Abstract
The effects of tRNA, RF1 and RRF on trans-translation by tmRNA were examined using a stalled complex of ribosome prepared using a synthetic mRNA and pure Escherichia coli translation factors. No endoribonucleolytic cleavage of mRNA around the A site was found in the stalled ribosome and was required for the tmRNA action. When the A site was occupied by a stop codon, alanyl-tmRNA competed with RF1 with the efficiency of peptidyl-transfer to alanyl-tmRNA for trans-translation inversely correlated to the efficiency of translation termination. The competition was not affected by RF3. A sense codon also serves as a target for alanyl-tmRNA with competition of aminoacyl-tRNA. The extent of inhibition was decreased with the length of the 3′-extension of mRNA. RRF, only at a high concentration, slightly affected peptidyl-transfer for trans-translation, although it did not affect the canonical elongation. These results indicate that alanyl-tmRNA does not absolutely require the truncation of mRNA around the A site but prefers an mRNA of a short 3′-extension from the A site and that it can operate on either a sense or termination codon at the A site, at which alanyl-tmRNA competes with aminoacyl-tRNA, RF and RRF.
INTRODUCTION
A special RNA called tmRNA or SsrA RNA has been found in a wide variety of eubacteria and in some chloroplasts and mitochondria (1–3). tmRNA having both tRNA and mRNA properties plays an important role in a quality control system during protein synthesis. When a ribosome stalls on a problematic mRNA, presumably at the 3′ end of a truncated mRNA lacking a stop codon, tmRNA charged with alanine enters the A site of the ribosome to act first as an alanyl-tRNA (4,5). Then it acts as an mRNA to direct the addition of a short peptide tail (6). This co-translation, known as trans-translation, terminates at a stop codon that follows the tmRNA reading frame, releasing tagged polypeptide and rescuing the stalled ribosome (7–9). The C-terminal tag-peptide is recognized by several ATP-dependent proteases, resulting in degradation of tagged polypeptide (10). In addition, trans-translation has been shown to facilitate the degradation of truncated mRNA by releasing a stalled ribosome and thereby allowing 3′- to 5′-exonucleases to access the 3′ end of free mRNA (11).
Since tmRNA comprises two functional domains, a tRNA domain partially mimicking tRNA (12) and an mRNA domain including the coding region for tag-peptide surrounded by four characteristic pseudoknot structures (13), an elaborate interplay of the two domains should be required for trans-translation. Although the mode of tmRNA action for trans-translation has been extensively studied, it has not been clarified how tmRNA finds the ribosome of stalled translation. Initially, the target for the tmRNA system has been assumed to be the ribosome with a stalled translation in which the P site but not A site is occupied by mRNA owing to the lack of its 3′-portion (7). Tandem rare codons on an artificial mRNA can also be a target for trans-translation (14). Full-length and C-terminally truncated polypeptides with a tag-sequence at the C-terminus have also been identified as endogenous products of trans-translation in the cell (15,16), and trans-translation efficiently occurs at an inefficient UGA termination codon preceded by rare arginine codons (17). These in vivo data suggest that trans-translation can occur on either a sense or nonsense codon at the A site when translation is stalled. However, a recent finding of bacterial toxin that cleaves an mRNA specifically at the A site in the ribosome stalled upon amino acid starvation provides a new concept that mRNA of stalled translation is targeted initially by these A site-specific endoribonucleases and subsequently by alanyl-tmRNA for trans-translation (18,19).
In prokaryotes, either RF1 or RF2 recognizes a stop codon and catalyzes the hydrolysis of peptidyl-tRNA to release the polypeptide from the ribosome (20). After this event, RF3 stimulates the dissociation of RF1 or RF2 from the ribosome (21), and then RRF, with the help of EF-G, allows the ribosome to enter a new round of translation (22,23). In the present study, a stalled complex of the ribosome with a polyphenylalanyl-tRNA at the P site was prepared from ribosomes programmed with synthetic mRNAs and other pure translation factors from Escherichia coli. Using this stalled complex of ribosome, the competition between alanyl-tmRNA and RF1 at a nonsense codon was examined. Ivanova et al. (24) have recently shown the competition at a nonsense codon, and such a competition was also observed in our system. We further examined the competition between alanyl-tmRNA and valyl-tRNA or RRF at a sense codon with different lengths of the 3′-extension of mRNAs. We found that alanyl-tmRNA can enter an A site for trans-translation at either the sense or termination codon with competition among aminoacyl-tRNA, RF1 and RRF, even without cleavage of mRNA at the A site. We also found that alanyl-tmRNA prefers a shorter 3′-extension of mRNA from the A site.
MATERIALS AND METHODS
Preparation of RF1
The gene encoding E.coli RF1, prfA, ligated into a pKK223-3 plasmid was cloned into E.coli strain JM109. Eight grams of cells that overproduced RF1 were suspended in 16 ml of buffer A [50 mM Tris–HCl (pH 7.5) and 10 µg/ml of 4-(2-aminoethyl)benzenesulfonyl fluoride] and lysed by sonication. The debris was removed by centrifugation at 9000 r.p.m. (Hitachi R10A2) for 10 min, 20 000 r.p.m. (Hitachi P65A) for 30 min and 80 000 r.p.m. (Hitachi RP80AT) for 30 min. The supernatant was fractionated by DEAE–Sepharose column chromatography with a linear gradient of KCl from 0 to 200 mM in buffer A. The fractions containing RF1 were pooled and applied to an Ni-NTA column (Pharmacia). RF1 was eluted by a linear gradient of imidazole from 0 to 20 mM. The protein concentration was measured by Bradford's method using BSA as a standard (25). The purity was analyzed by electrophoresis on an SDS polyacrylamide gel (26). The polypeptide-chain releasing activity of RF1 was determined by measuring free formyl-[3H]methionine released from formyl-[3H]methionyl-•UUCAUGUAA•ribosome complex as described by Grentzmann and Kelly (27).
Preparation of RF3
The gene encoding E.coli RF3, prfC, ligated into a pKK223-3 plasmid was cloned into E.coli strain JM109. Eight grams of cells that overproduced RF3 were suspended in 16 ml of buffer B [50 mM Tris–HCl (pH 7.5), 5 µM GDP and 10 µg/ml of 4-(2-aminoethyl)benzenesulfonyl fluoride] and were lysed by sonication. The debris was removed by centrifugation three times at 9000 r.p.m. (Hitachi R10A2) for 10 min, 20 000 r.p.m. (Hitachi P65A) for 30 min and 80 000 r.p.m. (Hitachi RP80AT) for 30 min. The supernatant was then applied to DEAE–Sepharose, and the proteins were eluted with a linear gradient of KCl from 25 to 250 mM in buffer B. The fractions containing RF3 were pooled, adjusted to be pH 6.0 with 20 mM sodium phosphate, and applied to a CM–Sepharose column (Tosoh). RF3 was eluted with a linear gradient of NaCl from 0 to 200 mM. The GTPase activity of RF3 was measured in the presence of ribosome. The hydrolysis of [32P]GTP into [32P]GDP was measured by thin-layer chromatography (28).
Preparation of RRF
The gene encoding E.coli RRF, frr, ligated into a pKK223-3 plasmid was cloned into E.coli strain JM109. Eight grams of cells that overproduced RRF were suspended in 16 ml of buffer A and lysed by sonication. The debris was removed by centrifugation at 9000 r.p.m. (Hitachi R10A2) for 10 min, 20 000 r.p.m. (Hitachi P65A) for 30 min and 80 000 r.p.m. (Hitachi RP80AT) for 30 min. The supernatant was then applied to Q-Sepharose Fast Flow (Pharmacia). The flow-through fraction containing RRF was adjusted to be 1.5 M ammonium sulfate and applied to Phenyl Sepharose 6 Fast Flow (Pharmacia). RRF was eluted with a linear gradient of ammonium sulfate from 1.5 to 0.5 M. The activity of RRF was confirmed by monitoring the conversion from polysomes to monosomes in the presence of EF-G by sucrose density gradient centrifugation (23).
Preparation of tmRNA and SmpB
E.coli tmRNA was induced from an overproducing strain by the addition of 1.0 mM Isopropyl-β-d-thiogalactopyranoside (IPTG) and was purified as described previously (29). The nucleic acid fraction was extracted with phenol and then subjected to ethanol precipitation. After performing phenol extraction and ethanol precipitation, the resulting fraction was subjected to differential isopropylalcohol precipitations to roughly remove DNA, followed by incubation with RNase-free DNase I (Pharmacia). tmRNA was purified by electrophoresis on a 5% polyacrylamide gel containing 7M urea.
C-terminally His6-tagged SmpB was induced from an overproducing strain by the addition of 1.0 mM IPTG and was purified as described previously (30).
Preparation of the stalled complex of ribosome
Ribosomes were prepared from E.coli W3110 (28,31). E.coli EF-G and EF-Tu with the addition of the sequence of His-tag at the C-terminal were cloned and overproduced in E.coli. They were purified using an Ni-NTA column (32,33) and their ribosome-dependent GTPase activities were confirmed by thin-layer chromatography (28).
E.coli tRNAPhe (Sigma) was aminoacylated with [14C]phenylalanine by E.coli phenylalanyl-tRNA synthetase. Polyphenylalanines were synthesized using ribosome programmed with synthetic mRNAs. Translation reaction mixture (50 µl) contained 5 pmol ribosome, 100 pmol mRNA, 100 pmol EF-Tu, 3 pmol EF-G, 10 pmol [14C]phenylalanyl-tRNAPhe, 2 mM spermidine, 1 mM ATP, 0.2 mM GTP, 80 mM Tris–HCl (pH 7.5), 30 mM ammonium chloride, 5 mM magnesium acetate and 2.5 mM DTT. After different periods of incubation at 37°C, a 10 µl aliquot of the mixture was withdrawn and put in 5 ml of hot 5% trichloroacetic acid, preheated at 90°C for 5 min. Then 14C-labeled phenylalanines incorporated into a polypeptide, which were in the acid-insoluble fraction, were quantified by using a liquid scintillation counter (LSC-3500; Aloka). The phenylalanine incorporation was saturated by incubation at 37°C for 10 min, and the solution was then used as a stalled complex of ribosome for further studies.
Trans-translation and translation using the stalled complex of ribosome
tmRNA was aminoacylated with [3H]alanine by alanyl-tRNA synthetase from E.coli (4,34). Alanine was incorporated from alanyl-tmRNA into a polyphenylalanine peptide chain by incubating the stalled complex of ribosome with a tmRNA mixture containing 5 pmol [3H]alanyl-tmRNA and 30 pmol SmpB. After different periods of incubation at 37°C, an aliquot of the mixture was withdrawn and put in 5 ml of hot 5% trichloroacetic acid to stop the reaction, and the radioactivity of the acid-insoluble fraction was measured by using a liquid scintillation counter.
E.coli tRNAVal (Sigma) was aminoacylated with [3H]valine by valyl-tRNA synthetase from E.coli (35). Valine incorporation into polypeptide was determined by mixing the stalled complex of ribosome with a valyl-tRNAVal mixture containing 1 pmol [3H]valyl-tRNAVal, 0.1 pmol EF-G and 20 pmol EF-Tuz. After incubation at 37°C for 30 s, an aliquot of the mixture was withdrawn and put in 5 ml of hot 5% trichloroacetic acid to stop the reaction and the radioactivity of the acid-insoluble fraction was measured by using a liquid scintillation counter.
RESULTS
In vitro translation and peptidyl-transfer to alanyl-tmRNA for trans-translation
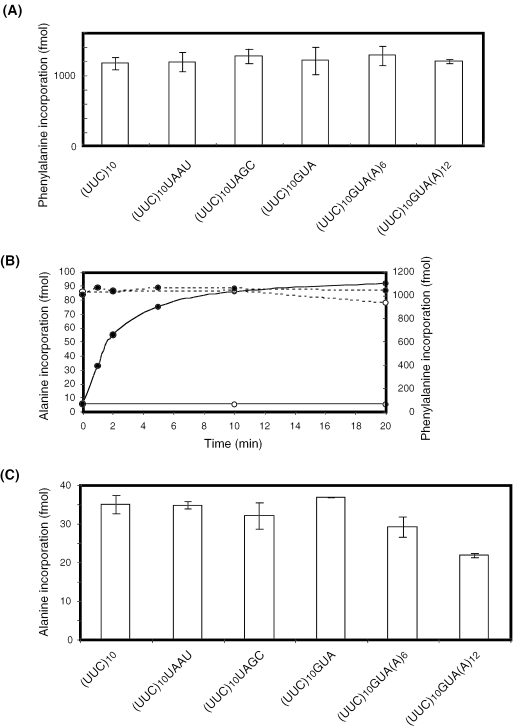
Using pure translation factors, [14C]phenylalanine was incorporated into polyphenylalanine from (UUC)10 or its derivatives having a varying sequence of 3′-extension. After incubation of the mixture at 37°C for 10 min, the phenylalanine incorporation was saturated (data not shown). The levels of phenylalanine incorporation at 10 min from six kinds of mRNAs were similar as shown in Figure 1A, and the solution was used as a stalled complex of ribosome for further studies.
Figure 1.
In vitro polyphenylalanine synthesis directed by (UUC)10-based mRNAs and alanine incorporation by tmRNA from the stalled complex of ribosome. (A) Fifty microliters of translation reaction mixture contained 5 pmol ribosome, 100 pmol mRNA, 200 pmol EF-Tu, 3 pmol EF-G, 10 pmol [14C]phenylalanyl-tRNAPhe, 2 mM spermidine, 1 mM ATP, 0.2 mM GTP, 80 mM Tris–HCl (pH 7.5), 30 mM ammonium chloride, 5 mM magnesium acetate and 2.5 mM DTT. After incubation at 37°C for 10 min, a 10 µl aliquot was withdrawn from the reaction mixture and put in 5 ml of hot 5% trichloroacetic acid. (B) Sixty microliters of stalled complex of ribosomes programmed with (UUC)10GUA prepared in (A) was incubated at 37°C with 20 µl of tmRNA mixture containing 4 pmol [3H]alanyl-tmRNA with (solid circle) or without (open circle) 60 pmol SmpB. After incubation at the indicated time point, a 10 µl aliquot was withdrawn and put in 5 ml of hot 5% trichloroacetic acid. The incorporated alanines (solid line) and phenylalanines (dotted line) were quantified. (C) Fifteen microliters of stalled complex of ribosome programmed with each kind of mRNA was incubated with 5 µl of tmRNA mixture containing 1 pmol pre-charged [3H]alanyl-tmRNA and 15 pmol SmpB. After incubation at 37°C for 30 s, a 15 µl aliquot was withdrawn and put in 5 ml of hot 5% trichloroacetic acid. The incorporated alanines were quantified.
The stalled complex of ribosome was then incubated with 3H-labeled alanyl-tmRNA mixture. Alanine was gradually incorporated only in the presence of SmpB, a tmRNA binding protein (30,36), and the alanine incorporation was almost saturated after 10 min of incubation (Figure 1B). During this period, the amount of phenylalanines incorporated did not change (Figure 1B, dotted lines). At a saturated time point, the ratio of alanines to phenylalanines incorporated was ∼0.09. The alanine incorporation at 30 s from six kinds of mRNAs is shown in Figure 1C. When mRNA having the longest 3′-extension, (UUC)10GUA(A)12, was used, alanine incorporation was significantly less than the alanine incorporations using the other kinds of mRNAs.
Competition between RF1 and alanyl-tmRNA
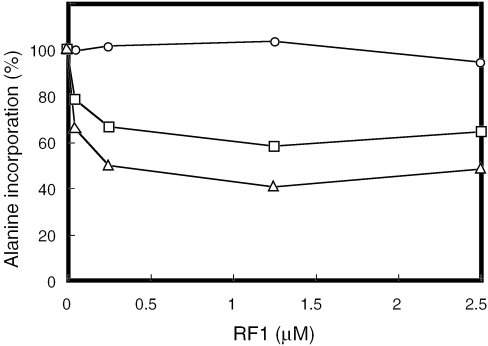
The efficiency of termination depends on the stop codon and its 3′-adjacent (+4) nucleotide. UAAU and UAAG serve as strong stop signals, while UGAC and UAGC give poor termination efficiency (37). In the present study, two different contexts of stop codons UAAU and UAGC that are strong and weak stop signals for RF1, respectively, were used as mRNAs to examine the competition between RF1 and alanyl-tmRNA. Various amounts of purified RF1 were added to the tmRNA mixture with a fixed amount of 3H-labeled alanyl-tmRNA. Each of the tmRNA mixtures containing various amounts of RF1 was incubated with the stalled complex of ribosome at 37°C for 1 min. During this process, phenylalanine incorporation was not affected by RF1. By using mRNA without a stop signal sequence, RF1 had no influence on the alanine incorporation by alanyl-tmRNA (Figure 2). When using (UUC)10UAAU and (UUC)10UAGC as mRNAs containing a stop signal for RF1, however, alanine incorporation was significantly affected by RF1, the effect being much greater when (UUC)10UAAU was used. When using the same amounts of tmRNA and RF1 in the reaction, the percentages remaining of alanine incorporation from the stalled complex of ribosome programmed with (UUC)10UAAU and (UUC)10UAGC were 66 and 78%, respectively. Apparently, the efficiency of peptidyl-transfer for trans-translation in the presence of RF1 depended on the preference of the stop signal for RF1. These results indicate that alanyl-tmRNA competes with RF1 at a termination codon with the efficiency of trans-translation inversely correlated to the efficiency of translation termination.
Figure 2.
Effect of RF1 on alanine incorporation by tmRNA. Stalled complex of ribosomes (30 µl) programmed with (UUC)10 (circle), (UUC)10UAGC (rectangle) or (UUC)10UAAU (triangle) mRNA was incubated in 10 µl of tmRNA mixture containing 2 pmol pre-charged [3H]alanyl-tmRNA, 30 pmol SmpB and various concentrations of RF1. After incubation at 37°C for 1 min, a 10 µl aliquot was withdrawn and put in 5 ml of hot 5% trichloroacetic acid. Alanine incorporation in the reaction without RF1 is regarded as 100%. The experiments showed a good reproducibility and the error was within 10%.
RF3 did not affect the inhibition of RF1 on peptidyl-transfer for trans-translation at a termination codon
RF3 has been shown to stimulate the dissociation of RF1 from the ribosome after peptidyl-tRNA hydrolysis (21). It accentuates the strength of the stop signal specified by the 4 nt stop signal. Therefore, RF3 was added to the reaction mixture. However, no significant difference was found between alanine incorporation or between phenylalanine incorporation in the presence of RF3 and absence of RF3 (Figure 3). This result indicates that RF3 neither enhances nor inhibits the step of peptidyl-transfer for trans-translation at a termination codon.
Figure 3.
Effect of RF1 on alanine incorporation by tmRNA in the presence of RF3. Stalled complex of ribosomes (30 µl) programmed with a (A) (UUC)10-based mRNA (used as control), (B) (UUC)10UAGC or (C) (UUC)10UAAU mRNAs was incubated with 10 µl of tmRNA mixture containing 2 pmol [3H]alanyl-tmRNA, 30 pmol SmpB, various concentrations of RF1 and 0 (solid circle) or 20 pmol RF3 (open circle). After incubation at 37°C for 1 min a 10 µl aliquot was withdrawn and put in 5 ml of hot 5% trichloroacetic acid. Alanine incorporation in the reaction without release factors is regarded as 100%. The experiments showed a good reproducibility and the error was within 10%.
Competition between RRF and alanyl-tmRNA
The effect of RRF on alanine incorporation by tmRNA was examined by the addition of RRF to the [3H]alanyl-tmRNA mixture. After incubation of this mixture for 30 s with the stalled complex of ribosome programmed with six kinds of mRNAs, alanine incorporation was measured. Alanine incorporation was decreased when the reaction mixture contained 0.5 and 5.0 µM of RRF, which were 10 and 100 times greater than the amount of tmRNA, respectively (Figure 4). In the case of a smaller amount of RRF, no reduction of alanine incorporation was detected (data not shown). The patterns of inhibition by RRF for all kinds of mRNAs were similar. Since phenylalanine incorporation was not decreased by the addition of RRF, the observed inhibition by RRF was not due to the decrease in the stalled complex of ribosome. These results indicate that alanyl-tmRNA competes with RRF to enter the A site of the stalled ribosome either in the presence or absence of mRNA in the A site.
Figure 4.
Effect of RRF on alanine incorporation by tmRNA. Stalled complex of ribosome (15 µl) was incubated with 5 µl of tmRNA mixture containing 1 pmol pre-charged [3H]alanyl-tmRNA, 15 pmol SmpB and 0 (white), 0.5 (gray) or 5.0 (black) µM of RRF. After incubation at 37°C for 30 s, a 15 µl aliquot was withdrawn and put in 5 ml of hot 5% trichloroacetic acid. The incorporation of alanine in the reaction without RRF is regarded as 100%.
Effect of alanyl-tmRNA on valine incorporation
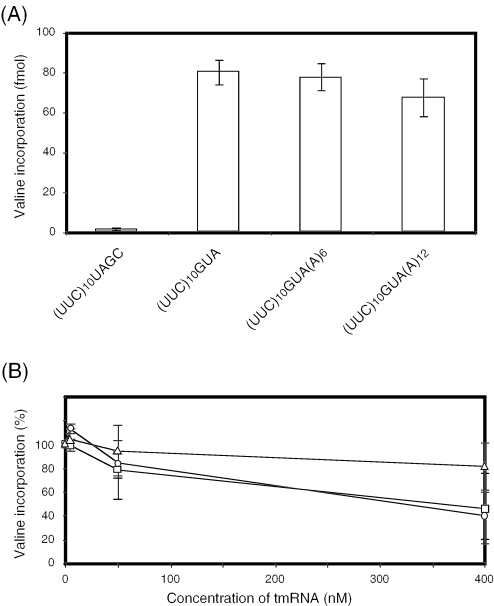
To study the competition between tRNA and tmRNA at a sense codon, the effect of alanyl-tmRNA on the incorporation of valine using mRNAs containing a GUA codon were observed. After a saturating level of polyphenylalanine was synthesized as explained above, the [3H]valyl-tRNAVal mixture was added. After incubation at 37°C for 30 s, valine incorporation was measured. As expected, [3H]valine was significantly incorporated when using (UUC)10GUA, (UUC)10GUA(A)6 or (UUC)10GUA(A)12 as an mRNA, while valine incorporation was not observed when using (UUC)10UAGC lacking a valine codon (Figure 5A). Similar levels of valine incorporation were observed from three mRNAs having a GUA codon with different lengths of the 3′-extension from the GUA codon. This result indicates that aminoacyl-tRNA, unlike alanyl-tmRNA (Figure 1C), enters the A site of a translating ribosome regardless of the length of the 3′-extension of mRNA from the A site.
Figure 5.
In vitro valine incorporation from (UUC)10-dependent polyphenylalanine–ribosome complex and effect of tmRNA on valine incorporation. (A) Stalled complex of ribosomes (15 µl) programmed with a (UUC)10GUA-based mRNA or (UUC)10UAGC (used as control) was incubated with 5 µl of the mixture containing 1 pmol pre-charged [3H]valyl-tRNAVal, 0.1 pmol EF-G and 20 pmol EF-Tu. (B) Stalled complex of ribosome (15 µl) programmed with (UUC)10GUA (circle), (UUC)10GUA(A)6 (rectangle) or (UUC)10GUA(A)12 (triangle) was incubated with 5 µl of the mixture containing 1 pmol pre-charged [3H]valyl-tRNAVal, 0.1 pmol EF-G, 20 pmol EF-Tu, 15 pmol SmpB and various concentrations of alanyl-tmRNA. After incubation at 37°C for 30 s, a 15 µl aliquot was withdrawn and put in 5 ml of hot 5% trichloroacetic acid. Valine incorporation in the reaction without alanyl-tmRNA is regarded as 100% (B).
Various concentrations of alanyl-tmRNA with 15 pmol SmpB were added to the [3H]valyl-tRNAVal mixtures. These mixtures were then combined with the stalled complex of ribosome. After incubation at 37°C for 30 s, valine incorporations were measured (Figure 5B). At the same concentrations of valyl-tRNAVal and alanyl-tmRNA (50 nM), valine incorporation was reduced by 10–20%. When the concentration of alanyl-tmRNA was increased to 400 nM, 8 times higher than that of valyl-tRNAVal, valine incorporation was decreased more. Alanyl-tmRNA had the smallest inhibitory effect when mRNA having the longest (12 nt) extension from the GUA codon was used. These results indicate that the length of 3′-extension of mRNA from the A site for alanyl-tmRNA is critical for entrance into the stalled complex of ribosome more than that for aminoacyl-tRNA.
RRF did not affect valine incorporation
Various concentrations of RRF were added to the [3H]valyl-tRNAVal mixtures. These mixtures were then mixed with the stalled complex of ribosome. After incubation at 37°C for 30 s, incorporation of [3H]valine was measured. No significant change in valine incorporation from any type of mRNA used was observed by the addition of 50 µM RRF, a 1000-fold higher concentration than that of valyl-tRNAVal (Figure 6). This result indicates that RRF neither enhances nor inhibits substantially the entrance of aminoacyl-tRNA into the A site of the translating ribosome.
Figure 6.
Effect of RRF on valine incorporation. Stalled complex of ribosome (15 µl) programmed with (UUC)10GUA (circle), (UUC)10GUA(A)6 (rectangle) or (UUC)10GUA(A)12 (triangle) was incubated with 5 µl of the mixture containing 1 pmol [3H]valyl-tRNAVal, 0.1 pmol EF-G, 20 pmol EF-Tu and various concentrations of RRF. After incubation at 37°C for 30 s, a 15 µl aliquot was withdrawn and put in 5 ml of hot 5% trichloroacetic acid. Valine incorporation in the reaction without RRF is regarded as 100%.
Cleavage of mRNAs was not found from the stalled complex of ribosome
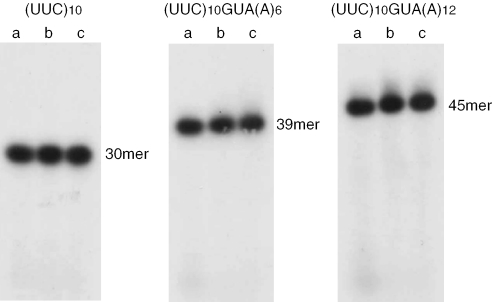
mRNA labeled with 32P at the 5′ end was used for synthesizing the stalled complex of ribosome. Then mRNA in the stalled complex of ribosome was examined by PAGE. To increase the population of mRNA involved in the stalled complex of ribosome, the concentrations of mRNA and ribosome in the mixture of the stalled complex formation were decreased and increased, respectively (Figure 7). In this system, >10% of mRNA was involved in the stalled complex of ribosome, judging from the amount of alanine incorporated. If (UUC)10GUA(A)6 or (UUC)10GUA(A)12 were cleaved at the A site of the stalled complex of ribosome, a band of 30–33mer should appear. However, no band was detected around this position (Figure 7, lane b). Besides, no significant change in the band pattern was observed when the ribosome was removed from the reaction mixture (Figure 7, lane a). We also confirmed that the addition of alanyl-tmRNA did not change the band pattern (Figure 7, lane c). Even after 10-fold longer exposure, no band of cleavage was detected in any lane of Figure 7. These results indicate that no endoribonucleolytic cleavage of mRNA occurs around the A site in the stalled ribosome in our in vitro system and that alanyl-tmRNA can accept peptide from peptidyl-tRNA for trans-translation without any cleavage of mRNA around the A site in the stalled ribosome.
Figure 7.
Detection of the degradation of mRNA in the stalled complex of ribosome. The 5′ end of synthetic mRNA was labeled with γ-[32P]ATP by T4 polynucleotide kinase (TaKaRa). The amount of mRNAs was photometrically quantified at a wavelength of 260 nm. The stalled complex of ribosome was formed in a 20 µl of reaction mixture containing 0 pmol (lane a) or 30 pmol ribosome (lanes b and c), 3 pmol 32P-labeled-mRNA, 80 pmol EF-Tu, 3 pmol EF-G, 10 pmol [14C]phenylalanyl-tRNAPhe, 0 pmol (lanes a and b) or 5 pmol [3H]alanyl-tmRNA (lane c), 2 mM spermidine, 1 mM ATP, 0.2 mM GTP, 80 mM Tris–HCl (pH 7.5), 30 mM ammonium cloride, 5 mM magnesium acetate and 2.5 mM DTT. After incubation at 37°C for 10 min, mRNA was electrophoresed on 12% polyacrylamide gel and visualized on an X-ray film.
DISCUSSION
Poly(U)-dependent in vitro trans-translation systems using the E.coli S30 fraction (8) and using pure translation factors (38) have already been developed. In the present study, we developed an in vitro trans-translation system coupled with translation directed by a (UUC)10-based synthetic mRNA using pure translation factors. Compared with poly(U), (UUC)10 has an advantage for specifying the reading frame. In our system, peptidyl-transfer to alanyl-tmRNA for trans-translation efficiently occurred in the presence of mRNA, ribosome, aminoacyl-tRNA, EF-Tu, EF-G, alanyl-tmRNA and SmpB. Using this system, we studied the competitions at a codon in the A site of the stalled ribosome between alanyl-tmRNA and RF, between alanyl-tmRNA and aminoacyl-tRNA and between alanyl-tmRNA and RRF. We found that trans-translation by tmRNA can operate on sense and termination codons at the A site at which alanyl-tmRNA competes with aminoacyl-tRNA and RF, respectively, and that the peptidyl-transfer step of trans-translation does not absolutely require an mRNA truncated at the A site but prefers an mRNA of a shorter 3′-extension from the A site. The addition of RF3 had almost no effect on the competition between RF1 and alanyl-tmRNA at a nonsense codon.
Classically, tmRNA has been assumed to target a ribosome with a stalled translation in which the P site but not A site is occupied by mRNA owing to the truncation of its 3′-portion. Trans-translation can also occur at tandem rare codons on an artificial mRNA (14). Full-length and C-terminally truncated polypeptides with a tag sequence at the C-terminus have also been identified as endogenous products of trans-translation in the cell (15,16). These in vivo data suggest that trans-translation can occur when translation elongation or termination is stalled, although whether these tagged-polypeptides are produced from an intact or truncated mRNA in the cell is unknown. It has been shown recently that bacterial toxins such as RelE or ChpAK/MazF cleave an mRNA specifically at the A site in the stalled ribosome, providing a new concept that mRNA of stalled translation is targeted initially by these A site-specific endoribonucleases and subsequently by tmRNA for trans-translation (18,19). More recently, cleavage of mRNA at the A site has been detected even in cells lacking these bacterial toxins and some cellular ribonucleases, raising the possibility that the ribosome itself is directly involved in the cleavage of mRNA at the A site (39–41). The present in vitro results clearly showed that cleavage of mRNA around the A site did not occur in the stalled ribosome and that, notwithstanding this, peptidyl-transfer for trans-translation efficiently occurred. It might be true that mRNA in the A site of the stalled ribosome in the cell is also a target for some kinds of nucleases, and indeed RelE has been shown to greatly enhance the efficiency of trans-translation (24). At the A site codon of the intact mRNA in the stalled ribosome in the cell, aminoacyl-tRNAs, release factors, alanyl-tmRNA and A site-specific nucleases would always compete with one another, and consequently alanyl-tmRNA would enter the ribosome both before and after the cleavage of mRNA around the A site. The competition should significantly be influenced by the activities or levels of these molecules in the cell, which fluctuate with change in the physiological conditions; bacterial toxins are activated upon starvation of amino acids (18,19), while tmRNA is induced under some stressful conditions (42).
The present study showed the competition between tmRNA and a release factor at a termination codon, which has been suggested by recent in vivo (43) and in vitro (41) studies. We also showed that the efficiency of peptidyl-transfer for trans-translation depends on the context of the stop signal. Furthermore, the competition with aminoacyl-tRNA at a sense codon was also shown. Apparently tmRNA prefers a shorter 3′-extension as shown in Figure 1C. Besides, the preference for the length of the 3′-extension of mRNA from the A site was different between alanyl-tmRNA and cognate aminoacyl-tRNA. The entrance of alanyl-tmRNA into the A site may be more critical to the length of the 3′-extension of mRNA than that of aminoacyl-tRNA.
A crystal structure of the ribosome in complex with an mRNA and tRNAs has shown that the downstream tunnel of the small subunit covers the downstream of mRNA encompassing positions +7 to +15 (44). Thus, the downstream tunnel is covered almost entirely by (UUC)10GUA(A)12, partially by (UUC)10GUA(A)6 and not by (UUC)10GUA. mRNA with a short 3′-extension may be transiently detached from the A site owing to a weak interaction with the downstream tunnel, whereas mRNA with a long 3′-extension can be fixed to the A site owing to a sufficient interaction with the downstream tunnel. Consequently, the present observation described above can be rationalized, assuming that the region at the A site or its proximity in the small subunit required for interaction with alanyl-tmRNA is sequestered extensively by mRNA with a long 3′-extension but only weakly by mRNA with a short 3′-extension. Interaction between tmRNA and the proximity of the A site for trans-translation has been suggested (45,46), although such an interaction has not been found in a complex of tmRNA/SmpB/ribosome/EF-Tu/GTP/kirromycin, presumably reflecting the initial binding of the tmRNA/SmpB/EF-Tu/GTP complex with the ribosome (47).
RRF together with EF-G facilitates ribosome recycling after the release of polypeptides by promoting the dissociation of deacylated tRNA in the P site and/or the dissociation of the 50S subunit from the posttermination complex (22,48). Several structural studies have shown that RRF, unlike tRNA, locates across the A site of the 50S subunit, although it is a structural mimic of tRNA (49–51). Apparently, the sites of RRF, aminoacyl-tRNA and alanyl-tmRNA on the ribosome overlap each other. The empty A site of the posttermination complex in which the P site is occupied by a deacyl-tRNA is a preferable substrate for RRF (52). Our results obtained by using a stalled complex of ribosome with a GUA codon at the A site suggest that RRF does not substantially interfere with the binding of cognate aminoacyl-tRNA to the A site during the canonical elongation processes. This is consistent with an earlier finding that the affinity of the cognate EF-Tu/GTP/aminoacyl-tRNA ternary complex to the A site is much higher than that of RRF (53). In the present study, we further demonstrated that RRF inhibits alanine incorporation by alanyl-tmRNA, although only at very high concentration. It is consistent with a previous finding that RRF binds, albeit with very low affinity, to the ribosome in which the P site is occupied by a peptidyl-tRNA (53). The efficiency of peptidyl-transfer for trans-translation (Figure 1C) was dependent on the length of the 3′-extension of mRNA on the stalled ribosome, whereas the inhibition of peptidyl-transfer for trans-translation by RRF was not (Figure 4). It has been shown in vivo that drop-off of mRNA from the stalled ribosome is enhanced by RRF in combination with peptidyl-tRNA hydrolase (54–56). The recognition of the stalled ribosomes with different lengths of 3′-extension of mRNA by RRF has yet to be investigated.
The present study demonstrates in vitro that the trans-translation system can efficiently operate on a stalled ribosome having an mRNA with a short 3′-extension with competition of elongation or termination of translation. Several other factors, such as A site-specific nucleases or peptidyl-tRNA hydrolase, should also be involved in releasing a stalled ribosome having an intact mRNA with a long 3′-extension in vivo.
Acknowledgments
We thank Dr T. Yokogawa for giving us the overproducing strain for methiony-tRNA transformylase and the staff of Gene Research Center of Hirosaki University for the use of the facility. This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture, Japan to A.M. and H.H. (no. 14035201), a grant-in-aid for scientific research from the Japan Society for the Promotion of Science to A.M. and H.H. (no. 17380061) and a research grant from Hirosaki University to A.M. and H.H. Funding to pay the Open Access publication charges for this article was provided by The Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gueneau de Novoa P., Williams K.P. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004;32:D104–D108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwieb C., Gorodkin J., Knudsen B., Burks J., Wower J. tmRDB (tmRNA database) Nucleic Acids Res. 2003;31:446–447. doi: 10.1093/nar/gkg019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacob Y., Seif E., Paquet P.O., Lang B.F. Loss of the mRNA-like region in mitochondrial tmRNAs of jakobids. RNA. 2004;10:605–614. doi: 10.1261/rna.5227904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ushida C., Himeno H., Watanabe T., Muto A. tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res. 1994;22:3392–3396. doi: 10.1093/nar/22.16.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komine Y., Kitabatake M., Yokogawa T., Nishikawa K., Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu G.F., Reid G.E., Zhang J.G., Moritz R.L., Simpson R.J. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- 7.Keiler K.C., Waller P.R., Sauer R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 8.Himeno H., Sato M., Tadaki T., Fukushima M., Ushida C., Muto A. In vitro trans translation mediated by alanine-charged 10Sa RNA. J. Mol. Biol. 1997;268:803–808. doi: 10.1006/jmbi.1997.1011. [DOI] [PubMed] [Google Scholar]
- 9.Muto A., Ushida C., Himeno H. A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci. 1998;23:25–29. doi: 10.1016/s0968-0004(97)01159-6. [DOI] [PubMed] [Google Scholar]
- 10.Withey J.H., Friedman D.I. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto Y., Sunohara T., Jojima K., Inada T., Aiba H. SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA. 2003;9:408–418. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felden B., Hanawa K., Atkins J.F., Himeno H., Muto A., Gesteland R.F., McCloskey J.A., Crain P.F. Presence and location of modified nucleotides in E.coli tmRNA. Structural mimicry with tRNA acceptor branches. EMBO J. 1998;17:3188–3196. doi: 10.1093/emboj/17.11.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nameki N., Felden B., Atkins J.F., Gesteland R.F., Himeno H., Muto A. Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E.coli tmRNA. J. Mol. Biol. 1999;286:733–744. doi: 10.1006/jmbi.1998.2487. [DOI] [PubMed] [Google Scholar]
- 14.Roche E.D., Sauer R.T. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche E.D., Sauer R.T. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J. Biol. Chem. 2001;276:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- 16.Fujihara A., Tomatsu H., Inagaki S., Tadaki T., Ushida C., Himeno H., Muto A. Detection of tmRNA-mediated trans-translation products in Bacillus subtilis. Genes Cells. 2002;7:343–350. doi: 10.1046/j.1365-2443.2002.00523.x. [DOI] [PubMed] [Google Scholar]
- 17.Hayes C.S., Bose B., Sauer R.T. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:3440–3445. doi: 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen K., Zavialov A.V., Pavlov M.Y., Elf J., Gerdes K., Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 19.Christensen S.K., Pedersen K., Hansen F.G., Gerdes K. Toxin–antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura Y., Ito K., Isaksson L.A. Emerging understanding of translation termination. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- 21.Freistroffer D.V., Pavlov M.Y., MacDougall J., Buckingham R.H., Ehrenberg M. Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janosi L., Hara H., Zhang S., Kaji A. Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv. Biophys. 1996;32:121–201. doi: 10.1016/0065-227x(96)84743-5. [DOI] [PubMed] [Google Scholar]
- 23.Pavlov M.Y., Freistroffer D., Heurgue-Hamard V., Buckingham R.H., Ehrenberg M. Release factor RF3 abolishes competition between release factor RF1 and ribosome recycling factor (RRF) for a ribosome binding site. J. Mol. Biol. 1997;273:389–401. doi: 10.1006/jmbi.1997.1324. [DOI] [PubMed] [Google Scholar]
- 24.Ivanova N., Pavlov M.Y., Felden B., Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 25.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Grentzmann G., Kelly P.J. Ribosomal binding site of release factors RF1 and RF2. A new translational termination assay in vitro. J. Biol. Chem. 1997;272:12300–12304. doi: 10.1074/jbc.272.19.12300. [DOI] [PubMed] [Google Scholar]
- 28.Himeno H., Hanawa-Suetsugu K., Kimura T., Takagi K., Sugiyama W., Shirata S., Mikami T., Odagiri F., Osanai Y., Watanabe D., et al. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 2004;32:5303–5309. doi: 10.1093/nar/gkh861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nameki N., Tadaki T., Muto A., Himeno H. Amino acid acceptor identity switch of Escherichia coli tmRNA from alanine to histidine in vitro. J. Mol. Biol. 1999b;289:1–7. doi: 10.1006/jmbi.1999.2754. [DOI] [PubMed] [Google Scholar]
- 30.Hanawa-Suetsugu K., Takagi M., Inokuchi H., Himeno H., Muto A. SmpB functions in various steps of trans-translation. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaha G., Stelzl U., Spahn C.M., Agrawal R.K., Frank J., Hiehaus K.H. Preparation of functional ribosomal complexes and effect of buffer conditions on tRNA positions observed by cryoelectron microscopy. Methods Enzymol. 2000;317:292–309. doi: 10.1016/s0076-6879(00)17021-1. [DOI] [PubMed] [Google Scholar]
- 32.Boon K., Vijgenboom E., Madsen L.V., Talens A., Kraal B., Bosch L. Isolation and functional analysis of histidine-tagged elongation factor Tu. Eur. J. Biochem. 1992;210:177–183. doi: 10.1111/j.1432-1033.1992.tb17406.x. [DOI] [PubMed] [Google Scholar]
- 33.Chen B.P., Hai T. Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene. 1994;139:73–75. doi: 10.1016/0378-1119(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 34.Putney S.D., Sauer R.T., Schimmel P.R. Purification and properties of alanine tRNA synthetase from Escherichia coli. A tetramer of identical subunits. J. Biol. Chem. 1981;256:198–204. [PubMed] [Google Scholar]
- 35.Tamura K., Nameki N., Hasegawa T., Shimizu M., Himeno H. Role of the CCA terminal sequence of tRNAVal in aminoacylation with valyl-tRNA synthetase. J. Biol. Chem. 1994;269:22173–22177. [PubMed] [Google Scholar]
- 36.Karzai A.W., Susskind M.M., Sauer R.T. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole E.S., Brown C.M., Tate W.P. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu Y., Ueda T. The role of SmpB protein in trans-translation. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- 39.Hayes C.S., Sauer R.T. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 40.Sunohara T., Jojima K., Yamamoto Y., Inada T., Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004;10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunohara T., Jojima K., Tagami H., Inada T., Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 2004;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 42.Muto A., Fujihara A., Ito K., Matsuno J., Ushida C., Himeno H. Requirement of transfer-messenger RNA (tmRNA) for the growth of Bacillus subtilis under stresses. Genes Cells. 2000;5:627–636. doi: 10.1046/j.1365-2443.2000.00356.x. [DOI] [PubMed] [Google Scholar]
- 43.Collier J., Binet E., Bouloc P. Competition between SsrA tagging and translational termination at weak stop codons in Escherichia coli. Mol. Microbiol. 2002;45:745–754. doi: 10.1046/j.1365-2958.2002.03045.x. [DOI] [PubMed] [Google Scholar]
- 44.Yusupova G.Z., Yusupov M.M., Cate J.H., Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 45.Lee S., Ishii M., Tadaki T., Muto A., Himeno H. Determinants on tmRNA for initiating efficient and precise trans-translation. Some mutations upstream of the tag-encoding sequence of Escherichia coli tmRNA shift the initiation point of trans-translation in vitro. RNA. 2001;7:999–1012. doi: 10.1017/s1355838201010342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konno T., Takahashi T., Kurita D., Muto A., Himeno H. A minimum structure of aminoglycosides that causes an initiation shift of trans-translation. Nucleic Acids Res. 2004;32:4119–4126. doi: 10.1093/nar/gkh750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valle M., Gillet R., Kaur S., Henne A., Ramakrishnan V., Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 48.Peske F., Rodnina M.V., Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Lancaster L., Kiel M.C., Kaji A., Noller H.F. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- 50.Gao N., Zavialov A.V., Li W., Sengupta J., Valle M., Gursky R.P., Ehrenberg M., Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Wilson D.N., Schluenzen F., Harms J.M., Yoshida T., Ohkubo T., Albrecht R., Buerger J., Kobayashi Y., Fucini P. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujiwara T., Ito K., Yamami T., Nakamura Y. Ribosome recycling factor disassembles the post-termination ribosomal complex independent of the ribosomal translocase activity of elongation factor G. Mol. Microbiol. 2004;53:517–528. doi: 10.1111/j.1365-2958.2004.04156.x. [DOI] [PubMed] [Google Scholar]
- 53.Hirokawa G., Kiel M.C., Muto A., Kawai G., Igarashi K., Kaji H., Kaji A. Binding of ribosome recycling factor to ribosomes, comparison with tRNA. J. Biol. Chem. 2002;277:35847–35852. doi: 10.1074/jbc.M206295200. [DOI] [PubMed] [Google Scholar]
- 54.Heurgue-Hamard V., Karimi R., Mora L., MacDougall J., Leboeuf C., Grentzmann G., Ehrenberg M., Buckingham R.H. Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J. 1998;17:808–816. doi: 10.1093/emboj/17.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herr A.J., Wills N.M., Nelson C.C., Gesteland R.F., Atkins J.F. Drop-off during ribosome hopping. J. Mol. Biol. 2001;311:445–452. doi: 10.1006/jmbi.2001.4899. [DOI] [PubMed] [Google Scholar]
- 56.Singh N.S., Varshney U. A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Res. 2004;32:6028–6037. doi: 10.1093/nar/gkh924. [DOI] [PMC free article] [PubMed] [Google Scholar]