Abstract
Most of the known alternative splice events have been detected by the comparison of expressed sequence tags (ESTs) and cDNAs. However, not all splice events are represented in EST databases since ESTs have several biases. Therefore, non-EST based approaches are needed to extend our view of a transcriptome. Here, we describe a novel method for the ab initio prediction of alternative splice events that is solely based on the annotation of Pfam domains. Furthermore, we applied this approach in a genome-wide manner to all human RefSeq transcripts and predicted a total of 321 exon skipping and intron retention events. We show that this method is very reliable as 78% (250 of 321) of our predictions are confirmed by ESTs or cDNAs. Subsequent analyses of splice events within Pfam domains revealed a significant preference of alternative exon junctions to be located at the protein surface and to avoid secondary structure elements. Thus, splice events within Pfams are probable to alter the structure and function of a domain which makes them highly interesting for detailed biological investigation. As Pfam domains are annotated in many other species, our strategy to predict exon skipping and intron retention events might be important for species with a lower number of ESTs.
INTRODUCTION
The great majority of human multi-exon genes are estimated to express several alternative splice forms (1,2). Alternative splicing mainly contributes to proteome complexity (3,4) and protein isoforms may differ in function or subcellular localization (5–7). Numerous diseases are caused by mis-splicing (8) and a change of the normal splicing pattern is thought to contribute to cancer development (9). Thus, alternative splicing is a very important step during the processing of a pre-mRNA.
Almost all large-scale bioinformatics studies of alternative splicing use the wealth of information stored in expressed sequence tag (EST) databases and most alternative splice forms are detected by the alignment of EST sequences to the genome and to other ESTs/cDNAs (10–14). Despite more than six million human ESTs in dbEST (release December 2004), not all existing splice variants are represented in these databases owing to several reasons. Firstly, the expression level of a transcript must be sufficiently high to be sampled as an EST. Therefore, low expressed splice forms are underrepresented. However, minor splice forms can be very important. For example, a minor splice variant of the RAC1 gene produces Rac1b which constitutes a large portion of activated Rac1 proteins in a cell and might play a role in tumorgenesis (15). Secondly, alternative splicing can be highly specific for a tissue or a cell type, a developmental stage or an external stimulus (16). Thus, such splice forms can only be detected if ESTs are sampled from the right tissue, at the right time and under the right condition. Moreover, tissue distribution of ESTs is strongly biased with the brain having the highest number of ESTs (17). Additionally, low expressed variants have a tendency to be tissue specific (18) which makes their detection even more difficult. Thirdly, ESTs are biased towards the ends of transcripts, especially towards the 3′ end. For example, the first exons of CFTR or NRXN2 are not covered by a single EST, whereas their 3′-untranslated region (3′-UTR) is covered by 31 and 13 ESTs, respectively. Fourthly, many ESTs are sampled from tumor libraries. In some cases, this led to gene annotations based on tumor specific transcripts, although another predominant splice form is expressed in normal tissue (9). Finally, owing to the single read nature, ESTs are error-prone and false positive predictions may be included in alternative splice databases (2).
Apart from ESTs, microarrays with specific exon–exon junction probes have been used to find alternative exons in a genome-wide scale (1). Specific microarrays have also been used to detect a variety of alternative splice events including exon skipping, alternative donor/acceptor sites and mutually exclusive exons by searching for tissue-specific changes in the responses of certain microarray probes (19). Despite the power of microarrays, the main problem remains since it is very hard to test all combinations of tissues, developmental stages and external stimuli. Furthermore, events like intron retention and alternative donor/acceptor sites or additional exons that are located in introns (relative to the given exon structure of the gene for which probes are designed) can only be detected if intronic probes are included in the microarray design. Consequently, our current view of alternative splicing is still incomplete and non-EST based methods for the prediction of splice variants are needed to complete our knowledge of the human transcriptome.
Recently, Sorek et al. (20) described a non-EST based method which uses characteristic features of alternative exons to discriminate between constitutive and alternative ones. The most discriminative single-feature is a high conservation of alternative exons and their flanking intron regions in mouse (21). Additional features are an exon size divisible by three, differences in tri-mer counts and the composition of the splice sites (22). Comparative genomics were also successfully used to predict exon skipping events in Drosophila (23). Yeo et al. (24) described an approach ACESCAN that is able to identify conserved exon skipping events in both human and mouse. This approach also uses exonic and intronic conservation as well as splice site scores, exon and intron lengths, and oligonucleotide composition. Ohler et al. (25) demonstrated that even alternative exons that are completely missed in current gene annotations can be discovered by applying a pair hidden Markov model algorithm to orthologous human–mouse introns. These studies demonstrate that a classifier based on characteristic genomic features can reliably predict exon skipping events ab initio.
Here, we present a different approach that is able to predict exon skipping as well as intron retention events. This method uses only information about protein domain families (Pfam) (26) and, thus, it is independent of the existence of orthologous sequences. Furthermore, we report the results of a genome-wide application of this approach and demonstrate with EST/cDNA searches and experiments that our predictions are very reliable. We show that owing to the inclusion of alternative exons, inserts within Pfam domain structures have a significant preference to avoid secondary structure elements. Since many Pfam domains can be identified in a large number of species, our approach might be especially important for genomes with a lower EST coverage than the human genome.
MATERIALS AND METHODS
Algorithm
Briefly described, for the prediction of alternative splice events we used an algorithm that extends the Viterbi algorithm in order to allow exon skipping and intron retention during the computation of the Pfam alignment. It is an optimal dynamic programming procedure that finds the hypothetical splice form with the highest score for a given Pfam. All ATG codons are considered to be putative start codons, which allows us to find reading frames that start within introns or in the annotated 5′-UTR of the given transcript. Frameshifts are handled by working at the mRNA level in all three reading frames simultaneously. The algorithm only needs the pre-mRNA sequence and the positions of the donor and acceptor sites. More details and the complete recurrence equations are described in (27). Here, we used a modified version that outputs only hypothetical splice forms that are not nonsense-mediated mRNA decay (NMD) candidates (no stop codon 50 nt upstream the last exon–exon junction) (28). We accepted only splice forms that increase the Pfam score by at least 10.
Genome-wide prediction
All transcripts were taken from the RefSeq annotations in the UCSC Genome Browser (assembly hg17 with annotation August 2004, http://hgdownload.cse.ucsc.edu/goldenPath/hg17/database/refGene.txt.gz). We discarded single exon transcripts and all transcripts with an erroneous open reading frame (ORF) or ambiguous characters in their sequence. We translated all transcripts and using hmmpfam from the HMMER package (http://hmmer.wustl.edu/) we searched the human Pfam database version 14 (ftp://ftp.sanger.ac.uk/pub/databases/Pfam/current_release/Pfam_fs.gz) for all Pfams that match the proteins with an E-value of 10 or less. These lists of Pfams were given to our algorithm. We only considered predictions with a Pfam score above the ‘gathering cut-off’ value as given in the Pfam database. All numbers and statistics refer to unique genes, that means, if a gene has two RefSeq transcripts and the same prediction is made for both, we counted only one transcript.
Data evaluation
The EST/cDNA search for exon skipping was done with a 60 nt search string from the flanking exons (30 nt from each side) and BLAST against dbEST (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/est_human.gz) and against cDNA sequences downloaded from GenBank (December 2004). The EST/cDNA search for exon inclusion was done with a search query that comprises the exon in the middle and again 60 nt from the flanking exons. Differences in the Pfam score were evaluated with hmmpfam using the ‘gathering cut-off’ scores. Orthologous mouse exons were found using the Ensembl genome browser (http://www.ensembl.org/). To check whether the mouse intron contains an orthologous exon or not, we used a local alignment (water program from the Emboss package) and checked the presence of splice sites. We aligned the orthologous exon pairs with the needle program from the Emboss package (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/).
Test if the algorithm identifies known alternative exons
To find exons that are skipped in a RefSeq transcript and whose inclusion results in a lower score, we first extracted a set of RefSeq exon pairs that together encode a Pfam domain. Then, using BLAST we searched for EST hits with two separate high scoring segment pairs and kept those ESTs that included an alternative exon between the two RefSeq exons. We discarded all those cases where the inclusion of these exons does not result in a score decrease by at least 10. Then, we used the algorithm with the splice sites of all exons of the RefSeq transcript and the splice sites of the alternative exon. The predicted splice form was compared with the RefSeq transcript.
Location of alternative and constitutive exon junctions in Pfam domains
We considered all confirmed single peptide-cassette exons that insert a sequence into a Pfam domain. We searched for proteins with a known 3D structure that encode such a domain using the Pfam websites (http://www.sanger.ac.uk/Software/Pfam/) and used the resulting proteins as a template. If available, we used a human protein. The pdb2pfam function of the Pfam websites was used to compute the secondary structure and the surface accessibility from the known domain structure. Then, we compared the Pfam alignment of the RefSeq protein with the Pfam alignment of the protein with the known structure to find the positions of the alternative and constitutive exon junctions. If the exon–exon junction splits a codon, only this amino acid was marked as the exon junction. If the exon junction is located between two codons, we marked both neighboring amino acids. The secondary structure assignment from the eight DSSP states (H, G, I, E, T, S, C and B) was done as described in (29): H, G, I helix, E sheet and T, S, C, B non-regular with the correction that the combined occurrence of states BC is converted to EE.
Experimental verification with RT–PCR
We designed primers that flank the predicted alternative exon(s) or intron using Primer3 (Supplementary Table 6). RT–PCR was done on pooled cDNA from different tissues obtained by mixing equal volumes of cDNAs from the HUMAN MTC Panels I and II (BD Biosciences, Palo Alto). PCR setup was 3 µl of template, 10 pmol of each primer in a volume of 25 µl using ReadyToGo PCR beads (Amersham). Cycling conditions were 1 cycle of denaturation at 95°C for 30 s, followed by 35 cycles of denaturing at 92°C for 30 s, annealing at 59°C for 30 s and extension at 72°C for 60 s; 1 cycle of final extension at 72°C for 5 min. We separated PCR products on a 1.5% agarose gel and sequenced them. We inspected the sequence traces for overlaps at the exon–exon boundaries and electropherograms for the presence of products of the expected size. For verification we cloned (pCR-TOPO2.1, Invitrogen) and sequenced the respective products.
RESULTS
Effect of alternative exons on Pfam domains
Our strategy to predict alternative splice events is solely based on the annotation of Pfam domains. To investigate the differences in the contribution to Pfam domains between alternative and constitutive exons, we constructed a set of 213 alternative and 5 728 constitutive exons that are contained in the human RefSeq annotation of the UCSC Genome Browser. We only considered ‘peptide-cassette’ exons that do not introduce a frameshift or a premature termination codon (PTC) when skipped. Each exon encodes a complete Pfam domain or a part of it. We consider an exon as constitutive if it has at least six ESTs that show inclusion and no EST that shows skipping. An alternative exon is skipped in at least three ESTs. Then, we compared the Pfam score between the proteins with and without these exons. From the 5 728 constitutive exons only 99 (1.7%) result in an increase of the Pfam score if they are not included. In contrast, from the 213 alternative exons 34 (16%) result in a Pfam score increase when skipped (Fisher's exact test: P < 0.0001). Furthermore, the average score increase of 13.4 for the 34 alternative exons is considerably higher than the average increase of 2.9 for the 99 constitutive ones (t-test: P < 0.0001). Therefore, we searched for a minimum score increase that leads to a further separation of constitutive and alternative exons. We decided to use 10 as a threshold value since this constraint is fulfilled by only 6 (6% of 99) of the constitutive exons but by 19 (56% of 34) of the alternative exons. Thus, only 0.1% (6 of 5 728) of the constitutive but 9% (19 of 213) of the alternative exons result in a Pfam score increase of at least 10 when they are skipped. This suggests that a genome-wide search for such exons can be used to predict alternative exons with a high specificity.
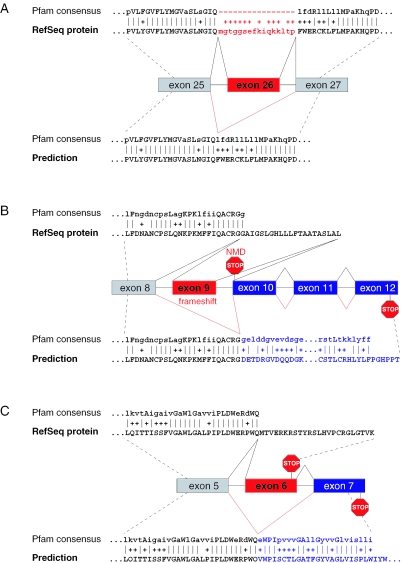
Up to now, we have only considered peptide-cassette exons. Most of these exons are aligned to gaps in a Pfam alignment and exon skipping will increase the score since the number of gaps is reduced (Figure 1A). A special case is the creation of a domain by exon skipping if both sequence parts alone are not recognized as parts of a Pfam (30). However, exons that are not peptide-cassettes can also result in a Pfam score increase. Firstly, the skipping of such an exon can lead to a frameshift and the new protein sequence downstream can encode a longer C-terminus of a Pfam domain or a completely new domain (Figure 1B). Secondly, the skipping of an exon that encodes PTCs can elongate the reading frame (Figure 1C). Such exons most likely are alternative ones since Pfam domains usually have a high sequence specificity and, thus, it is very unlikely that the protein sequence in the other frame or downstream of the PTC has a high similarity to a Pfam domain just by chance. Apart from exon skipping, a retained intron can also encode a new part of the domain or result in a frameshift, thus, increasing the score. Therefore, we extend our strategy to include skipping of non-peptide-cassette exons and retention of introns.
Figure 1.
Effect of alternative exons on Pfam domains. (A) SLC4A5 (NM_033323): exon 26 disrupts the Pfam domain PF00955 as shown by the gaps in the alignment. Skipping of the exon increases the Pfam score from 1183 to 1208. (B) CASP2 (NM_001224): inclusion of exon 9 results in a reading frame with a stop codon in exon 10 and this transcript should induce NMD. The skipping of exon 9 leads to a frameshift and a new C-terminal part of the Caspase domain PF00656 (score increase from 174 to 317). (C) PIGF (NM_173074): exon 6 encodes an in-frame stop codon 33 nt upstream of the last exon–exon junction which should not elicit NMD. Skipping of exon 6 results in a new C-terminus encoded by exon 7 and a score increase for PF06699 from 299 to 362. Alternative exons are depicted in red, exons that become coded in the predicted splice form are depicted in blue. Pfam alignments for the RefSeq protein are shown at the top, for the predictions at the bottom. All exon skipping events are supported by several ESTs.
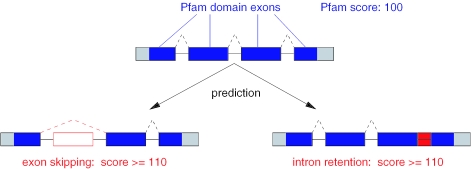
Outline of the approach
Our approach can be summarized as follows. Given the exon structure of a transcript and its pre-mRNA sequence, we search for exon skipping and intron retention events that increase the Pfam score for the respective protein by at least 10 (Figure 2). Without the input of additional splice sites, like alternative donors or acceptors, only the prediction of exon skipping and intron retention events is possible.
Figure 2.
Schematic representation of the approach for non-EST based prediction of exon skipping and intron retention events. Given are four coding exons and the respective intron sequences. Assume a Pfam domain is encoded by exons 1–4 and the respective Pfam score is 100. For the prediction, hypothetical novel splice variants are checked to find those with a higher Pfam score by at least 10. Exons are shown as boxes; dashed lines indicate the splicing patterns; open red box: skipped exon; and filled red box: retained intron.
Previously we have developed an efficient algorithm that computes the hypothetical splice form with the maximal score for a given Pfam domain (27). This algorithm allows (one or more) exons to be skipped and introns to be retained during the computation of the dynamic programming matrix. Usage of the algorithm allows a computationally more efficient search than considering single exon skipping and intron retention events or combinations of them individually. To test if the algorithm is able to identify alternative exons, we constructed two test sets of alternative exons that are skipped in a RefSeq transcript and retained in other EST/cDNA sequences. These test sets consisted of 202 peptide-cassette and of 195 non-peptide-cassette exons. Inclusion of these exons resulted in a Pfam score decrease of at least 10. Then, given the exon structure of the transcript with the alternative exon, we used the algorithm to find the splice form with the highest Pfam score. In 392 (99%) cases (200 peptide-cassette and 192 non-peptide-cassette exons) the predicted splice form was equal to the RefSeq transcript. This includes 18 cases where more than one exon was skipped in a RefSeq transcript (4 cases with two consecutive exons and 14 cases with two non-consecutive ones). In the five remaining cases, other exons were skipped in addition to the expected exon which gives an even higher score. Thus, all alternative exons in our test sets can be found by this algorithm.
Genome-wide application
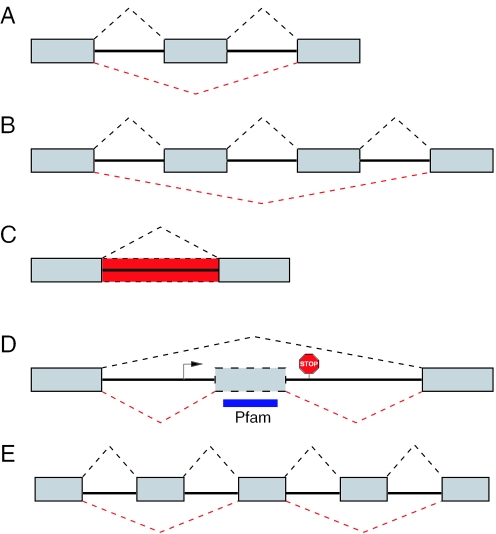
To predict exon skipping and intron retention events in the human genome, we applied this approach to all RefSeq transcripts (August 2004). We only considered novel splice forms that are not candidates for NMD (28) since the rationale behind our strategy is that the novel splice variant is expressed to be translated into a functional protein. In this genome-wide scan, we predicted alternative exons and introns for 309 RefSeq transcripts. We distinguish five cases: (i) the skipping of a single exon, (ii) the skipping of multiple consecutive exons, (iii) the retention of an intron, (iv) hidden exon events, and (v) complex events as any combination of (i)–(iv) (Figure 3). These results are summarized in Table 1. Detailed information about all predictions is given as Supplementary Data.
Figure 3.
Classification of predicted alternative splice events. (A) Skipping of a single exon. (B) Skipping of multiple consecutive exons (shown here for two exons). (C) Intron retention (shown as red dashed box). (D) Hidden exon: the algorithm found an ORF (start and stop codon are indicated) within an intron that encodes a Pfam domain (blue box) which indicates the existence of a hidden exon (gray dashed box). (E) Complex events involve any combination of (A–D). Here, we show an example with two skipped exons. Exons are shown as gray boxes and dashed lines indicate the splicing patterns.
Table 1.
Summary of the genome-wide scan
| Number of predictions | Confirmeda | Different event confirmedb | Unconfirmed | ||||
|---|---|---|---|---|---|---|---|
| Single exon skipping | 183 | 119 | 65% | 25 | 14% | 39 | 21% |
| Multiple exon skipping | 57 | 14 | 25% | 16 | 28% | 27 | 47% |
| Intron retention | 67 | 37 | 55% | 28 | 42% | 2 | 3% |
| Hidden-exon event | 5 | – | – | 5 | 100% | – | – |
| Complex event | 9 | – | – | 6 | 67% | 3 | 33% |
| Sum | 321 | 170 | 53% | 80 | 25% | 71 | 22% |
aExactly the predicted event is confirmed.
bA different event is confirmed (alternative donor/acceptor, inclusion of an exon that is skipped in the given transcript, alternative transcription start); most of these events involve frameshifts.
Single skipped exons
We predicted a total of 183 single RefSeq annotated exons to be alternative (Supplementary Table 1). To check if known alternative exons are contained in this set, we searched dbEST (December 2004) and cDNAs from GenBank and found exon skipping evidence for 119 (65%) of those. Three further exons are skipped in addition to alternative donor or acceptor usage of one neighboring exon. If the score is increased by introducing a frameshift there are generally several possibilities to introduce the frameshift. Our algorithm is only able to handle frameshifts caused by exon skipping; however, the same frameshift might be introduced by the usage of alternative donor/acceptor sites or the inclusion of exons that are skipped in the RefSeq transcript. Therefore, we examined frameshift predictions in detail and found that in 22 cases the EST confirmed frameshift is not caused by exon skipping but by a different splice event. Remarkably, the target reading frame of the predicted shift is always identical to the confirmed one. Thus, a frameshift prediction should be taken as a strong hint that a frameshift event exists in the vicinity of the skipped exon. These 22 predictions are not considered further. Altogether only 39 (21%) predictions remain that can not be confirmed by existing expressed sequences.
Then, we compared the number of ESTs that match the upstream and downstream exon of confirmed and unconfirmed predictions to see whether the exon skipping events in both groups have an equal chance to be detected. The downstream exon of the 119 confirmed alternatives is covered on average by 81 ESTs, which is four times higher than the average coverage of 20 for the unconfirmed predictions (median 14 versus 5). The upstream exon has similar EST counts in both groups (average 77 versus 13). This suggests that insufficient EST coverage may be the reason for the current lack of confirmation. Furthermore, we found that the unconfirmed exons are on average 688 nt further upstream of the 3′ mRNA end. Given the average EST length of 530 nt and that most ESTs are sampled from the 3′ end, this may contribute to their lower EST coverage.
To check which percentage of single exon skippings can be expected by chance, we randomly chose 2 828 Pfam domain exons. To exclude exons with an EST coverage too low for detection of skipping events, we only considered exons with at least 20 hits for the upstream and downstream exon which gives a median coverage of 48 (note, this is very conservative compared with 14 matches to the downstream exon of confirmed single exon skipping events). We only found for 15% (424 of 2.828 exons) EST/cDNA evidence for exon skipping. In contrast, 75% (119 of 158, excluding 25 with a different confirmed event) of the predicted single exons are EST/cDNA confirmed. This indicates that our predictions are significantly enriched in real alternative exons (Fisher's exact test: P < 0.0001).
We compared the number of ESTs/cDNAs that contain or miss a confirmed single exon. On average these exons are skipped in 39 cases and included in only 8 (5:1 skipping-inclusion ratio) which contributes to the high overall confirmation rate for predicted single-exon events. However, the inclusion in several transcripts and at least one RefSeq demonstrates that these exons are real. Alternative exons with a low inclusion rate are often not conserved in mouse and such exons are the result of exon creation or loss (18). Therefore, we searched for their existence in the mouse genome by considering the exons as well as introns of the orthologous mouse loci. For 15 single exons we failed to identify either an orthologous mouse gene or the exons that flank the single exon. For the remaining 104 exons, we only found an orthologous mouse exon for 45 (43%) which is in agreement with (18). In recent studies, Sorek et al. (20) and Yeo et al. (24) predicted a total of 952 and 2.092 exons to be alternative, respectively. Only 18% (21 of 119) of the confirmed single exons predicted here are contained in this combined exon set which may be attributed to the fact that 42% (19 of 45) of the orthologous human–mouse exon pairs have sequence identities of <95% [this cut-off was used in (20)]. Moreover, the exons predicted by Yeo et al. (24) have a tendency not to overlap InterPro domains which is in contrast to most of our predictions. Thus, the exons addressed by our Pfam based approach and the comparative methods have different characteristics and both approaches complement each other.
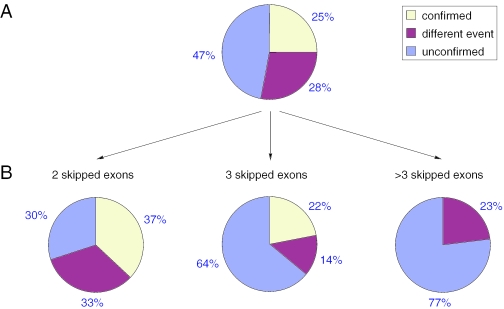
Multiple skipped consecutive exons
In the genome-wide scan, we predicted 57 multiple exon skipping events and found EST/cDNA evidence for 14 of them (Supplementary Table 2). Similar to single exons, another 16 frameshift predictions are confirmed by different splice events, and the remaining 27 predictions are unconfirmed (Figure 4A). Again EST coverage of the downstream exon is lower for the unconfirmed predictions (average 42 versus 23 and median 25 versus 7). Of all 57 predictions 30 events have two skipped exons, 14 three skipped exons and 13 more than three exons. We found that no prediction with more than three exons is confirmed and that the percentage of unconfirmed predictions increases with the number of skipped exons (Figure 4B). Thus, it is conceivable that some predictions are false positives and that the Pfam score is increased just by chance. This holds especially for predictions with many skipped exons since the number of possible exon–exon combinations goes up. Indeed, we found that the average Pfam score increase for the unconfirmed predictions is lower than for the confirmed predictions (19 versus 28) which suggests that an increase of the threshold value with the number of skipped exons should eliminate many false positive predictions.
Figure 4.
Percentage of confirmed multiple-exon skipping events. (A) Percentage of confirmed, confirmed by a different splice event and unconfirmed predictions with more than one skipped exon. (B) Percentage of confirmed, confirmed by a different splice event and unconfirmed predictions divided into the number of skipped exons.
Retained introns
We predicted 67 intron retention events and found EST/cDNA evidence for 65 (97%) of them (Supplementary Table 3). Only ESTs with a spliced intron upstream or downstream were accepted to reduce the possibility of partially spliced ESTs. We found that 36 (54% of 67) of these introns do not have consensus splice sites (GT-AG or GC-AG). These introns can be the result of annotation or mapping errors of the RefSeq transcripts or the consequence of allele-specific splicing (31) since some of those have splice sites that diverge from the consensus in only a single mutation (e.g. an AA instead of an AG acceptor site). Therefore, some of the predicted events do not involve real introns which may contribute to this extremely high confirmation rate. However, 25 (81%) retention events of the remaining 31 introns with consensus splice sites are confirmed by other RefSeq transcripts which indicates that they are real and not artifacts.
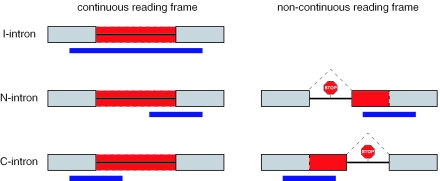
We classified predicted intron retentions into three groups (Figure 5). In case of ‘I-introns’ the internal region of a Pfam is encoded by the intron and both neighboring exons also contribute to the domain. ‘N-introns’ encode a novel N-terminal domain part and, thus, only the downstream exons add to the Pfam. Likewise, ‘C-introns’ encode a novel C-terminal Pfam part and only the upstream exons contributes to the domain. Of the 67 predictions, 23 are I-introns and all are experimentally confirmed. Of the 15 N-intron predictions, 13 are confirmed by at least partial EST matches to one intron–exon boundary. Eleven of them do not have a continuous ORF which is a strong indication for the existence of alternative acceptor sites further upstream of the Pfam encoding exons. Indeed, 10 of those have a confirmed alternative acceptor and we found one alternative transcription start. The remaining two N-introns with a continuous reading frame are confirmed by EST matches. Finally, all of the 29 predicted C-intron retention events are confirmed (12 intron retentions and 17 alternative donors). Interestingly, a PTC owing to the retention of the last intron can not trigger NMD and these splice events result in protein isoforms with an altered Pfam domain at their C-terminus.
Figure 5.
Classification of intron retention events. I-introns have a continuous reading frame and both neighboring exons also encode the Pfam domain. For N- and C-introns only the downstream and upstream exon encode the Pfam domain, respectively, and they may not have a continuous reading frame. Exons are shown as gray boxes with solid lines, introns as a line and a retained intron as a box with dashed lines. The position of the Pfam domain is shown as a blue box below the gene structure. Stop codons in the non-continuous reading frames and splicing of an intron (dashed line) are indicated.
Hidden exon events
In the genome-wide scan, we also found seven predictions that involve introns containing an ORF that encodes the complete or a part of a Pfam without the neighboring exons (Supplementary Table 4). Thus, it is possible that an exon which is skipped in the given transcript is hidden in this intron. Therefore, we examined these hits and for five of them we found EST confirmation of hidden exons. For example, intron 5 of the NM_013954 transcript of NOX1 contains seven alternative exons that encode parts of the ‘Ferric reductase like transmembrane component’ domain (PF01794). These exons are included in another transcript of NOX1 (NM_007052). Manual inspection of the remaining two unconfirmed predictions (NM_152476 intron 10, NM_206894 intron 5) with the Ensembl genome browser revealed that these RefSeq transcripts falsely span two non-overlapping genes and that the predicted intronic parts are exons of the downstream genes. Thus, these two cases are due to annotation errors and were excluded.
Complex events
We also predicted nine complex events (Supplementary Table 5). In each case the given transcript is a clear NMD candidate and our prediction aims at maintaining a reading frame. For six of the nine cases manual inspection revealed other splice events like the multiple usage of alternative donor and acceptor sites. For example, our prediction for the NMD candidate NM_152247 of CPT1B is to skip exon 22 and 26 to restore the reading frame. Instead of skipping two exons, an alternative acceptor 5 nt upstream of the beginning of exon 22 and an alternative donor 169 nt downstream of exon 26 is used in another transcript of CPT1B (NM_152246) to produce a non-NMD splice form.
Experimental verification of unconfirmed predictions
Using RT–PCR with primers from the flanking exons, we tested 11 randomly chosen unconfirmed single-exon skipping events in a pool of 16 human tissues (Supplementary Table 6). In 27% (3 of 11) the predicted exon skipping was observed (DHRS exon 7, CDH2 exon 11 and MYO9 exon 6). Since multiple exon skipping events have a lower EST confirmation rate compared with single exon events and no case of a four-exon skipping is EST confirmed, we selected three two-exon, one three-exon and two four-exon skippings for experimental verification. Furthermore, the two unconfirmed N-introns were tested. We did not observe the expected splice variants for these eight predictions. Alternatively to the predicted retention of intron 1 for ZFP37 (NM_003408), transcription and/or translation can start in the first intron which would have the same consequences for the Pfam domain, but this putative transcript can not be amplified using a forward primer for the annotated exon 1. In general, our experiments may suffer from some of the problems mentioned above for ESTs, since specific splice events can be restricted to narrow windows in space and time.
Location of alternative peptide-cassette exons within Pfam domain structures
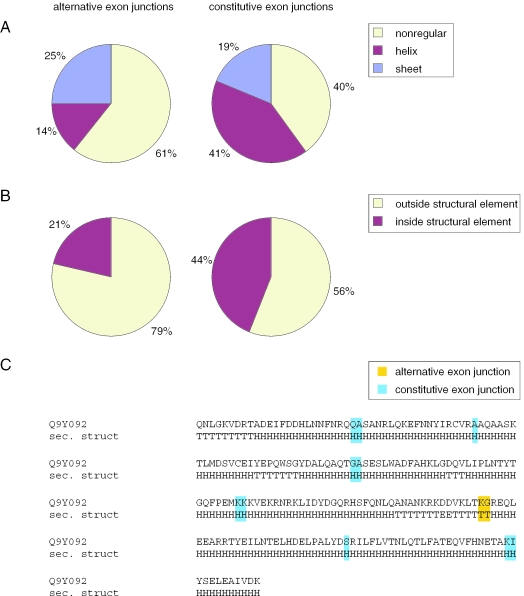
Alternative splicing has a tendency to coincide with domain boundaries and to avoid the interior of functional and structural domains (5,32). Since our single exon events might interfere with the Pfam domain structure as indicated by the low inclusion rate, we were interested in finding out where confirmed peptide-cassette exons are located with respect to the secondary structure and protein surface. The secondary structures and the surface accessibility of residues were computed from known 3D structures of Pfam domains. Since in each case the structure does not include the alternative exon, in the following, we consider the location of the exon–exon junction of both neighboring exons. We mapped 28 alternative exon junctions and, as a control group, 80 constitutive exon junctions to these secondary structures. The residue at the exon junction was classified as being located in an alpha-helix, in a beta-sheet or in a non-regular element. We found a significant difference between the alternative and constitutive junctions (χ2 = 6.76, df = 2 and P = 0.034) with a striking preference of alternative junctions for non-regular elements and the avoidance of helices (Figure 6A). To rule out that this result is biased by inaccuracies in the secondary structure assignment which is sometimes problematic at the end of structural elements, we considered a broader context (±1 residue) around the exon junction. We classified the context to be ‘inside a structural element’ if all three residues are either in a helix or in a sheet. If the three residues of the context are in two different structural elements or if all are inside a non-regular element, the context is classified as ‘outside a structural element’. Again we found a significant preference of the alternative exon junctions to be located outside structural elements (χ2 = 4.39, df = 1 and P = 0.0362) (Figure 6B). An interesting example is the BAR domain that consists of four long helices. While the constitutive junctions of all exons of BIN1 that encode this domain are located within these helices, the position of the alternative junction is in the loop between two helices (Figure 6C). Furthermore, alternative junctions have a tendency to be located at the protein surface (±1 residue context, average 2.96 versus 2.36, higher values indicate exposed residues). This clearly shows that alternative exon junctions are non-randomly distributed within Pfam domain structures. The preferred position at the surface and between secondary structure elements argues against a destructive role of most of these splice events.
Figure 6.
Location of alternative and constitutive exon junctions within Pfam domains. (A) Analysis of residues at the exon junction with respect to location in a helix, in a sheet or in non-regular elements. (B) Analysis of the ±1 amino acid context around exon junctions. (C) BAR domain PF03114 as an example: The exon junctions of BIN1 (NM_139345) are mapped to the secondary structure of the BAR domain using the known structure (PDB 1uru) of CG8604-PA (Swiss-Prot Q9Y092) as a template. While all constitutive exon junctions are located within helices, the alternative junction is in the loop. If the exon junction splits a codon, only this amino acid is highlighted and otherwise, if the junction is between two codons, both residues are highlighted. H: helix, E: sheet, T: non-regular.
DISCUSSION
We describe an approach that uses information about Pfam domains to predict exon skipping and intron retention events ab initio. Only the genomic sequence and gene structure annotation are required. Our approach is able to predict alternative exons regardless whether their size is divisible by three or not and is independent of the existence of orthologs in another species. We have shown that this approach can reliably identify exon skipping and intron retention events ab initio and that it complements existing comparative methods. Our approach has two limitations. First, it is restricted to the regions of a gene that encode Pfam domains. However, Pfam is one of the most comprehensive descriptions of functional domains as Pfam domains match 75% of all proteins in Swiss-Prot/TrEMBL and cover 53% of all residues (26). Apart from Pfam domains, the general approach can use other functional motif descriptions such as those contained in the InterPro database. Additionally the constant growth of these databases will lead to a higher coverage and more predictions. Second, our approach is restricted to cases where the Pfam score is increased because it is unlikely that this occurs just by chance. Many splice events result in a deletion of functional domains which decrease the overall Pfam score. Such events cannot be predicted by this approach since a strategy that arbitrarily predicts an exon to be alternative will also result in a lower Pfam score.
In this study, we considered a total of 18 572 human RefSeq transcripts and made a prediction for 307 (1.7%) of them. We only predicted exon skipping and intron retention events as no other putative, alternative splice sites are given. However indirectly, for a number of predictions that result in a frameshift, we found an alternative donor/acceptor site or an exon that is skipped in the given transcript. These alternative splice events cause the same frameshift that is predicted by our algorithm. Therefore, we evaluated the prediction if the positions of the additional splice sites are given. In all examples tested, the algorithm uses the additional splice sites and produces a splice form that equals the known transcript (data not shown). Moreover, C-intron retentions and hidden exon predictions were only found for the last intron in the transcript, since most of them do not have a continuous reading frame, and we excluded hypothetical splice forms that are NMD candidates. Numerous of these events in other introns can be found by relaxing the NMD criterion. Again, such events can be predicted if the corresponding alternative splice sites are included. It seems promising to include other splice sites, e.g. those derived from suboptimal exons which can be found by gene prediction programs such as Genscan (33). This will increase the number of predictions with alternative donor/acceptor sites as well as exons that are hidden in introns and whose inclusion is not seen in available expressed sequences.
We have shown that sequence inserts inside Pfam domains prefer to be located at the protein surface and strongly avoid a position within secondary structure elements which is in line with their negative impact on a Pfam domain (based on the score), their low inclusion level and their low conservation in mouse. This suggests that most of these inserts alter the domain structure and function which is in contrast to other alternative splice variants that delete an entire Pfam domain or an essential part of it (5). A probable evolutionary scenario is the exonization of a part of an intron followed by a selection pressure assuring that the novel exon is rarely included to produce enough amount of the functional protein. If the inclusion of this exon has no drastic consequences for the domain structure, it might aquire an important regulatory function that can be used by the cell. Indeed, we found examples in the literature where such splice events have important functional consequences. For example, a splice form of TRAF2 with a seven amino acid insert into a Ring finger domain acts as a dominant negative inhibitor of TNFR2-dependent NFκB activation (34). Alternatively spliced inserts modulate the structure of loops at a protein interaction surface of Neurexin Iβ which influences the binding of protein ligands (35). However, even small inserts may result in a change of the overall protein fold. For example, insertion of nine residues into the C2 domain of Piccolo owing to inclusion of exon 15 leads to a rearrangement of the β-sheets which explains the drastic differences in Ca2+ affinity for both splice forms (36). A 17 amino acid insert for UAP1 modifies the architecture of the active site and alters substrate specificity (37). We believe that many of the splice forms found in this study are biologically interesting as they affect a protein domain and presumably alter its structure and function.
Intron retention seems to be a rare splice event with an estimated frequency of 6% (38) and they are difficult to detect because of unspliced or partially spliced ESTs. Most of the intron retentions predicted here contribute to Pfam domains and the retention is confirmed by the existence of another RefSeq transcript. Therefore, they are probable to represent important alternative splice forms (39). Since nearly all predicted intron retention and hidden exon events are EST confirmed, we conclude that intronic ORFs encoding Pfam domains are very likely to become exonic in another transcript.
Owing to a high number of human ESTs and intensive biomedical research, the human transcriptome presumably is the best characterized one. In contrast, the number of ESTs is much lower for other species, e.g. chicken has <532 000 ESTs and Drosophila <383 000. Even in the well-annotated genome of Caenorhabditis elegans there are thousands of genes without EST/cDNA support (40). As alternative splicing is assumed to be equally frequent in other species (41), ab initio prediction should be very useful for species with low EST numbers. Therefore, we believe that the application of our approach to other organisms will lead to the discovery of numerous novel alternative splice events.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Ivonne Görlich for the expert technical assistance and Anke Busch for critical reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Friedrich-Schiller-University Jena.
Conflict of interest statement. None declared.
REFERENCES
- 1.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 3.Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 4.Hiller M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nature Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- 5.Kriventseva E.V., Koch I., Apweiler R., Vingron M., Bork P., Gelfand M.S., Sunyaev S. Increase of functional diversity by alternative splicing. Trends Genet. 2003;19:124–128. doi: 10.1016/S0168-9525(03)00023-4. [DOI] [PubMed] [Google Scholar]
- 6.Resch A., Xing Y., Modrek B., Gorlick M., Riley R., Lee C. Assessing the impact of alternative splicing on domain interactions in the human proteome. J. Proteome Res. 2004;3:76–83. doi: 10.1021/pr034064v. [DOI] [PubMed] [Google Scholar]
- 7.Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 9.Xu Q., Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brett D., Hanke J., Lehmann G., Haase S., Delbruck S., Krueger S., Reich J., Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- 11.Clark F., Thanaraj T.A. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 2002;11:451–464. doi: 10.1093/hmg/11.4.451. [DOI] [PubMed] [Google Scholar]
- 12.Mironov A.A., Fickett J.W., Gelfand M.S. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modrek B., Resch A., Grasso C., Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zavolan M., Kondo S., Schonbach C., Adachi J., Hume D.A., Hayashizaki Y., Gaasterland T. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matos P., Collard J.G., Jordan P. Tumor-related alternatively spliced Rac1b is not regulated by Rho-GDP dissociation inhibitors and exhibits selective downstream signaling. J. Biol. Chem. 2003;278:50442–50448. doi: 10.1074/jbc.M308215200. [DOI] [PubMed] [Google Scholar]
- 16.Stamm S. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet. 2002;11:2409–2416. doi: 10.1093/hmg/11.20.2409. [DOI] [PubMed] [Google Scholar]
- 17.Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Modrek B., Lee C.J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nature Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 19.Le K., Mitsouras K., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C. Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data. Nucleic Acids Res. 2004;32:e180. doi: 10.1093/nar/gnh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorek R., Shemesh R., Cohen Y., Basechess O., Ast G., Shamir R. A non-EST-based method for exon skipping prediction. Genome Res. 2004;14:1617–1623. doi: 10.1101/gr.2572604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorek R., Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dror G., Sorek R., Shamir R. Accurate identification of alternatively spliced exons using support vector machine. Bioinformatics. 2005;21:897–901. doi: 10.1093/bioinformatics/bti132. [DOI] [PubMed] [Google Scholar]
- 23.Philipps D.L., Park J.W., Graveley B.R. A computational and experimental approach toward a priori identification of alternatively spliced exons. RNA. 2004;10:1838–1844. doi: 10.1261/rna.7136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl Acad. Sci. USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohler U., Shomron N., Burge C.B. Recognition of unknown conserved alternatively spliced exons. PLoS Comp. Biol. 2005;1:e15. doi: 10.1371/journal.pcbi.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiller M., Backofen R., Heymann S., Busch A., Glaesser T.M., Freytag J.C. Efficient prediction of alternative splice forms using protein domain homology. In Silico Biol. 2004;4:195–208. [PubMed] [Google Scholar]
- 28.Maquat L.E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nature Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 29.Rost B., Sander C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 30.Hiller M., Huse K., Platzer M., Backofen R. Creation and disruption of protein features by alternative splicing - a novel mechanism to modulate function. Genome Biol. 2005;6:R58. doi: 10.1186/gb-2005-6-7-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nembaware V., Wolfe K.H., Bettoni F., Kelso J., Seoighe C. Allele-specific transcript isoforms in human. FEBS Lett. 2004;577:233–238. doi: 10.1016/j.febslet.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 32.Homma K., Kikuno R.F., Nagase T., Ohara O., Nishikawa K. Alternative splice variants encoding unstable protein domains exist in the human brain. J. Mol. Biol. 2004;343:1207–1220. doi: 10.1016/j.jmb.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 33.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 34.Brink R., Lodish H.F. Tumor necrosis factor receptor (TNFR)-associated factor 2A (TRAF2A), a TRAF2 splice variant with an extended RING finger domain that inhibits TNFR2-mediated NF-kappaB activation. J. Biol. Chem. 1998;273:4129–4134. doi: 10.1074/jbc.273.7.4129. [DOI] [PubMed] [Google Scholar]
- 35.Rudenko G., Nguyen T., Chelliah Y., Sudhof T.C., Deisenhofer J. The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing. Cell. 1999;99:93–101. doi: 10.1016/s0092-8674(00)80065-3. [DOI] [PubMed] [Google Scholar]
- 36.Garcia J., Gerber S.H., Sugita S., Sudhof T.C., Rizo J. A conformational switch in the Piccolo C2A domain regulated by alternative splicing. Nature Struct. Mol. Biol. 2004;11:45–53. doi: 10.1038/nsmb707. [DOI] [PubMed] [Google Scholar]
- 37.Peneff C., Ferrari P., Charrier V., Taburet Y., Monnier C., Zamboni V., Winter J., Harnois M., Fassy F., Bourne Y. Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J. 2001;20:6191–6202. doi: 10.1093/emboj/20.22.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thanaraj T.A., Stamm S. Prediction and statistical analysis of alternatively spliced exons. Prog. Mol. Subcell. Biol. 2003;31:1–31. doi: 10.1007/978-3-662-09728-1_1. [DOI] [PubMed] [Google Scholar]
- 39.Galante P.A., Sakabe N.J., Kirschbaum-Slager N., de Souza S.J. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei C., Lamesch P., Arumugam M., Rosenberg J., Hu P., Vidal M., Brent M.R. Closing in on the C. elegans ORFeome by cloning TWINSCAN predictions. Genome Res. 2005;15:577–582. doi: 10.1101/gr.3329005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brett D., Pospisil H., Valcarcel J., Reich J., Bork P. Alternative splicing and genome complexity. Nature Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.