guanine nucleosides/tides except 8-D-2′-dG are identical.
| 1 |
| 2 |
| 3 |
| 4 |
Figure 4.
(A) ESR spectrum showing SO4•− and Cl2•− formation in γ-irradiated (2.5 kGy) sample of a nucleoside (5′-D,D-2′-dG) in the presence of K2S2O8 in a 7 M N2-saturated LiCl/D2O glass. (B) Spectrum of the sample in (A) after annealing to 125 K for ∼10 min. (C) After further annealing at 150 K for 4 min. (D) After annealing for another 6 min (i.e. total 10 min) at 150 K. Only the spectrum of G•+ is observed at this point. All ESR spectra were recorded at 77 K.
2′-dG and its deuterated derivatives
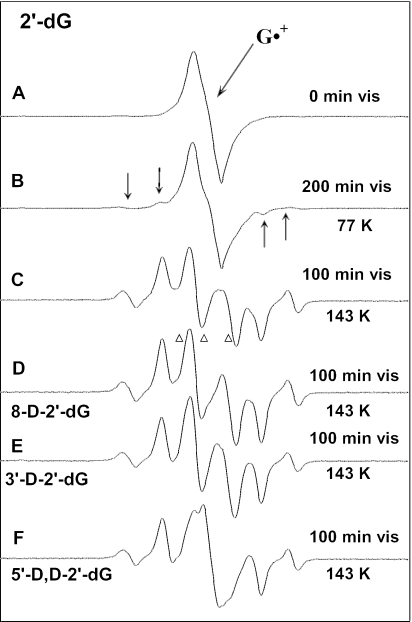
In Figure 5A, we show the ESR spectrum of G•+ in 2′-dG, after preparation from reaction with Cl2•−. Figure 5B shows the spectrum after 200 min of visible light illumination at 77 K. After illumination, four new weak line components, which have previously been assigned to C3′• (8–19), are visible in the wings of the spectrum (arrows). Figure 5C shows the spectrum of a sample prepared identically to that in Figure 5A, after 100 min of illumination at 143 K followed by subtraction of the spectrum of G•+ (5A) as ∼10% of the intensity of the original spectrum. The resulting spectrum (5C) is attributed entirely to sugar radicals. Computer analysis of this spectrum using the benchmark spectra for C1′•, C5′• and C3′• shown in Figure 1A–C, respectively, indicates it is a composite from C1′• (∼10%), C5′• (∼55%) and C3′• (∼35%) (Table 4).
Figure 5.
(A) Spectrum of G•+ in 2′-dG before illumination; (B) after visible illumination at 77 K of G•+ in 2′-dG. Arrows indicate four outer line components from C3′•. (C) After visible illumination at 143 K of a new sample of G•+ in 2′-dG, showing a nearly complete conversion to sugar radicals (Table 4). A central doublet assigned to C5′• is present. (D) Spectrum after visible illumination at 143 K of G•+ in 8-D-2′-dG. A central doublet from C5′• is present. (E) Spectrum after visible illumination at 143 K of G•+ in 3′-D-2′-dG. The central doublet from C5′• is also present here, but with a slightly smaller splitting than that observed in 2′-dG (Figure 5C) and in 8-D-2′-dG (Figure 5D). (F) After visible illumination at 143 K of G•+ in 5′-D,D-2′-dG. The central doublet from C5′• has collapsed to a singlet. All ESR spectra are recorded at 77 K. The sample in (A) is red–violet; those in (C–F) were colorless.
Table 4.
| Compound | Temperature (K)c | Percent convertedd | C1′•e | C3′•e | C5′•e |
|---|---|---|---|---|---|
| 2′-dG | 143 | 90 | 10% | 35% | 55% |
| 77 | 30 | 10 | 40 | 50 | |
| 5′-dGMP | 143 | 95 | 95 | 5 | – |
| 77 | 30 | 15 | 30 | 55 | |
| 3′-dGMP | 143 | 85 | 40 | – | 60 |
| 77 | 15 | 40 | – | 60 | |
| dsDNA (ice) | 143 | ∼50 | 100 | – | – |
| 77 | ∼50 | 100 | – | – |
aPercentage expressed to ±5% relative error.
bAll glassy samples are at the native pH of 7 M LiCl (ca 5). For DNA, the pH of the aqueous solution before freezing was 7.
cTemperature at which guanine radical cation was illuminated.
dPercentage of guanine radical cation that converts to sugar radicals. The total spectral intensities before and after illumination were the same, within experimental uncertainties.
eEach calculated as percentage of total sugar radical concentration; these sum to 100%.
In earlier work, we suggested that the central doublet in spectrum 5C, now assigned to C5′•, might be due to 8-HO-G• (15). However, using 8-D-2′-dG handled, irradiated and illuminated identically as 2′-dG, the spectrum shown in Figure 5D results. The central ca. 19 G doublet present in 2′-dG is still present; had the doublet been due to 8-HO-G•, it would have collapsed to a singlet in the deuterated compound. Because of the smaller magnetic moment of deuterons, they show couplings that are only 15% (1/6.514) that of protons in the same environment. Thus, the results with the C-8 deuterated compound show that the doublet cannot originate with 8-HO-G•.
After photo-excitation of G•+ in 3′-D-2′-dG at 143 K and subtraction of the remaining G•+ (15% of the original spectrum), the resulting spectrum is shown in Figure 5E. In this case, the overall features of the spectrum are the same as for 2′-dG and 8-D-2′-dG. This confirms the C3′ proton is not the source of the central doublet splitting. We do note the doublet splitting is observed to be slightly smaller (∼17 G) than that in 2′-dG and 8-D-2′-dG (∼19 G).
The spectrum found after photo-excitation of G•+ in 5′-D,D-2′-dG at 143 K (Figure 5F) and subtraction of the remaining G•+ (15% of the original spectrum) is shown in Figure 5E. As can be seen, the central (∼19 G) doublet observed in 2′-dG and 8-D-2′-dG is converted to a singlet. It should be noted that the singlet in Figure 5F does not originate with any remaining G•+, since it differs considerably in shape from the G•+ spectrum (see Supplementary Data S1); most importantly, the characteristic visible absorption associated with G•+ is lost after 100 min of photo-excitation.
The collapse of the doublet to a singlet only in 5′-D,D-2′-dG and not in 3′-D-2′-dG restricts the choice of radicals giving rise to the doublet. In principle, the doublet could be due to an alpha-hydrogen on C5′• or a beta-hydrogen on C4′•; no other radical should have a significant hyperfine coupling to a C5′ proton. The results for deuteration at C3′ show that the doublet remains. This is more consistent with C5′• than C4′•. We therefore assigned the doublet in spectra 5C–5E and the corresponding singlet in 5F to C5′•.
ESR line components assigned to C3′• and C1′• are not affected by the deuterium substitutions in 3′-D-2′-dG and 5′-D,D-2′-dG (Figure 5C, E and F); this result is consistent with these assignments, since deuterium substitution at C3′ and at C5′ should have no significant effect on the spectra of these two radicals. Samples of trideuterated 2′-dG, i.e. 2′-dG dideuterated at C5′, and also deuterated at C8 (8-D-5′-D,D-2′-dG) were also investigated (see Supplementary Data S1). The sharper singlet from C-8 deuterated G•+ is easily distinguished from the broad singlet from C5′• in the 5′-D,D-2′-dG and further confirms the collapse of the doublet to a singlet.
From the yields of C5′• in 5′-protonated versus 5′-deuterated 2′-dG, we have estimated a small deuterium isotope effect for the formation of C5′• of ∼1.5 ± 0.3. From the yields of C3′• in 3′-protonated versus 3′-deuterated 2′-dG, we have estimated a small deuterium isotope effect for the formation of C3′• of ∼1.3 ± 0.3.
Wavelength dependence of sugar radical formation from photoexcitation of G•+ in 2′-dG
An examination of the dependence of the wavelength of light used to illuminate the radical cation (G•+) of 2′-dG on the relative yields of the resulting sugar radicals was performed. No significant dependence on the wavelength of light was found (Table 2).
5′-dGMP
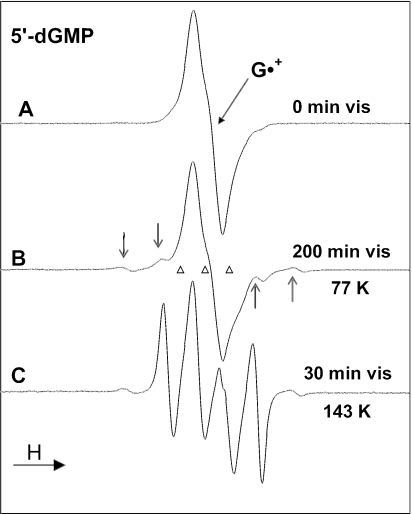
In Figure 6A, we show the ESR spectrum of G•+ (77 K) from 5′-dGMP produced through oxidation by Cl2•−. Figure 6B shows the ESR spectrum after 200 min of visible light illumination at 77 K. Computer analysis using the benchmark spectra in Figure 1 and the G•+ spectrum from Figure 6A indicates that ∼30% of the original G•+ is converted to sugar radicals; in this sugar radical cohort the spectral composition is C1′• (∼15%), C3′• (30%) and C5′• (55%) (Table 4). Warming this sample to 143 K without any photo-excitation in the dark results only in small changes in hyperfine couplings and sharpening of the C1′• and C3′• spectra (data not shown). This effect is ascribed to structural relaxation of the radicals.
Figure 6.
(A) ESR spectrum from G•+ in 5′-dGMP. (B) Spectrum after photo-excitation at 77 K with visible light, giving a small amount of C3′• (arrows show four outer components from C3′•) and C1′• (Table 4). (C) After photo-excitation, at 143 K, of a fresh sample of G•+, showing 95% conversion of G•+ to sugar radicals, primarily C1′• (prominent quartet) (see text and Table 4). The sample in (A) is red–violet whereas that in (C) is colorless.
On photo-excitation of a new sample of G•+ in 5′-dGMP at 143 K, spectrum 6C is observed; near complete conversion of G•+ to C1′• is observed after only 30 min of illumination. After subtraction of G•+ as 5% of spectrum 6C, the remaining sugar radical spectrum originates with ∼95% C1′• and ∼5% C3′•. The ∼19 G doublet from C5′• is not present at any observable level. After subtraction of the appropriate amount of C3′•, the spectrum in Figure 1A results. The g-values and hyperfine couplings of the C1′• spectrum thus obtained agree well with those in the literature (8–19) and, hence, Figure 1A is used as a benchmark spectrum for C1′•.
3′-dGMP and its deuterated derivative
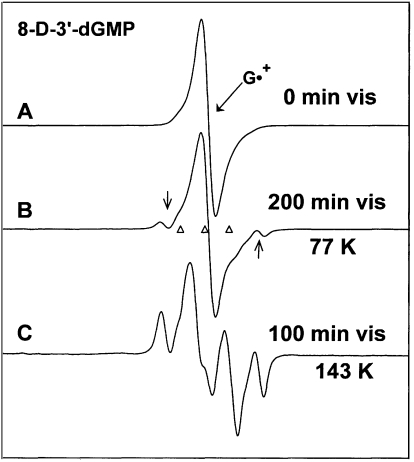
3′-dGMP and 8-D-3′-dGMP were investigated to determine the effect of phosphate position on radical formation and to ascertain whether or not there is any contribution of the base adduct radical 8-HO-G• to the ESR spectra observed. Identical results were found for these two compounds, except that the spectrum from G•+ is sharper in 8-D-3′-dGMP than in 3′-dGMP. Only the results for 8-D-3′-dGMP are presented. Figure 7A shows the ESR spectrum of G•+ in 8-D-3′-dGMP at 77 K. The presence of a deuteron at C8 rather than a proton reduces the ca. 8 G proton hyperfine coupling (41) to an unresolved 1.2 G deuterium splitting and, thereby, significantly sharpens the ESR spectrum of G•+ relative to those observed in Figures 5A and 6A. After 200 min of visible light illumination at 77 K, the spectrum in Figure 7B is observed. Computer analysis using the spectrum of G•+ (Figure 7A) and the benchmark spectra in Figure 1 indicates the spectrum in Figure 7B is made up of contributions from G•+ (85%), C1′• (6%) and C5′• (9%) (Table 2); 3′-dGMP gave the same results after illumination at 77 K. In earlier work (15), we reported a small contribution from C3′• (∼10%) for a similarly prepared sample, but now believe that was in error.
Figure 7.
(A) ESR spectrum of G•+ in 8-D-3′-dGMP. The deuteron at C8 causes this spectrum to be sharper than other G•+ spectra shown. (B) After visible light illumination at 77 K. Two C1′• line components are visible (arrows). (C) After illumination, at 143 K, of a fresh sample of G•+, showing a composite spectrum from (C5′•) (central doublet, 50%), C1′• (quartet, 35%) and G•+ (15%). Subtractions of the C1′• and the G•+ spectra from (7C) result in spectrum 1B, the benchmark for C5′•.
The ESR spectrum found in a separate sample after 100 min of visible illumination of G•+ in 8-D-3′-dGMP at 143 K is shown in Figure 7C. Prominent line components from C1′• and an intense central ca. 19 G doublet from C5′• are visible in the spectrum. Analysis using the spectrum in Figure 7A and the benchmark spectra in Figure 1 indicates the spectrum in Figure 7C contains contributions from C1•′ (35%), C5′• (50%) and G•+ (15%). The doublet assigned to C5′•, created by subtracting the spectra of C1• and G•+ from Figure 7C is shown in Figure 1B. Samples of 3′-dGMP gave an identical final spectrum as that found in Figure 7C for 8-D-3′-dGMP; this again confirms that the radicals giving rise to the spectrum are on the sugar moiety.
pH dependence on photo-excitation in G•+
The effect of visible light illumination at 143 K on G•+ in 2′-dG was investigated at different pHs (see Supplementary Data S2). Photo-conversion of G•+ to sugar radicals occurs in the pH range 2–6; on the other hand, at pH ∼7–11, no significant photo-conversion is found. In G•+, the pKa of the N1-hydrogen is 3.9 (27–31), at pH 7 and above the N1-hydrogen is almost entirely dissociated (reaction 5). Thus, deprotonation at N1 in G•+ essentially prevents sugar radical formation on illumination. In dsDNA, G•+ is partially deprotonated (29,40); hence, the partial deprotonation of the one-electron oxidized guanine base in dsDNA may explain the somewhat lower yield of C1′•, relative to the model nucleosides/tides investigated (Table 4).
| 5 |
TD-DFT calculations
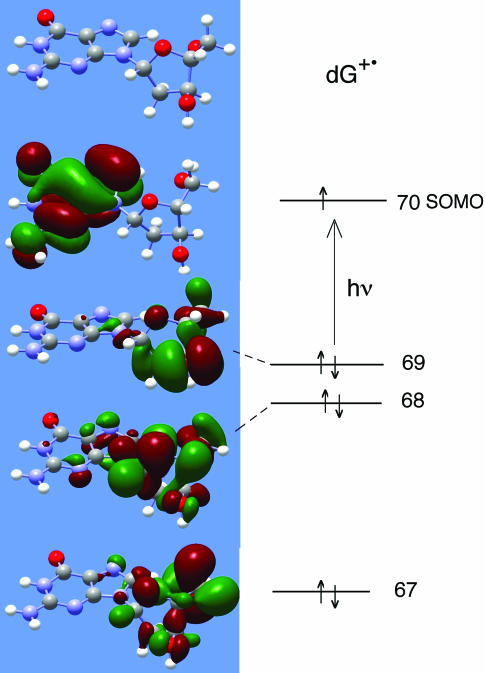
Our proposed mechanism of action is that on photo-excitation the various sugar radicals are produced by charge and spin delocalization into the sugar moiety of the nucleoside/tide (15). Subsequent fast deprotonation from the sugar results in the formation of a neutral sugar radical. To aid in our understanding of this mechanism we have performed TD-DFT calculations of the excited states and transition energies using B3LYP functionals and a 6–31G(d) basis set (37–39, 42–44) for G•+ in 2′-dG; the TD-DFT method has been shown to produce results superior to other methods for this type of calculation (44). The structure of G•+ was first geometry optimized [B3LYP, 6–31G(d)]. The first 12 states predicted throughout the UVA-visible region are shown in Table 3. All 12 include transitions between lower lying MOs 58–69 and the SOMO 70. TD-DFT calculations were also performed with the much larger 6–311++G(d,p) basis set, with very similar results for energies (each was higher by only 0.05 eV on average), similar wavelengths of the transitions and similar oscillator strengths; thus, our conclusions are not affected by the basis set size (see Supplementary Table S1). However, more mixing of MOs in the states occurs with the larger basis set, making the calculation less visually instructive.
Table 3.
TD-DFT b3lyp 6–321G* calculated electronic transitions for 2′-dG•+
| ΔE (eV) | λ (nm) | fa | Transition (density) | Delocalization into sugarb |
|---|---|---|---|---|
| 1.41 | 881 | 0.0009 | 68β→70β (0.98)c | 3 |
| 1.48 | 836 | 0.0003 | 69β→70β (0.99) | 3 |
| 1.70 | 728 | 0.0001 | 66β→70β (0.96) | 0 (none) |
| 1.83 | 679 | 0.0000 | 67β→70β (0.95) | 4 (full) |
| 2.10 | 588 | 0.0001 | 64β→70β (0.80) | 2 |
| 2.51 | 494 | 0.0067 | 63β→70β (0.79) | 2 |
| 2.80 | 444 | 0.0122 | 62β→70β (0.88) | 0 |
| 2.91 | 426 | 0.0001 | 65β→70β (0.72) | 3 |
| 3.36 | 368 | 0.0137 | 58β→70β (0.69) | 2 |
| 61β→70β (−0.59) | 3 | |||
| 3.41 | 363 | 0.0664 | 59β→70β (0.66) | 1 |
| 60β→70β (–0.58) | 3 | |||
| 3.61 | 343 | 0.0021 | 61β→70β (0.72) | 3 |
| 3.67 | 337 | 0.0015 | 60β→70β (0.72) | 3 |
aOscillator strength.
bEstimate of degree of hole delocalization from the base onto the sugar suggested by the initial MO (58β through 69β): 0. Nearly all remains on the Guanine base, 1. Shared between base and sugar ring favoring base, 2. Equally shared between base and sugar ring, 3. Shared between base and sugar ring favoring sugar, 4. Nearly all transferred to the sugar ring.
c68β→70β represents the transition between the 68th MO and the SOMO (70th MO) in dG•+. β refers to the orbital for the beta electron spin. The number in parentheses is the 1-particle RhoCI excited state density for that transition. Only the major contributor(s) to each state are given.
The inner filled MOs often have substantial contributions from atomic orbitals on the sugar ring; we have ranked in Table 3, on a 0–4 scale, each of these MOs according to its extent of delocalization onto the sugar moiety. The fact that transitions with significant delocalization are found throughout the UV-visible region is in good agreement with our results with model compounds that show no significant wavelength dependence for sugar radical formation. DNA, however, shows no sugar radical formation when light with wavelengths ≥520 nm is used. We speculate that this may be due to base stacking which allows for other energy transfer mechanisms, such as photo-induced hole and energy transfer through the stacked bases in the DNA [(32) and references therein].
In Figure 8, we show four molecular orbitals for G•+ in 2′-dG computed by TD-DFT (6–31G*, B3LYP) and visualized via Gaussview. As expected, the SOMO for G•+ indicates the hole is localized on the guanine base. As seen in the MOs in Figure 8 and indicated in Table 3, many of the filled inner MOs have most of the MO localized on the sugar ring. As a consequence, significant contribution of these MOs to singly excited states transfers the hole from the guanine ring to the sugar ring. In several MO's (61β, 67β) there is a large contribution of orbitals at C5′. This is of interest since positive charge localized at C5′ would tend to promote deprotonation from this site and may explain the predominance of formation C5′• in some cases. Finally, we note that the transition oscillator strengths are weakest for systems in which the lower energy MO is solely on the sugar ring (e.g. 67β → 70β) and strongest for systems in which the lower energy MO is on both the base and the sugar (e.g. 58β→70β).
Figure 8.
Molecular orbitals and energy level filling diagram for G•+ in 2′-dG. MOs were computed by TD-DFT (6–31G*, B3LYP) and visualized via Gaussview. The SOMO shows the expected MO for G•+ with the hole localized on the guanine base. A number of inner shell MOs are localized on the sugar ring, as shown (Table 3).
DISCUSSION
Our results clearly demonstrate that visible illumination of guanine radical cations at elevated temperatures results in high yields of sugar radicals. Table 4 summarizes the yields of these sugar radicals at both 77 and 143 K for the compounds investigated.
Identification C1′•, C3′• and C5′•
Throughout this work, a central doublet of ca. 19 G in guanine nucleosides/tides is attributed to C5′•. The collapse of the ca. 19 G doublet for 5′-D,D-2′-dG is in accord with its assignment to C5′• (Figure 5). A similar collapse of a doublet to a singlet would occur in C4′• if the only major hydrogen hyperfine coupling present in C4′• were to one of the C5′ protons. However, investigations of C4′• report hyperfine couplings inconsistent with collapse of a ca. 19 G doublet into a singlet on C5′ dideuteration. This is found both at room temperature in solution (45), and in a number of single crystal investigations (12,13,18,46) at a variety of temperatures. A single crystal investigation of C5′• in 2′-deoxyadenosine monohydrate reports a single anisotropic C5′ hydrogen hyperfine coupling with aiso = ca. 19 G and a small (5 G) coupling to a C4′ hydrogen (12,13); this is entirely consistent with our assignment for C5′•.
It is also noteworthy that deuteration at C3′ and dideuteration at C5′ in 2′-dG have no effect on the spectra and couplings assigned to C3′• and C1′• (Figure 5). This is entirely consistent with these assignments since significant hyperfine couplings from C5′ protons are not expected for C3′• and C1′•. Theoretical calculations of the hyperfine coupling constants of the individual radicals are also consistent with each of these assignments (21–26).
The status of C4′• is of interest, because we have not observed it, even though calculations show it is energetically as stable as C1′• and C3′• (21–26). One possibility is that C4′• may be present as an underlying broad spectrum and, as a result, be hidden under the more resolved line components of other radicals (18). It is also well known that C4′• is unstable toward beta-phosphate elimination with the formation of a highly reactive enol-ether radical cation intermediate (7,47–51). This latter species is highly oxidizing and readily reforms the guanine radical cation via electron transfer (47–51). The activation energy and rate of beta-phosphate elimination from C4′• in model systems is known (47); from these values the half life of the C4′• radical in nucleotides can be estimated to be on the order of only a few seconds at 143 K. Thus, C4′• may be too short lived to be observed in 3′- and 5′-dGMP, owing to facile beta-phosphate elimination. However, this would not explain the absence of C4′• in 2′-dG where no phosphate is present.
Effect of phosphate groups on radical formation from photoexcitation of G•+ in model compounds
We have previously presented evidence that the presence of a phosphate group (instead of a hydroxyl group) at C3′ and C5′ deactivates the site to radical formation (15). This was supported by previous theoretical calculations for the relative stability of sugar phosphate radicals that indicated a destabilization of the resultant radical on phosphate substitution (22,23). In our current work, the experimental data indicate that the effect involves more than energy considerations. As can be seen in Table 4, a phosphate group at C3′ or C5′ does discourage radical formation at C3′ or C5′, respectively, on photo-excitation of G•+ at 143 K, consistent with energy considerations. However, at 77 K, phosphate substitution at C5′ does not reduce the percentage of C5′• found (Table 4), suggesting that sugar radical formation is driven by the charge distribution in the excited state. For dsDNA the fact that only C1′• is observed via photo-excitation of G•+ (Figure 3) (15), is consistent with phosphate group deactivation of the C3′ and C5′ sites toward radical formation. It is, of course, likely that other factors, such as the DNA conformation, come into play. For example, single-stranded DNA may exhibit different behavior from the double-stranded form (under investigation).
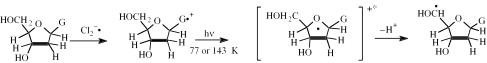
Mechanism of sugar radical formation
Based on our earlier work (15), and the current experimental and theoretical studies, our proposed mechanism of action is that positive charge and unpaired spin density delocalization occurs from the DNA base into the sugar phosphate moiety via light-induced electronic excitation (from inner MOs to the SOMO). This is followed by a rapid deprotonation from those sugar sites on which charge and spin is concentrated (Scheme 3) resulting in a neutral sugar radical.
Scheme 3.
Mechanism of sugar radical formation using formation of C5′• as an example.
The electronic component of the excitation is supported by our TD-DFT calculations, which show that transitions in the 1.5–3.7 eV energy range (UVA-visible) for G•+ in 2′-dG produce singly excited states leading to transfer of charge and unpaired spin density from the guanine ring to the sugar ring. We note that as the sugar conformation changes, the MOs and their contributions to transitions will be altered (Table 3). A conformationally driven change in the charge/spin distribution in the excited dGMP radical cation may explain why 5′-dGMP produces mostly C5′• at 77 K, but C1′• at 143 K in which molecular relaxation is allowed. The deprotonation from the sugar group in the excited radical cation will be followed by relaxation of the resulting neutral sugar radical and its surrounding solvation shell, coupled with diffusion of a proton away from the radical site; these processes will be augmented by the lower viscosity of the glass at 143 K. The increased solvent rotational/translational mobility at 143 K will also provide proton accepting sites that are not available at 77 K. Clearly, the sugar sites of highest positive charge density will have the greatest tendency toward deprotonation and these sites are not necessarily correlated with the energy of the resulting radical. Thus, radical formation may, in some circumstances, be partially or largely under kinetic rather than thermodynamic control.
In dsDNA in ices, unlike the deoxyribonucleoside/tide systems in glasses, we do not observe any increase in the conversion of G•+ to C1′• at 143 K as opposed to 77 K. In DNA, there are already available, at 77 K, multiple local sites on the DNA itself for proton attachment as well as a structure that allows for facile proton diffusion. As a consequence, the increase in temperature provides few new opportunities for deprotonation. For dsDNA, previous work has shown that G•+ is in equilibrium with G(−H)• through a proton transfer from the guanine N1-H to N3 of cytosine (27,29,38). In our pH study with nucleosides, G(−H)• in mononucleotides does not form sugar radicals by illumination even at 143 K.
Our TD-DFT calculations are consistent with the observed lack of wavelength dependence on sugar radical formation in nucleosides/tides since they show that there are electronic transitions allowed which delocalize spin and positive charge (the hole) to the sugar moiety throughout the full spectral region (Figure 8). In dsDNA, however, a wavelength dependence of sugar radical formation is found since only in the range of 310–480 nm does photo-induced formation of C1′• from G•+ occur. Since base stacking in DNA is well known to permit photo-induced transfer of charge and spin to nearby bases, such hole transfer may compete with hole transfer to the adjoining deoxyribose moiety (30,50–52). To produce the observed wavelength dependence, hole transfer to adjacent DNA bases as a deactivation mechanism would need to be most effective in the 1.5–2.5 eV range, with transfer to the sugar competitive at higher energies.
CONCLUSION
This work shows that guanine radical cations in DNA and model systems readily convert to deoxyribose radicals via photo-excitation. The use of elevated temperatures which more closely correspond to biological conditions is shown to lead to near complete conversion in model systems. The mechanism observed here and in our earlier work (15) has not been recognized in the past and may bear on the conclusions reached in some earlier works [(14,32) and the references therein]. For example, in a study of photolysed 5′-dCMP, it was hypothesized that the radical cations produced by laser two-photon ionization produced sugar radicals via ground-state deprotonation at C1′ or hydrogen abstraction from C5′ (14). Formation of sugar radicals via excitation of the cytosine radical cation, as observed here for the guanine radical cation, is a plausible alternative explanation. In addition, many investigations of hole transfer through DNA are based on competition of the hole transfer with the quenching reaction of the hole (G•+) with water, forming 8-HO-G• and ultimately 8-oxo-G (32,33,52–54). Since continuous irradiation is often employed during these experiments, it is clear from our work that photo-excited G•+ can also be quenched by the formation of C1′•. We also note that C1′• in DNA results in a site, which, as 8-oxo-G, is susceptible to piperidine-induced DNA-strand cleavage (5–7). If, as we suggest, formation of C1′• is competitive with water addition, then hole transfer distances could be effectively limited by high light intensities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by the NIH NCI Grant (Grant no. R01CA045424). A.A. is grateful to the authorities of the Rajdhani College and the University of Delhi for leave to work on this research program. J.K. was supported by the Dershwitz Summer Fellowship. The authors thank Omicron Biochemicals Inc., for its generous assistance in providing isotopically labeled nucleotides. The authors are also grateful to Prof. J. Chattopadhyaya (Uppsala University, Sweden) for valuable guidance in synthesizing the deuterated derivatives at C-8 in the guanine moiety. Funding to pay the Open Access publication charges for this article was provided by the NIH NCI Grant (Grant no. R01CA045424).
Conflict of interest statement. None declared.
APPENDIX
C1′• benchmark
The benchmark spectrum for C1′• (Figure 1A) was obtained from 5′-dGMP by producing G•+ using Cl2−• (as described in Materials and Methods) and illuminating with visible light for 30 min to obtain spectrum 6C, which is a composite spectrum originating from 95% C1′• and 5% C3′•. The small amount of C3′• is then subtracted out using a simulation produced with the parameters in Table 1 (Figure 1E). The C1′• quartet spectrum is a result of two hydrogen hyperfine couplings, A(1βH) = 16 G, A(1βH) = 32 G.
C5′• benchmark
The benchmark spectrum for C5′• (Figure 1B) is obtained from 8-D-3′-dGMP by visible illumination of G•+ and subtraction of the spectrum of C1′• as 40% of the original spectrum. The C1′• spectrum used for the subtraction was simulated using the hyperfine coupling constants A(1βH) = 15.5 G, A(1βH) = 35 G, g = 2.0029 and a line width of 4 G; this simulation matches the four line components for C1′• which are clearly visible in the original ESR spectrum. The benchmark is a ca. 19 G doublet at g = 2.0025 (8–19).
C3′• benchmark
The g-value for C3′• is ∼2.0032 in all systems studied here but the measured couplings for C3′• vary slightly with compound and temperature. The benchmark spectrum for C3′• in Figure 1C is produced by visible illumination of G•+ at 143 K, followed by subtraction of the C5′• benchmark (Figure 1B) as 55%, C1′• as 7% and G•+ as 3% from the original spectrum. A simulation (Figure 1D) using the hyperfine couplings and g-value given in Table 1 and 6 G line width matches this spectrum very well. The benchmark spectrum for C3′• in Figure 1E is based on an experiment in which G•+ in 5′-dGMP is photolyzed with a xenon lamp (filtered to 340–370 nm) for 300 min at 77 K, at which point the four outer lines of C3′• are visible, followed by subtraction of the C5′• benchmark (Figure 1B) as 28%, C1′• benchmark as 5% and G•+ as 47% from the original spectrum. It was also simulated using A(1βH) = 20 G, A(1βH) = 30 G, A(1βH) = 41 G with a 7 G line width and g = 2.0032, as shown in Figure 1F.
REFERENCES
- 1.Kanaar R., Hoeijmakers J.H., van Gent D.C. Molecular mechanisms of DNA double strand break repair. Trends Cell. Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 2.Frankenberg D., Frankenberg-Schwager M., Blöcher D., Harbich R. Evidence for DNA double-strand breaks as the critical lesions in yeast cells irradiated with sparsely or densely ionizing radiation under oxic or anoxic conditions. Radiat. Res. 1981;88:524–532. [PubMed] [Google Scholar]
- 3.Ward J.F. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 4.Frankenberg-Schwager M., Kirchermeier D., Greif G., Baer K., Becker M., Frankenberg D. Toxicology. Vol. 212. 2005. Cisplatin-mediated DNA double-strand breaks in replicating but not in quiescent cells of the yeast Saccharomyces cerevisiae; pp. 175–184. [DOI] [PubMed] [Google Scholar]
- 5.Pogozelski W.K., Tullius T.D. Oxidative strand scission of nucleic acids: routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 6.Tronche C., Goodman B.K., Greenberg M.M. DNA damage induced via independent generation of the radical resulting from formal hydrogen atom abstraction from the C1(-position of a nucleotide. Chem. Biol. 1998;5:263–271. [PubMed] [Google Scholar]
- 7.von Sonntag C. The Chemical Basis of Radiation Biology. London: Taylor and Francis; 1987. pp. 167–294. [Google Scholar]
- 8.Sieber K., Hüttermann J. Matrix-isolation of H-induced free radicals from purines in acidic glasses. Int. J. Radiat. Biol. 1989;55:331–345. doi: 10.1080/09553008914550391. [DOI] [PubMed] [Google Scholar]
- 9.Weiland B., Huttermann J., Malone M.E., Cullis P.M. Formation of C1′ located sugar radicals from X-irradiated cytosine nucleosides and -tides in BeF2 glasses and frozen aqueous solutions. Int. J. Radiat. Biol. 1996;70:327–336. doi: 10.1080/095530096145067. [DOI] [PubMed] [Google Scholar]
- 10.Weiland B., Hüttermann J. Free radicals from X-irradiated ‘dry’ and hydrated lyophilized DNA as studied by electron spin resonance spectroscopy: analysis of spectral components between 77 K and room temperature. Int. J. Radiat. Biol. 1998;74:341–358. doi: 10.1080/095530098141483. [DOI] [PubMed] [Google Scholar]
- 11.Shukla L.I., Pazdro R., Becker D., Sevilla M.D. Sugar radicals in DNA: isolation of neutral radicals in gamma-irradiated DNA by hole and electron scavenging. Radiat. Res. 2005;163:591–602. doi: 10.1667/rr3347. [DOI] [PubMed] [Google Scholar]
- 12.Sevilla M.D., Becker M.D. ESR studies of radiation damage to DNA and related biomolecules. In: Gilbert B.C., Davies M.J., Murphy D.M., editors. Royal Society of Chemistry Specialist Periodical Report. Electron Spin Resonance. Vol. 19. 2004. pp. 243–278. [Google Scholar]
- 13.Bernhard W.A., Close D.M. DNA damage dictates the biological consequences of ionizing irradiation: the chemical pathways. In: Mozumdar A., Hatano Y., editors. Charged Particle and Photon Interactions with Matter Chemical, Physicochemical and Biological Consequences with Applications. New York, Basel: Marcel Dekker, Inc.; 2004. pp. 431–470. [Google Scholar]
- 14.Malone M.E., Cullis P.M., Symons M.C.R., Parker A.W. Biphotonic photoionization of cytosine and its derivatives with UV radiation at 248 nm: an EPR study in low-temperature perchlorate glasses. J. Phys. Chem. 1995;99:9299–9308. [Google Scholar]
- 15.Shukla L.I., Pazdro R., Huang J., DeVreugd C., Becker D., Sevilla M.D. The formation of DNA sugar radicals from photoexcitation of guanine cation radicals. Radiat. Res. 2004;161:582–590. doi: 10.1667/rr3167. [DOI] [PubMed] [Google Scholar]
- 16.Becker D., Bryant-Friedrich A., Trzasko C., Sevilla M.D. Electron spin resonance study of DNA irradiated with an argon-ion beam: evidence for formation of sugar phosphate backbone radicals. Radiat. Res. 2003;160:174–185. doi: 10.1667/rr3037. [DOI] [PubMed] [Google Scholar]
- 17.Becker D., Razskazovskii Y., Callaghan M.U., Sevilla M.D. Electron spin resonance of DNA irradiated with a heavy-ion beam ([16]O[8+]): evidence for damage to the deoxyribose phosphate backbone. Radiat. Res. 1996;146:361–368. [PubMed] [Google Scholar]
- 18.Close D.M. Where are the sugar radicals in irradiated DNA? Radiat. Res. 1997;147:663–673. [PubMed] [Google Scholar]
- 19.Debije M.G., Bernhard W.A. Electron paramagnetic resonance evidence for a C3′ sugar radical in crystalline d(CTCTCGAGAG) X-irradiated at 4 K. Radiat. Res. 2001;155:687–692. doi: 10.1667/0033-7587(2001)155[0687:eprefa]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sänger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. pp. 18–21. 48, 55, 62–63. [Google Scholar]
- 21.Miaskiewicz K., Osman R. Theoretical study on the deoxyribose radicals formed by hydrogen abstraction. J. Am. Chem. Soc. 1994;116:232–238. [Google Scholar]
- 22.Colson A.-O., Sevilla M.D. Elucidation of primary radiation damage in DNA through application of ab initio molecular orbital theory. Int. J. Radiat. Biol. 1995;67:627–645. doi: 10.1080/09553009514550751. [DOI] [PubMed] [Google Scholar]
- 23.Colson A.-O., Sevilla M.D. Structure and relative stability of deoxyribose radicals in a model DNA backbone: ab initio molecular orbital calculations. J. Phys. Chem. 1995;99:3867–3874. [Google Scholar]
- 24.Parr K.D., Wetmore S.D. The properties of DNA C4′-centered sugar radicals: the importance of the computational model. Chem. Phys. Lett. 2004;389:75–82. [Google Scholar]
- 25.Wetmore S.D., Boyd R.J., Eriksson L.A. A comprehensive study of sugar radicals in irradiated DNA. J. Phys. Chem. B. 1998;102:7674–7686. [Google Scholar]
- 26.Guerra M. Anomalous phase shift between the angular dependence of the β 5′-H hfs constants produced by the phosphate groups in C4′ deoxyribose radicals: an electronic rather than a structural effect. Phys. Chem. Chem. Phys. 2001;3:3792–3796. [Google Scholar]
- 27.Seidel C.A.M., Schulz A., Sauer M.H.M. Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem. 1996;100:5541–5553. [Google Scholar]
- 28.Fukuzumi S., Miyao H., Ohkubo K., Suenobu T. Electron-transfer oxidation properties of DNA bases and DNA oligomers. J. Phys. Chem. A. 2005;109:3285–3294. doi: 10.1021/jp0459763. [DOI] [PubMed] [Google Scholar]
- 29.Steenken S., Jovanovic S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 30.Milligan J.R., Aguilera J.A., Ward J.F. Redox equilibrium between guanyl radicals and thiocyanate influences base damage yields in gamma irradiated plasmid DNA. Estimation of the reduction potential in guanyl radicals in plasmid DNA in aqueous solution at physiological ionic strength. Int. J. Radiat. Biol. 2001;77:1195–1205. doi: 10.1080/09553000110083988. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi K., Tagawa S. Direct observation of guanyl radical cation deprotonation in duplex DNA using pulse radiolysis. J. Am. Chem. Soc. 2003;125:10213–10218. doi: 10.1021/ja036211w. [DOI] [PubMed] [Google Scholar]
- 32.Schuster G.B., editor. Topics In Current Chemistry: Long Range Charge Transfer in DNA I and II. Berlin: Springer; 2004. pp. 27–115. [Google Scholar]
- 33.Shukla L.I., Adhikary A., Pazdro R., Becker D., Sevilla M.D. Formation of 8-oxo-7,8-dihydroguanine-radicals in gamma-irradiated DNA by multiple one-electron oxidations. Nucleic Acids Res. 2004;32:6565–6574. doi: 10.1093/nar/gkh989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang X., Yu P., LeProust E., Gao X. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker D., La Vere T., Sevilla M.D. ESR detection at 77 K of the hydroxyl radical in the hydration layer of gamma-irradiated DNA. Radiat. Res. 1994;140:123–129. [PubMed] [Google Scholar]
- 36.La Vere T., Becker D., Sevilla M.D. Yields of •OH in gamma-irradiated DNA as a function of DNA hydration: hole transfer in competition with •OH formation. Radiat. Res. 1996;145:673–680. [PubMed] [Google Scholar]
- 37.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Montgomery J.A., Jr, Vreven T., Kudin K.N., Burant J.C., et al. Gaussian 03. Pittsburgh PA: Gaussian, Inc.; 2003. [Google Scholar]
- 38.Shukla M.K., Leszczynski J. TDDFT investigation on nucleic acid bases: Comparison with experiments and standard approach. J. Comput. Chem. 2004;25:768–778. doi: 10.1002/jcc.20007. [DOI] [PubMed] [Google Scholar]
- 39.Shukla M.K., Leszczynski J. Interaction of water molecules with cytosine tautomers: an excited-state quantum chemical investigation. J. Phys. Chem. A. 2002;106:11338–11346. [Google Scholar]
- 40.Candeias L.P., Steenken S. Electron-transfer in di (deoxy) nucleoside phosphates in aqueous-solution—rapid migration of oxidative damage (via adenine) to guanine. J. Am. Chem. Soc. 1993;115:2437–2440. [Google Scholar]
- 41.Hiraoka W., Kuwabara M., Sato F. OH-induced free radicals in purine nucleoside monophosphates: e.s.r. and spin-trapping. Int. J. Radiat. Biol. 1989;55:51–58. doi: 10.1080/09553008914550061. [DOI] [PubMed] [Google Scholar]
- 42.Aquino A.J.A., Lischka H., Hättig C. Excited-state intramolecular proton transfer: a survey of TDDFT and RI-CC2 excited-state potential energy surfaces. J. Phys. Chem. A. 2005;109:3201–3208. doi: 10.1021/jp050288k. [DOI] [PubMed] [Google Scholar]
- 43.Sobolewski A.L., Domcke W., Dedonder-Lardeux C., Jouvet C. Excited-state hydrogen detachment and hydrogen transfer driven by repulsive 1πσ* states: a new paradigm for nonradiative decay in aromatic biomolecules. Phys. Chem. Chem. Phys. 2002;4:1093–1100. [Google Scholar]
- 44.Tsolakidis A., Kaxiras E. A TDDFT study of the optical response of DNA bases, base pairs, and their tautomers in the gas phase. J. Phys. Chem. A. 2005;109:2373–2380. doi: 10.1021/jp044729w. [DOI] [PubMed] [Google Scholar]
- 45.Peukert S., Batra R., Giese B. ESR evidence for a heterolytic C,O-bond cleavage in models of 4′-DNA radicals. Tetrahedron Lett. 1997;38:3507–3510. [Google Scholar]
- 46.Hole E.O., Nelson W.H., Sagstuen E., Close D.M. Free radical formation in single crystals of 2′-deoxyguanosine 5′-monophosphate tetrahydrate disodium salt: an EPR/ENDOR study. Radiat. Res. 1992;129:119–138. [PubMed] [Google Scholar]
- 47.Whitted P.O., Horner J.H., Newcomb M., Huang X., Crich D. Heterolytic cleavage of a β-phosphatoxyalkyl radical resulting in phosphate migration or radical cation formation as a function of solvent polarity. Org. Lett. 1999;1:153–156. doi: 10.1021/ol990054h. [DOI] [PubMed] [Google Scholar]
- 48.Bernhard K., Geimer J., Canle-Lopez M., Reynisson J., Beckert D., Gleiter R., Steenken S. Photo- and radiation-chemical formation and electrophilic and electron transfer reactivities of enolether radical cations in aqueous solution. Chem. Eur. J. 2001;7:4640–4650. doi: 10.1002/1521-3765(20011105)7:21<4640::aid-chem4640>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 49.Crich D., Huang W.H. Dynamics of alkene radical cation/phosphate anion pair formation from nucleotide C4′ radicals. The DNA/RNA paradox revisited. J. Am. Chem. Soc. 2001;123:9239–9245. doi: 10.1021/ja010841l. [DOI] [PubMed] [Google Scholar]
- 50.Glatthar R., Spichty M., Gugger A., Batra R., Damm W., Mohr M., Zipse H., Giese B. Mechanistic studies in the radical induced DNA strand cleavage–formation and reactivity of the radical cation intermediate. Tetrahedron. 2000;56:4117–4128. [Google Scholar]
- 51.Giese B., Erdmann P., Giraud L., Göbel T., Petretta M., Schäfer T. Heterolytic C,O-bond cleavage of 4′-nucleotide radicals. Tetrahedron. 1994;35:2683–2686. [Google Scholar]
- 52.Giese B. Hole injection and hole transfer through DNA: the hopping mechanism. In: Schuster G.B., editor. Long Range Charge Transfer in DNA I. Topics In Current Chemistry. Vol. 236. Berlin, Germany: Springer; 2004. pp. 27–44. [Google Scholar]
- 53.Giese B. Long distance charge transport in DNA: the hopping mechanism. Acc. Chem. Res. 2000;33:631–636. doi: 10.1021/ar990040b. [DOI] [PubMed] [Google Scholar]
- 54.Delaney S., Barton J.K. Long-range DNA charge transport. J. Org. Chem. 2003;68:6475–6483. doi: 10.1021/jo030095y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.