Abstract
We have solved the crystal structure of the MHCII molecule, IAb, containing an antigenic variant of the major IAb-binding peptide derived from the MHCII IEα chain. The four MHC pockets at p1, p4, p6, and p9 that usually bind peptide side chains are largely empty because of alanines in the peptide at these positions. The complex is nevertheless very stable, apparently because of unique alternate interactions between the IAb and peptide. In particular, there are multiple additional hydrogen bonds between the N-terminal end of the peptide and the IAb α chain and an extensive hydrogen bond network involving an asparagine at p7 position of the peptide and the IAb β chain. By using knowledge of the shape and size of the traditional side chain binding pockets and the additional possible interactions, an IAb peptide-binding motif can be deduced that agrees well with the sequences of known IAb-binding peptides.
The crystal structures of a variety of MHCI (1–9) and MHCII (10–23) molecules have revealed their features that lead to stable peptide binding. Conserved amino acids of MHCI interact with the N terminus and C terminus of the peptide in virtually all isotypes and alleles. Between these termini, the peptide can vary in length and in the path it takes through the binding groove, often bulging out of the groove in the middle. In MHCII, the N and C termini of the peptide generally extend beyond the binding groove and do not interact with the MHCII molecule. Rather, H-bonding interactions between the peptide backbone and conserved amino acids of the MHCII α-helices force the peptide to take a similar, extended path through the groove of all MHCII isotypes and alleles.
The unique interactions that determine the allelic specificity of peptide binding seem to be controlled by the character of pockets within the interior of the binding groove that preferentially accept particular amino acid side chains. Because the MHC amino acids that line these pockets are among the most genetically variable, their depth, shape, and chemistry vary considerably among MHC alleles and isotypes, and they appear to be the most important factor in the specificity of peptide binding.
In the current study we have solved the crystal structure of the mouse MHCII molecule IAb with a bound immunogenic peptide variant of a dominant peptide derived from the MHCII Eα protein that is found in IAb in some strains of mice (24). Despite the high stability and immunogenicity of this complex, the four usual p1, p4, p6, and p9 peptide side chain binding pockets of IAb are largely unfilled, because alanines occur in the peptide at these positions. Rather, the stability of this peptide/MHCII complex can be attributed to the combination of the conserved interactions with the peptide backbone as well as extensive unique interactions of the IAb molecule with other peptide amino acids particularly at the N-terminal end and the p7 position of the peptide. These results suggest that IAb can achieve stable peptide binding in more than one way and may account for the previous difficulty in defining the IAb peptide-binding motif.
Materials and Methods
Preparation of Soluble IAb.
The production of soluble IAb containing a peptide attached to the β-chain N terminus via a flexible peptide linker has been described (25). In the original constructions, peptides included both the Eα-derived peptide, ASFEAQGALANIAVDKA (pEα), which occupies about 10% of natural IAb, and an immunogenic variant, ASFEAQKAKANKAVDKA (p3K), in which three positions in the peptide (underlined) had lysines substituted for the Eα peptide amino acids. For crystallization, the construct was altered to use a shortened form of p3K, FEAKGAKANKAVD, and to shorten the IAb α and β chains to the predicted ends of the α2 and β2 domains. The construct was cloned into a previously described dual promoter baculovirus transfer vector (26, 27) and introduced into BacVector 3000 version of AcNPV baculovirus (Novagen). High-titer virus stocks were prepared in Sf9 insect cells (Invitrogen). Soluble IAb–p3K was immunoaffinity-purified from the supernatant of High Five infected insect cells (Invitrogen) by using the IAb β-chain-reactive monoclonal antibody M5/114 (28). The eluted protein was concentrated and further purified by size exclusion chromatography using Superdex-200 (Amersham Pharmacia).
Crystallization.
The crystals of IAb–p3K were grown by the vapor diffusion method in hanging drops. Reproducible small crystals grew at room temperature under the following conditions: 16% PEG 8000/0.1 M Mes, pH 6.0/0.1 M KH2PO4, with a protein concentration of 4–5 mg/ml. Streaking seeding was used to improve the size of crystals. Crystals with a size of 0.4 × 0.4 × 0.05 mm were used for x-ray diffraction experiments. Crystals were soaked overnight in a cryoprotectant consisting of 25% trehalose in the mother liquor, and then flash-cooled in liquid nitrogen for data collecting.
Data Collection, Processing, and Detwinning.
A high-resolution data set (2.5 Å) was collected at 100 K for a single crystal on beam line SBC 19BM at the Advanced Photon Source at Argonne National Laboratories (Argonne, IL). The data were initially indexed and integrated with HKL2000 (29) in a C-centered orthorhombic lattice, but the data merged poorly (Rmerge = 22%). After the data were reindexed in the less symmetric space group P21, they merged well; however, the crystals were determined to be merohedral-twinned, as judged by cumulative intensity distribution calculated with the program truncate (30). We used the program detwin (30) with the twinning operator l, −k, h, and a twinning fraction of 0.31, to create the detwinned data set that was used to solve the structure. A Vm value (31) of 2.46 Å3/Da indicated four molecules per asymmetric unit with a solvent content of ≈50%. Collection statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P21 |
| Unit cell dimensions, Å | a = 65.04 |
| b = 274.18 | |
| c = 65.12 | |
| Unit cell angles, ° | β = 111.42 |
| No. of molecules in asymmetric unit | 4 |
| Resolution limits, Å | 40–2.5 (2.59–2.50)* |
| Merohedral twinning | |
| Operator | l, −k, h |
| Twinning fraction | 0.31 |
| Unique reflections | 54,497 (1,829) |
| Completeness, % | 74.8 (21.3) |
| Average redundancy | 2.2 |
| Average I/σ | 7.0 (4.3) |
| Rmerge, %† | 8.9 (35.2) |
| Refinement | |
| Resolution, Å | 40–2.5 (2.59–2.50) |
| Rejection criterion | F ≤ 0 |
| Total reflections | 54,427 (1,591) |
| Reflections used for Rfree | 2,621 (66) |
| Rworking, %‡ | 21.1 (27.7) |
| Rfree, %‡ | 24.5 (31.3) |
| Average B factors, Å2 | 20.8 |
| Ramachandran data | |
| % of residues§ in: | |
| Favored regions | 80.4 |
| Allowed regions | 18.9 |
| Disallowed regions | 0.6 |
| rms deviation | |
| Bonds, Å | 0.0105 |
| Angles, ° | 1.511 |
| B factor main chain [1.5],¶ Å2 | 2.9 |
| B factor side chain [2.0],¶ Å2 | 4.3 |
| Cross-validated coord. error, Å | 0.39 |
All data (outer shell).
Rmerge = Σ(|I − 〈I〉|)/Σ(I).
Rworking/Rfree = Σ∥Fo| − |Fc∥/Σ|Fo|.
Excluding Gly and Pro.
Target value.
Structure Determination.
The structure of the IAb–p3K complex was determined by molecular replacement method using the program amore (30). The coordinates of IAd–pHA (PDB accession code 2IAD; ref. 14) were used as the search model to find the initial molecular positions. The peptide and the water molecules were deleted in the model, and mismatched residues were mutated to alanines. Four clear molecular replacement solutions were found with high correlation coefficients (51.3%) and low R value (45.4%). The packing was analyzed by using the program o (32), and no serious clashes were found. The program suite cns (33) was then used for further refining. After initial rigid body refinement, subsequent rounds of refinement included energy minimization, B-factor refinement, and simulated annealing. After each round, the structure was inspected by using o (32) and adjusted manually to improve the fit to the electron density maps, eventually adding p3K and the missing IAb side chains to the structure. During refinement strong noncrystallographic symmetry (NCS) was applied to restrain the coordinates of four molecules. The NCS was released gradually and, on the final round of refinement, released completely. Eventually, 239 water molecules and 12 carbohydrate molecules were added into the refined structure. Except for a largely disordered loop involving amino acids 104–112 of the β chain of IAb and the linker between the peptide and the N terminus of the β chain, other residues were readily found in the 2Fo − Fc electron density maps. A Ramachandran plot was calculated with the program procheck (34). Structure determination statistics are summarized in Table 1.
Cell Lines, Peptides, and Mice.
The T cell hybridomas, B3K05–06 and B3K05–08, were produced from C57BL/6 mice, which were immunized as described (25). p3K and its variants were synthesized in the Molecular Resource Center of Nation Jewish Medical and Research Center. The IAb-expressing B cell hybridoma, LB-15.13 (35), was used as an antigen-presenting cell for these peptides. Antigen-induced IL-2 release from the T cell hybridomas was assayed as described (36, 37). Invariant chain knockout mice on the C57BL/10 background were bred in our facility at National Jewish Medical and Research Center from breeding stock obtained from Elizabeth Bikoff of Harvard University (38).
Results and Discussion
Structure of p3K Bound to IAb.
A soluble version of the IAb molecule with a covalently attached peptide was made as described in Materials and Methods. The peptide was a version of the MHCII IEα-derived peptide (pEα) that naturally occupies a large percentage of IAb in strains of mice that express both IAb and an IE molecule (24). In this version of the peptide (p3K), three positions in pEα were changed to Lys residues. We have shown previously that this peptide binds to IAb as stably as pEα does (39) and is very immunogenic in IAb-expressing mice (25, 39).
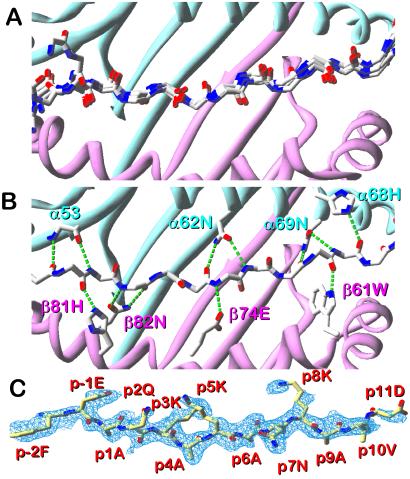
IAb–p3K molecule was crystallized and its structure was solved by molecular replacement to a resolution of 2.5 Å by using the mouse IAd molecule (14) as a model. As expected, the overall structure of the IAb molecule was very similar to that of other MHCII IA molecules, especially IAd (14, 15, 18, 19) (data not shown). We compared the backbone of the p3K peptide to those of peptides bound to other IA alleles after superimposing the molecules on the basis of the α1 and β1 domain backbones (Fig. 1A). The paths of the peptides through the binding groove were nearly identical. Consistent with this observation, we found that the predicted H-bonding network between the peptide backbone and conserved MHCII amino acids seen in other MHCII/peptide structures is present in the IAb-p3K structure as well (Fig. 1B).
Figure 1.
Structure of the p3K peptide bound to IAb. (A) A ribbon structure of the IAb α1/β1 domains is shown with the α1 domain in cyan and the β1 domain in magenta. A wire frame representation of the p3K peptide backbone is shown with coloring: carbon, white; oxygen, red; nitrogen, blue. Overlaid is a wire frame representation of six other peptides as they are bound to other IA molecules with the same color scheme. (B) IAb α1/β1 domains and p3K backbone as in A. Also shown are the backbone or side chains of eight conserved MHCII amino acids that make predicted H-bonds (green dotted lines) to the peptide backbone in IAb–p3K and in nearly all other published MHCII/peptide structures. (C) Electron density (Fo − Fc, peptide omit, 2.5σ) associated with p3K as it is bound to IAb. View is from the side looking through the β-chain α-helix. Coloring: carbon, yellow; oxygen, red; nitrogen, blue. All three figures were made with swiss pdb viewer (53).
These conserved backbone interactions determine the path of the peptide through the binding groove allowing the N and C termini to emerge from the ends. Theoretically, on the basis of these interactions alone, a peptide could bind in any register provided there were no steric clashes among side chains. In the MHCII structures reported so far, the register of the peptide in the binding groove appears to be determined by preferential interactions between peptide side chains and pockets in the interior of the MHCII binding groove. Aligning the peptides based on the amino acid closest to the peptide N terminus that fills a pocket (p1), well-defined pockets generally occur at positions p1, p4, p6, and p9. Surprisingly, we found that for p3K bound to IAb, the peptide contained an Ala at each of these positions (Fig. 1C), so that only the Ala methyl side chains were available to fill these pockets.
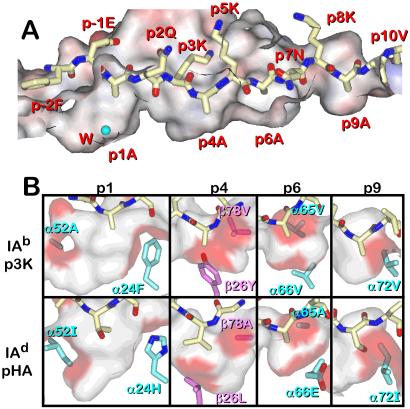
One explanation for alanines at these positions could have been that IAb lacked pockets at p1, p4, p6, and p9 capable of accepting amino acids with larger side chains. However, examination of the water-accessible surface of the IAb peptide-binding groove showed that for the most part this was not the case (Fig. 2A). The pocket at the p1 position is in fact very large and mostly hydrophobic, capable of accepting virtually any aliphatic or aromatic side chain, such as those found in the p1 position of many other MHCII/peptide complexes (10–13, 17, 19). In our structure, this mostly empty pocket contained a water molecule. The other three pockets were neutral to hydrophobic and somewhat shallow. Nevertheless, each was certainly capable of accepting side chains larger than that of alanine, such as those of Ser, Pro, or Val.
Figure 2.
The p1, p4, p6, and p9 pockets of IAb. (A) The solvent-accessible surface (electrostatic potential coloring) of a portion of the peptide binding groove of IAb is shown with a wire frame representation of p3K (carbon, yellow; oxygen, red; nitrogen, blue). In this side view the β-chain α-helix has been cut away. A water molecule in the p1 pocket is shown (cyan). Figure was created with weblab viewer (Accelrys, San Diego). (B) The p1, p4, p6, and p9 pockets of IAb–p3K and IAd/pHA are compared. A side view of a semitransparent solvent-accessible surface is shown for each pocket. For p1, p4, and p9, the view is from the β-chain side with the β-chain α-helix cut away. For p6, the view is from the α-chain side with the α-chain α-helix cut away. The surfaces are colored white except for that portion contributed by the side chains of amino acids that differ between IAb and IAd, which is colored red. The side chains of these allelic amino acids are also shown (α-chain carbon, cyan; β-chain carbon, magenta; oxygen, red; nitrogen, blue). Figure was produced with weblab viewer pro.
The IAb and IAd molecules are most related among the independent IA alleles. Two recent structures of IAd complexed with two different peptides also found no large amino acid side chains in the binding pockets, although some were larger than Ala (14). Therefore, we examined how similar the pockets of IAb or IAd were by comparing the IAb-p3K structure to that of IAd bound to a peptide from influenza hemagglutinin (pHA). Of the 80 amino acids in the α1 domain, 12 differ between IAb and IAd. Of the 94 amino acids in the β1 domain, 8 differ between IAb and IAd. Of these 20 differences, 7 significantly affect the size and/or shape of the peptide-binding pockets (Fig. 2B). The p1 pocket of IAd is not as large as that of IAb due primarily to the change of α52 Ala to Ile. On the other hand the p4 pocket of IAd is considerably larger than that of IAb because of changes at β26 (Val to Ala) and β78 (Tyr to Leu). The size of the p6 pocket is similar in both molecules but the shapes are different because of changes at α65 (Val to Ala) and α66 (Val to Glu). The p9 pockets of both molecules are very similar except for the slight difference caused by the change of Val to Ile at α72.
Overall, these differences predict that, were the pockets to be filled optimally by peptide side chains, these alleles of IA would bind very different peptides. However, with peptides containing relatively small side chains at p1, p4, p6, and p9, one could predict that either of these alleles could bind the same peptides. For example, p3K is derived from pEα, which also has four Ala residues in the same positions. pEα binds well to both IAb and IAd (40, 41). A peptide of ovalbumin, VHAAHAEINEAG, is presented by either IAb or IAd to the T cell hybridoma DO-11.10 (42, 43). Placing Ala, Ala, Ile, and Ala at the p1, p4, p6, and p9 positions, respectively, would be consistent with its binding to IAb or IAd. The structure of IAd was solved bound to a longer form of this peptide in a different register (14). However, the authors pointed out that this alternate register was also consistent with the IAd binding groove.
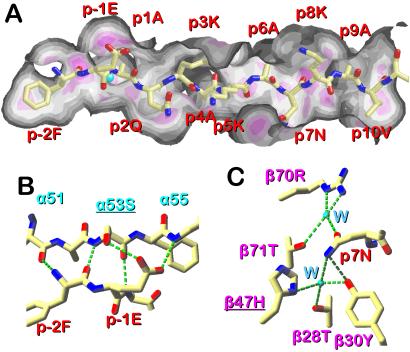
Our previous studies have shown that p3K binds very strongly to IAb and, when bound to soluble IAb, forms a very stable molecule (25, 39). How is this stability maintained in the face of virtually no occupancy of the conventional side chain binding pockets? One clue comes from the footprint of the p3K on the IAb molecule surface (Fig. 3A). In this figure, the MHC contact surface is color coded such that bright magenta shows areas of closest peptide contact fading to dark gray for areas of poorest contact. Despite the absence of side chains filling the binding pockets, there are extensive interactions between the p3K and IAb along the length of the binding groove. For example, strong van der Waals interactions can be predicted between IAb and side chains of p-2Phe, p2Gln, and p10Val. There are also numerous predicted H-bond interactions in addition to the conserved ones shown in Fig. 1B. At the N-terminal end of the peptide there are three additional H-bonds (Fig. 3B), an additional H-bond between the N of p-2Phe and the O of α52, creating a short parallel β-strand pair whose interaction is extended further by two H-bonds between the carboxylate of p-1Glu with the OH of α53Ser and the backbone N of α55. These interactions are not seen in the IAd structures.
Figure 3.
Extensive interaction of p3K with IAb. (A) The footprint of p3K on the solvent-accessible surface of the IAb molecule. The surface has been colored to reflect the distance of the peptide from the IAb surface: magenta, closest; white, intermediate; gray, farthest. Figure was created with protein explorer (54). (B) Details of the predicted H-bond interactions (green dotted lines) between p-2Phe and p-1Glu of p3K and the IAb α chain are shown. (C) Details of the predicted H-bond interactions (green dotted lines) between p7Asn and the IAb β chain are shown. Two water molecules (W) involved in the interactions are shown (cyan). View is from the C-terminal end of the peptide toward its N terminus. In B and C (produced with swiss pdb viewer) underlined amino acids differ between IAb and IAd.
Particularly interesting is a predicted large H-bond network between p7Asn and the IAb β chain involving two ordered water molecules (Fig. 3C). This network cross-links amino acids in the β-chain α-helix (β70Arg and β71Thr) to those on the β-strand floor (β28Thr, β30Tyr, and β47His). Because β47 of IAd is a Tyr, this precise network is not possible in IAd, but some other type of interaction may be possible, but was not seen in the published structures. In nearly all MHCII structures the p7 amino acid points laterally toward the β-chain α-helix. In some cases, its contribution to MHC interaction is minimal because of a small side chain (12–14, 23) or a rotamer that brings the side chain to the surface (12, 16, 20, 21). In a few cases, the p7 rotamer buries a portion of the side chain with considerable β-chain interaction (11, 15, 17, 18). The p7Asn of p3K is a particularly dramatic example of the latter.
Thus, overall the structure of IAb suggests that this molecule can uniquely compensate for the absence of side chains in the traditional p1, p4, p6, and p9 peptide-binding pockets by forming new interactions with the peptide, particularly at the N-terminal portion of the peptide and p7 position.
The IAb Peptide-Binding Motif.
Over the years, the sequences of naturally MHC-bound self peptides and immunogenic foreign peptides have been combined with crystal structures to define the peptide-binding motifs of various MHC molecules. For example, peptide studies predicted the properties of the p1, p6, and p9 pockets of IEk (44–46). The structure of IEk with bound peptides confirmed these conclusions and determined the properties of the p4 pocket (12, 23).
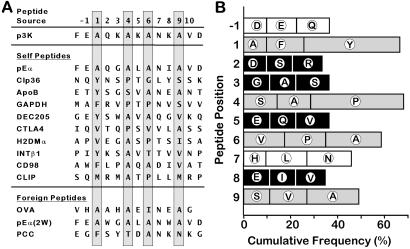
There are many studies of peptides bound to IAb (39, 40, 47, 48). However, these studies alone have not yet yielded a consensus on the peptide binding motif for IAb. We decided to revisit this question, combining this peptide sequence data with the properties of the IAb–p3K structure to predict the motif. We aligned the peptides based mainly on the size and properties of the p1, p4, p6, and p9 pockets, but also with the knowledge that amino acids at the p-1 and p7 position may contribute to peptide/MHC interactions. The previously unappreciated large p1 pocket, which appears able to accept any aromatic side chain, was particularly helpful in aligning the peptides. In fact, placing the favored small aliphatic or polar amino acid (e.g., Ala, Pro, Ser, Val) in the p4, p6, and p9 pockets often required an aromatic amino acid at p1. In a number of cases, this alignment led to the placement of an Asn at p7 and a Glu or Gln at p-1, allowing the additional interactions seen in the IAb–p3K structure. In some other cases, when deciding among more than one possible alignment satisfying the initial conditions, the one that showed either of these secondary interactions was selected. Fig. 4A shows the core sequences of some self and foreign peptides aligned by these principles. Fig. 4B summarizes the results for the alignment of a set of 64 self peptides from a study by Dongre et al. identifying peptides bound to IAb isolated from normal B cells and tumor cells lines of B cell and macrophage origin. In this figure the frequency of occurrence of the five most frequent amino acids at each position in the aligned peptides are shown from p1 to p9. The complete alignment of this peptide set is included in Table 2, which is published as Supporting Information on the PNAS web site, www.pnas.org.
Figure 4.
The IAb peptide binding motif. Sixty-four peptides found bound to IAb (48) were aligned by attempting to place an aliphatic/aromatic amino acid at p1 and amino acids with small aliphatic or neutral side chains at p4, p6, and p9. If more than one alignment was possible, secondary consideration was given to the placement of a Glu or Gln at p-1 and/or an Asn, Asp, Glu, or Gln at p6. (A) Alignment of 13 representative peptides showing the portion of the peptide from p-2 to p11 aligned to p3K. Self peptides (48): pEα = amino acids 54–66 of IEα chain; CLP36 = amino acids 140–152 of CLP-36; ApoB = amino acids 769–781 of apolipoprotein B; GAPDH = amino acids 228–242 of glyceraldehyde-3-phosphate dehydrogenase; DEC205 = amino acids 567–579 of DEC205 receptor; CTLA4 = amino acids 38–50 of CTLA-4; H2DMα = amino acids 140–152 of H-2DM α chain; INTβ1 = amino acids 781–793 of integrin β-1; CD98 = amino acids 210–222 of CD98 heavy chain; CLIP = amino acids 88–100 of invariant chain. Foreign peptides: OVA = aa 327–338 of chicken ovalbumin (42); pEα(2W) = pEα(Q57W,I63W) (25, 39); PCC = aa 43–55 of pigeon cytochrome c (47, 50). (B) Summary of the alignment of all self peptides. The frequency (%) of the five most frequent amino acids at each position from p-1 to p9 is shown. Bar is the frequency of the most frequent amino acid. Open bars, p1, p4, p6, and p9 anchors; gray bars, other MHC-interacting amino acids; solid bars, potential T cell-interacting amino acids.
Our alignment for the 64 self peptides predicts an aromatic amino acid as highly favored at p1. Tyr and Phe are predicted at p1 in 36 of the peptides (56%) versus only a 7% frequency of these amino acids in the mouse proteome (49). A small side chain is also common at p1, with Ala, Val, Ser, and Gly accounting for 16 of the remaining 28 peptides. Pro is most frequent at p4 (39% versus 6% in the proteome). Other amino acids lacking large side chains, Ala, Ser, Val, and Gly, are also frequent at p4 (30 of 39 of the remaining amino acids). Amino acids lacking large side chains are favored at p6 as well. Ala, Pro, and Val account for 58% of the p6 amino acids versus 19% in the proteome. Aromatic and charged amino acids are completely excluded from p4 and p6. A small side chain is also favored at p9, with Ala and Val accounting for 38% of the amino acids versus only 13% in the mouse proteome. However, the exclusion of other amino acids is not so striking as with p4 and p6.
Gln and Glu are slightly favored at p1, accounting for 27% of the amino acids versus 13% in the proteome; however, many other amino acids occur at this position, perhaps indicating that the backbone interactions at the N terminus of the peptide may be more important than the p1 side chain interactions. Asn is the most favored amino acid at p7, with a frequency of 19% versus only 3.5% in the proteome. Although Leu, as the second most frequent amino acid at p7, could not take part in the H-bonding network shown in Fig. 3C, immediately adjacent to this part of the β-chain wall of the binding groove is an extensive hydrophobic patch including the side chains of β67Ile and β61Trp that could easily be reached by the appropriate rotamer of Leu. Many different amino acids occur at p2, p3, p5, or p8, and the five most frequent do not share a particular size or chemistry, consistent with these being the predicted central αβ T cell antigen receptor contact amino acids.
As pointed out above, the four Ala residues of p3K are present in pEα from which it was derived. Also present in pEα is p-1Glu and p7Asn, so that it is reasonable to predict that pEα would bind to IAb in the same register as p3K. Because in strains of mice that express both IAb and the IEα chain pEα occupies about 10% of IAb (24), this complex also appears very stable.
We also analyzed several foreign peptides that have been well studied as immunogens in H-2b mice (Fig. 4A). For example, the alignment of the DO-11.10 reactive ovalbumin peptide described above that places amino acids with small side chains in the p1, p4, p6, and p9 pockets also puts a favorable Asn at p7. Another variant of pEα that is immunogenic in IAb-expressing mice and binds very well to IAb has Trp substituted at positions p2 and p8 (25, 39), but otherwise has the same potential interactions with IAb as p3K and pEα.
Finally, another very well analyzed peptide antigen presented by IAb consists of amino acids 43–58 of pigeon cytochrome c (50). Extensive mutational analysis of this peptide (47) is consistent with the alignment shown in Fig. 4A. For example, the stimulatory potency of the peptide was decreased by mutation of the p1Phe to Ala and increased by mutation of the p9Asn to Ala without changing T cell specificity, consistent with effects on IAb binding such that the best binding was seen by optimal side chains at either position. This alignment also places a favorable Thr at p4, an Ala at p6, and again an Asn at p7. The predicted surface-exposed p5Asp was shown to be important for T cell recognition. Interestingly the p7Asn was also suggested to be important for T cell recognition, because mutations at this position created new T cell epitopes. The IAb–p3K structure predicts that this Asn should not be readily accessible on the surface (Fig. 3), but because it interacts extensively with the highly exposed β71Arg, mutations in this Asn could very well lead to substantial surface changes resulting in apparently new epitopes.
Interaction of T Cells with IAb/p3K.
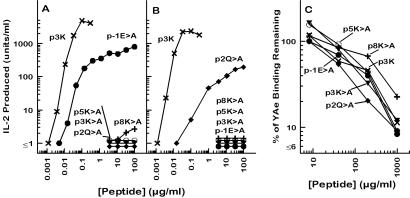
The structure of the 3K peptide bound to IAb reveals five upwardly pointing amino acids as potential candidates for recognition by the αβ T cell antigen receptors (TCR): the p-1Glu, p2Gln, p3Lys, p5Lys, and p8Lys. The side chains of these amino acids are at least partially exposed on the molecule surface and fall within the footprints seen of the αβTCRs on other MHCII/peptide complexes (51, 52). To test whether these are indeed important for recognition, we synthesized a set of mutant peptides in which each of these amino acids was mutated to Ala. The peptides were compared with p3K for recognition by two T cell hybridomas (25) specific for IAb + p3K (Fig. 5 A and B). The peptides were also tested for their ability to compete with pEα for binding to IAb on the surface of invariant chain knockout, IAb-bearing B cells (Fig. 5C). Mutation of any of the three central Lys residues at p3, p5, or p8 to Ala completely eliminated recognition by either hybridoma. Mutation of the Gln at p2 eliminated the response of one hybridoma and partially affected the other hybridoma. Reciprocally, mutation of the Glu at p-2 only partially reduced the response of the first hybridoma, but eliminated the response of the second. All of the peptides tested were able to bind to IAb based on their ability to compete with pEα for IAb binding. Therefore, all five of the predicted surface-exposed amino acids play a role in T cell recognition.
Figure 5.
p3K amino acids important for T cell recognition. The responses of two T cell hybridomas, B3K05-6 (A) and B3K05-8 (B), produced from T cells from p3K-immunized IAb mice (25), were tested for their responses to a set of p3K mutant peptides presented by the IAb-bearing cell line, LB-15.13 (35). The results are shown as the units of IL-2 produced vs. the concentration of peptide offered. (C) Spleen cells from IAb-bearing invariant chain knockout mice were incubated with pEα (20 μg/ml) for 4 h at 37°C. The level of binding of pEα to IAb was assessed by flow cytometry using the IAb/pEα-specific monoclonal antibody, YAe (24). Competition between pEα and p3K or its variants was assessed by the reduction in YAe staining when various concentrations of the competitor peptide were included in the incubation of the spleen cells with pEα. The results are shown as the percent of the mean fluorescence (corrected for autofluorescence) remaining measured with fluoresceinated YAe vs. the concentration of the competitor peptide. In all three panels the symbols are as follows: Unmutated p3K (×); p-1E>A (which indicates an E → A mutant) (●), p2Q>A (♦), p3K>A (▾), p5K>A (○), and p8K>A (+).
Concluding Remarks.
Models that attribute the allele/isotype specificity and differences in stability of peptide/MHCII complexes to the relative occupancy of pockets at p1, p4, p6, and p9 may be too simple. In IAb other specific interactions, especially at the N terminus and p7 position of the peptide, appear capable of compensating for poor interactions in these pockets to generate a very stable complex. Our findings point out the importance of combining structural and peptide sequence information in identifying peptide-binding motifs for MHCII molecules.
Supplementary Material
Acknowledgments
We are extremely grateful to Dr. Gongyi Zhang for advice during the course of these studies and help in the data collection. We also thank Sergey Korolev for his help with the synchrotron data collection and processing. The Structural Biology Center beam lines at the Advanced Photon Source of the Argonne National Laboratory are supported by the U.S. Department of Energy, Office of Biological and Environmental Research under contract W-31-109-ENG-38. We also thank Dr. Eleanor J. Dodson and the CCP4 newsgroup for advice on detwinning our data. This work was supported in part by U.S. Public Health Service Grants AI-17134, AI-18785, and AI-22295.
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1LNU).
References
- 1.Garrett T P, Saper M A, Bjorkman P J, Strominger J L, Wiley D C. Nature (London) 1989;342:692–696. doi: 10.1038/342692a0. [DOI] [PubMed] [Google Scholar]
- 2.Madden D R, Gorga J C, Strominger J L, Wiley D C. Nature (London) 1991;353:321–325. doi: 10.1038/353321a0. [DOI] [PubMed] [Google Scholar]
- 3.Matsumura M, Fremont D H, Peterson P A, Wilson I A. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 4.Fremont D H, Matsumura M, Stura E A, Peterson P A, Wilson I A. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 5.Guo H C, Jardetzky T S, Garrett T P, Lane W S, Strominger J L, Wiley D C. Nature (London) 1992;360:364–366. doi: 10.1038/360364a0. [DOI] [PubMed] [Google Scholar]
- 6.Madden D R, Gorga J C, Strominger J L, Wiley D C. Cell. 1992;70:1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- 7.Madden D R, Garboczi D N, Wiley D C. Cell. 1993;75:693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- 8.Bouvier M, Wiley D C. Science. 1994;265:398–402. doi: 10.1126/science.8023162. [DOI] [PubMed] [Google Scholar]
- 9.Fremont D H, Stura E A, Matsumura M, Peterson P A, Wilson I A. Proc Natl Acad Sci USA. 1995;92:2479–2483. doi: 10.1073/pnas.92.7.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stern L J, Brown J H, Jardetzky T S, Gorga J C, Urban R G, Strominger J L, Wiley D C. Nature (London) 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh P, Amaya M, Mellins E, Wiley D C. Nature (London) 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 12.Fremont D H, Hendrickson W A, Marrack P, Kappler J. Science. 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 13.Dessen A, Lawrence C M, Cupo S, Zaller D M, Wiley D C. Immunity. 1997;7:473–481. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 14.Scott C A, Peterson P A, Teyton L, Wilson I A. Immunity. 1998;8:319–329. doi: 10.1016/s1074-7613(00)80537-3. [DOI] [PubMed] [Google Scholar]
- 15.Fremont D H, Monnaie D, Nelson C A, Hendrickson W A, Unanue E R. Immunity. 1998;8:305–317. doi: 10.1016/s1074-7613(00)80536-1. [DOI] [PubMed] [Google Scholar]
- 16.Smith K J, Pyrdol J, Gauthier L, Wiley D C, Wucherpfennig K W. J Exp Med. 1998;188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murthy V L, Stern L J. Structure (London) 1997;5:1385–1396. doi: 10.1016/s0969-2126(97)00288-8. [DOI] [PubMed] [Google Scholar]
- 18.Latek R R, Suri A, Petzold S J, Nelson C A, Kanagawa O, Unanue E R, Fremont D H. Immunity. 2000;12:699–710. doi: 10.1016/s1074-7613(00)80220-4. [DOI] [PubMed] [Google Scholar]
- 19.Corper A L, Stratmann T, Apostolopoulos V, Scott C A, Garcia K C, Kang A S, Wilson I A, Teyton L. Science. 2000;288:505–511. doi: 10.1126/science.288.5465.505. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Li H, Martin R, Mariuzza R A. J Mol Biol. 2000;304:177–188. doi: 10.1006/jmbi.2000.4198. [DOI] [PubMed] [Google Scholar]
- 21.Lee K H, Wucherpfennig K W, Wiley D C. Nat Immunol. 2001;2:501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 22.Kersh G J, Miley M J, Nelson C A, Grakoui A, Horvath S, Donermeyer D L, Kappler J, Allen P M, Fremont D H. J Immunol. 2001;166:3345–3354. doi: 10.4049/jimmunol.166.5.3345. [DOI] [PubMed] [Google Scholar]
- 23.Fremont D H, Dai S, Chiang H, Crawford F, Marrack P, Kappler J. J Exp Med. 2002;195:1043–1052. doi: 10.1084/jem.20011971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy D B, Rath S, Pizzo E, Rudensky A Y, George A, Larson J K, Janeway C A., Jr J Immunol. 1992;148:3483–3491. [PubMed] [Google Scholar]
- 25.Rees W, Bender J, Teague T K, Kedl R M, Crawford F, Marrack P, Kappler J. Proc Natl Acad Sci USA. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozono H, Parker D, White J, Marrack P, Kappler J. Immunity. 1995;3:187–196. doi: 10.1016/1074-7613(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 27.Kozono H, White J, Clements J, Marrack P, Kappler J. Nature (London) 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharya A, Dorf M E, Springer T A. J Immunol. 1981;127:2488–2495. [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 31.Matthews B W. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 32.Kleywegt G J, Zou J Y, Kjeldgaard M, Jones T A. International Tables for Crystallography. Crystallography of Biological Macromolecules. F. Dordrecht, The Netherlands: Kluwer Academic; 2001. pp. 353–367. [Google Scholar]
- 33.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R, MacArthur M, Moss D, Thorton J. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Kappler J, White J, Wegmann D, Mustain E, Marrack P. Proc Natl Acad Sci USA. 1982;79:3604–3607. doi: 10.1073/pnas.79.11.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kappler J W, Skidmore B, White J, Marrack P. J Exp Med. 1981;153:1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 38.Bikoff E K, Huang L Y, Episkopou V, van Meerwijk J, Germain R N, Robertson E J. J Exp Med. 1993;177:1699–1712. doi: 10.1084/jem.177.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ignatowicz L, Rees W, Pacholczyk R, Ignatowicz H, Kushnir E, Kappler J, Marrack P. Immunity. 1997;7:179–186. doi: 10.1016/s1074-7613(00)80521-x. [DOI] [PubMed] [Google Scholar]
- 40.Rudensky A, Preston-Hurlburt P, al-Ramadi B K, Rothbard J, Janeway C A., Jr Nature (London) 1992;359:429–431. doi: 10.1038/359429a0. [DOI] [PubMed] [Google Scholar]
- 41.Hunt D F, Michel H, Dickinson T A, Shabanowitz J, Cox A L, Sakaguchi K, Appella E, Grey H M, Sette A. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 42.Shimonkevitz R, Colon S, Kappler J W, Marrack P, Grey H M. J Immunol. 1984;133:2067–2074. [PubMed] [Google Scholar]
- 43.Marrack P, Shimonkevitz R, Hannum C, Haskins K, Kappler J. J Exp Med. 1983;158:1635–1646. doi: 10.1084/jem.158.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evavold B D, Allen P M. Adv Exp Med Biol. 1992;323:17–21. doi: 10.1007/978-1-4615-3396-2_3. [DOI] [PubMed] [Google Scholar]
- 45.Marrack P, Ignatowicz L, Kappler J W, Boymel J, Freed J H. J Exp Med. 1993;178:2173–2183. doi: 10.1084/jem.178.6.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reay P A, Kantor R M, Davis M M. J Immunol. 1994;152:3946–3957. [PubMed] [Google Scholar]
- 47.Ogasawara K, Maloy W L, Beverly B, Schwartz R H. J Immunol. 1989;142:1448–1456. [PubMed] [Google Scholar]
- 48.Dongre A R, Kovats S, deRoos P, McCormack A L, Nakagawa T, Paharkova-Vatchkova V, Eng J, Caldwell H, Yates J R, 3rd, Rudensky A Y. Eur J Immunol. 2001;31:1485–1494. doi: 10.1002/1521-4141(200105)31:5<1485::AID-IMMU1485>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 49.Stoesser G, Baker W, van den Broek A, Camon E, Garcia-Pastor M, Kanz C, Kulikova T, Leinonen R, Lin Q, Lombard V, et al. Nucleic Acids Res. 2002;30:21–26. doi: 10.1093/nar/30.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogasawara K, Maloy W L, Schwartz R H. Nature (London) 1987;325:450–452. doi: 10.1038/325450a0. [DOI] [PubMed] [Google Scholar]
- 51.Reinherz E L, Tan K, Tang L, Kern P, Liu J, Xiong Y, Hussey R E, Smolyar A, Hare B, Zhang R, et al. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 52.Hennecke J, Carfi A, Wiley D C. EMBO J. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 54.Martz E. protein explorer Software, http://proteinexplorer.org. 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.