Abstract
Natural killer (NK) cells express inhibitory and activation receptors that recognize MHC class I-like molecules on target cells. These receptors may be involved in the critical role of NK cells in controlling initial phases of certain viral infections. Indeed, the Ly49H NK cell activation receptor confers in vivo genetic resistance to murine cytomegalovirus (MCMV) infections, but its ligand was previously unknown. Herein, we use heterologous reporter cells to demonstrate that Ly49H recognizes MCMV-infected cells and a ligand encoded by MCMV itself. Exploiting a bioinformatics approach to the MCMV genome, we find at least 11 ORFs for molecules with previously unrecognized features of predicted MHC-like folds and limited MHC sequence homology. We identify one of these, m157, as the ligand for Ly49H. m157 triggers Ly49H-mediated cytotoxicity, and cytokine and chemokine production by freshly isolated NK cells. We hypothesize that the other ORFs with predicted MHC-like folds may be involved in immune evasion or interactions with other NK cell receptors.
Viruses have evolved several means to down-regulate MHC class I expression to evade specific immunity (1). However, decreased MHC class I expression may enhance susceptibility to natural killer (NK) cells (2) that are normally prevented from killing by MHC class I-specific inhibitory receptors (3, 4). Herpesviruses counter with molecules that engage inhibitory receptors (5–7). Because inhibitory NK cell receptors block NK cell function by preventing signaling through activation receptors, this viral strategy suggested that NK cell activation receptors are critical to control virus infection.
One such NK cell activation receptor was recently revealed after a combination of genetic and immunological approaches. C57BL/6 mice are resistant to murine cytomegalovirus (MCMV), as manifested by splenic control of viral replication and survival, whereas susceptible strains (BALB/c and DBA/2) display high splenic viral titers and lethality. This phenotypic difference is controlled by an autosomal dominant locus termed Cmv1 (8) that was mapped to the NK gene complex (NKC) on distal mouse chromosome 6 (9) that contains gene clusters for NK cell receptors. Consistent with NK cell-mediated resistance, mAb elimination of NK cells converts resistance to susceptibility (10). Detailed genetic and physical mapping yielded an informative recombinant inbred mouse strain, BXD-8, that retains the resistant NKC haplotype but was susceptible. BXD-8 has a selective deletion in the gene for Ly49H, a putative NK cell activation receptor (11, 12). Furthermore, when Ly49H was perturbed by specific mAb, MCMV replication was uncontrolled, leading to lethality in otherwise resistant mice (11–13). These genetic and immunologic data provided strong evidence that Ly49H is required for NK cell-mediated resistance to MCMV infection.
Ly49H is a member of the Ly49 family of NK cell receptors and is expressed on about 50% of NK cells in C57BL/6 mice (14–16). Unlike the original Ly49 family member, the MHC class I-specific inhibitory receptor Ly49A, Ly49H lacks cytoplasmic immunoreceptor tyrosine-based inhibitory motifs (ITIMs) and instead contains a charged transmembrane residue that permits association with the immunoreceptor tyrosine-based activation motif (ITAM)-containing DAP12 [also known as killer cell-activating receptor-associated protein (KARAP)] molecule. Previous in vitro studies have demonstrated that cross-linking of Ly49H results in tyrosine phosphorylation of DAP12/KARAP and downstream activation events, including cytokine production and cytotoxicity (14, 15). However, in vivo studies revealed that there was no preferential activation of Ly49H+ NK cells soon after MCMV infection (17). Instead, there is first an early (day 1–2) “nonspecific” phase characterized by IFN-γ production and proliferation of NK cells, without regard to Ly49H expression, that could be due to global activation by systemic cytokines (18). It is not until somewhat later (day 4) that selective activation of Ly49H+ NK cells could be recognized, i.e., a “specific” phase in which there is preferential proliferation of Ly49H+ NK cells, and this proliferation is blocked by the anti-Ly49H mAb. These findings appeared to be discordant with the observation that NK cells from resistant mice are able to control MCMV replication in the early phase of infection, suggesting that the “nonspecific” early phase may be masking detection of specific activation of Ly49H+ NK cells by MCMV.
The previous in vivo analysis also suggested that Ly49H specifically recognizes MCMV-infected cells. Because the known ligands for Ly49 receptors are MHC class I molecules and other NKC-encoded molecules (NKG2D, CD94-NKG2) can recognize MHC class I-like structures (19), the Ly49H ligand was speculated to be an MHC-related molecule of host (11) or virus (11, 12) origin, although other host or viral molecules remained possible (11). The ligand may be constitutively expressed on normal cells, but only on virus-mediated down-regulation of MHC class I expression would NK cells be released from inhibition and kill the target, as predicted by the “missing-self” hypothesis (2). Alternatively, perhaps ligand expression could be induced, as for NKG2D ligands (19). Therefore, another issue in understanding innate NK cell control of viral infection was the nature of the ligand for Ly49H.
Herein, we used heterologous reporter cells to demonstrate that Ly49H is triggered by MCMV-infected cells. We examined the basic parameters of the putative ligand, including specificity and, by excluding most of the other possibilities, reached the interim conclusion that the ligand is encoded by the virus itself. By exploiting a bioinformatics approach, we determined that MCMV has at least 12 ORFs encoding molecules with putative MHC class I-like folds. A systematic examination of these and other candidate molecules led to the identification of m157 as the ligand for Ly49H. m157-transfected target cells are selectively killed by Ly49H+ NK cells and can quickly stimulate the activation of freshly isolated NK cells, suggesting that specific activation of Ly49H+ NK cells early in infection is masked in vivo.
Methods
Cells and Generation of BWZ Transfectants.
All cells and media are described in detail in supporting text (which is published as supporting information on the PNAS web site, www.pnas.org). The BWZ.36 cell line, generously provided by N. Shastri (University of California, Berkeley, CA) (20) was cultured in RPMI 1640 containing 10% FCS and 200 units/ml hygromycin (Calbiochem). The pMX, pMX-puro (puromycin-resistant) or pMX-internal ribosomal entry site (IRES) plasmid vectors (21), and the PLAT-E packaging cell line were generously provided by T. Kitamura, (University of Tokyo).
Reporter Assay with MCMV-Infected Cells.
Cells were infected with MCMV at a multiplicity of infection of 5 and agitated every 10 min for 60 min. γHV68 and HSV infections were performed similarly except in media containing 2% FCS and with 2 h adsorption time. When indicated, antibodies were added at a final concentration of 20 to 50 μg/ml, and mAb 2.4G2 (20 μg/ml) was added to prevent inadvertent Fc receptor mediated effects. For UV radiation-mediated inactivation, an aliquot of MCMV was exposed to a 30-W germicidal UV lamp at a distance of ≈7 cm for 30 min with agitation. The titer of the UV-inactivated virus was ≤102 plaque-forming units/ml (limit of detection) in a plaque assay with 3T12 fibroblasts. After coculture with MCMV-infected or m157-transfected cells, β-galactosidase (β-gal) activity was quantitatively assayed in BWZ transfectants with the substrate chlorophenol red β-d-galactoside (CPRG; Calbiochem) or qualitatively by fixation and staining with 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) as described (20). Photomicrographs were acquired by using a Nikon TMS-F microscope and a Nikon model 990 digital camera.
Flow Cytometric Analysis and Intracellular Staining.
For routine staining, cells were analyzed with a FACSCalibur instrument (BD Biosciences) as previously described (15). Data from 104 to 105 gated events were collected. For intracellular staining, freshly isolated B6.RAG1−/− splenocytes were cocultured for 6–8 h with uninfected or MCMV-infected IC-21 cells (1:1 ratio) in a 96-well plate (150,000 cells each). In a different experiment, we cocultured B6.RAG1−/− splenocytes with BaF3 transfectants (1:1 ratio). In all experiments, brefeldin A was added for the last 5–7 h. Cells were stained with anti-NK1.1-APC, anti-CD3-PerCP-Cy5.5 (PharMingen), and anti-Ly49H-biotin (clone 3D10), along with unlabeled mAb 2.4G2 to block Fc receptor binding, followed by streptavidin-phycoerythrin. Cells were fixed and permeabilized with the Cytofix/Cytoperm kit (PharMingen) according to the manufacturer's instructions. Intracellular IFN-γ was detected with XMG1.2-FITC (PharMingen) and intracellular activation-induced T cell-derived and chemokine-related cytokine (ATAC)/lymphotactin with mAb MTAC-2 (22). Cells were gated on the NK1.1+CD3− population.
Cloning of Ly49H Ligand.
PCR primers were designed to amplify each candidate ORF from 3 nt before and after the predicted ORF and contained restriction enzyme sites for directional cloning into the pMX-puro vector. ThermalAce DNA polymerase (Invitrogen) or Pfu DNA polymerase (Stratagene) were used to PCR amplify a cDNA library produced from MCMV-infected IC-21 cells. Readily generated amplicons were ligated into pMX-puro vector. To neutralize possible PCR errors, five independent clones of each amplicon were isolated, pooled, and used as a group to transfect PLAT-E cells. ORF transfectants were used to stimulate BWZ transfectants or generate retroviruses for infection of target cells. Puromycin-resistant cells expressing each ORF were cocultured with HD12 cells for 12–22 h, which were then fixed and stained with X-GAL. The sequence of multiple m157 clones was verified.
IL-2-Activated NK (LAK) and Cytotoxicity Assays.
Generation of LAK cells, sorted Ly49H+ and Ly49H− NK cells, and standard 4 h 51Cr-release assays were performed as previously described (15, 23). Alexa 488-labeled Fab fragments of mAb 3D10 were used for flow sorting of Ly49H subsets.
Results
Specific Activation of Ly49H by MCMV-Infected Cells.
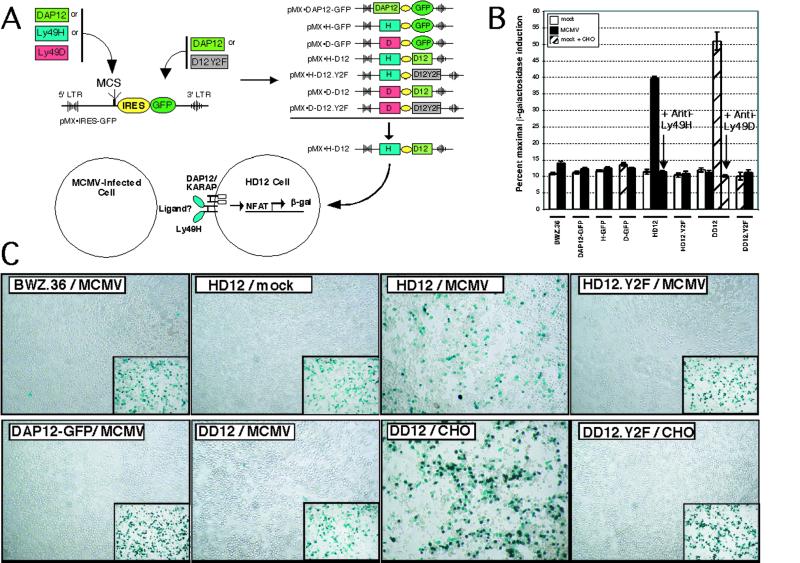
To test the hypothesis that Ly49H specifically recognizes MCMV-infected cells, we expressed Ly49H with DAP12/KARAP (referred to as HD12) or other control constructs in heterologous BWZ.36 cells (20), containing a stably integrated reporter construct for β-gal that is activated on ITAM signaling (Fig. 1A). As with T cell antigen receptor (TCR) stimulation in other BWZ transfectants (20), immobilized anti-Ly49H led to β-gal expression in HD12 cells but not in BWZ transfected with vector, green fluorescent protein (GFP), or DAP12/KARAP alone as indicated by β-gal substrates (data not shown). A related DAP12/KARAP-associated NK cell activation receptor, Ly49D, specifically recognizes Chinese hamster ovary (CHO) cells (24) but does not participate in MCMV immunity (11). Ly49D plus DAP12/KARAP transfectants (DD12) were not stimulated by anti-Ly49H (data not shown). Conversely, DD12 cells showed strong β-gal induction when stimulated with CHO cells (Fig. 1B,C), consistent with Ly49D ligand recognition, and suggesting that HD12 cells could be used as reporters to test ligand recognition by Ly49H.
Figure 1.
Specific activation of Ly49H-expressing reporter cells by MCMV-infected cells. (A) Schematic illustration of retroviral vector constructs and the HD12 reporter assay. cDNA cassettes for murine DAP12, Ly49H, or Ly49D were cloned into the multiple cloning site (MCS) upstream from the internal ribosomal entry site (IRES) of pMX-IRES-GFP to generate the control vectors DAP12-GFP, H-GFP, and D-GFP. The GFP cassette in the downstream position was replaced with either a wild-type or ITAM-deficient mutant murine DAP12 cassette to generate the pMX-HD12, pMX-HD12.Y2F, pMX-DD12, and pMX-DD12.Y2F expression vectors. Individual constructs were transfected into BWZ cells. For example, the pMX-HD12 construct was used to produce HD12 indicator cells in which Ly49H is associated with DAP12 (KARAP) that contains ITAMs (boxes). Activation through Ly49H and DAP12 leads to nuclear factor of activated T cells (NFAT)-induced production of β-gal. (B) Chlorophenol red β-d-galactoside analysis of BWZ.36 reporter lines after coculture with mock- or MCMV-infected primary bone marrow macrophages, as indicated. Ly49D-expressing lines (DD12 and DD12.Y2F) were additionally tested with CHO cells as indicated. For HD12 and DD12 lines, the effect of addition of mAbs 3D10 (anti-Ly49H) or 4E4 (anti-Ly49D) are indicated (arrows). Data are expressed as a percentage of maximal β−gal induction by phorbol 12-myristate 13-acetate (PMA) + ionomycin for each reporter-stimulator combination. (C) X-Gal staining of the indicated BWZ reporter lines after coculture with mock- or MCMV-infected IC-21 cells. (Insets) Maximal β-gal induction after stimulation with PMA + ionomycin. The Ly49D-expressing DD12 and DD12.Y2F lines were additionally stimulated with CHO cells, known to express a cognate ligand for Ly49D (24).
Indeed, MCMV but not mock-infected macrophage cells induced β-gal activity in HD12 cells (Fig. 1B,C). Although DD12 cells expressed comparable receptor levels (see supporting information), DD12 cells were not activated by MCMV-infected cells (Fig. 1 B and C). Furthermore, HD12 activation by MCMV-infected stimulators depended on DAP12/KARAP signaling, because the HD12.Y2F line, which expresses Ly49H with a signaling-deficient mutant form of DAP12/KARAP, showed no detectable β-gal activity. Soluble mAbs against Ly49H or Ly49D completely abrogated the activation of HD12 or DD12 reporter cells by MCMV-infected or CHO stimulators, respectively, indicating Ly49H (and Ly49D) specificity (Fig. 1B). Other control mAbs had no effect (data not shown). These data indicated that MCMV-infected cells directly and specifically activate the reporter cells through Ly49H, demonstrating that the ligand for Ly49H is expressed by MCMV-infected cells.
Multiple MCMV ORFs for Putative MHC Class I Molecules.
HD12 cells were not triggered by target cells stimulated with IFN-α or supernatants from MCMV-infected cells (see supporting information). Fixed infected cells stimulated HD12 cells, indicating that the ligand was expressed on the cell surface. However, stimulation occurred even with infected macrophages from H-2 Db/Kb/β2-microglobulin (β2m)-triple-deficient (3KO) mice, against the possibility that the ligand is a host MHC class I molecule. Infection with other herpes viruses known to infect rodent cells (herpes simplex, γHV68) failed to stimulate HD12 cells, implying that the ligand is MCMV specific. HD12 cells were activated by MCMV-infected cells as little as 8 h postinfection (infection plus activation time). These data suggested that the ligand for Ly49H is expressed on the surface of MCMV-infected cells with early phase kinetics and is most likely encoded by the virus itself.
To identify the putative MCMV-encoded ligand for Ly49H, we took a bioinformatics approach and examined the 230-kb MCMV genome for candidate ORFs (25). In general, herpesvirus genomes contain a central 100-kb region with genes closely related to each other whereas sequences at either end are unrelated (25–27). In MCMV, the end regions include the m02 and m145 gene families. However, we eliminated the m02 to m16 region because a mutant MCMV strain (SMsubm02-16) with a deletion spanning this region (28) stimulated HD12 cells (data not shown). We therefore examined the MCMV genome for other ORFs containing probable signal peptides, N-linked glycosylation sites, and hydrophobic transmembrane domains. We were particularly interested in possible MHC class I-related molecules, but only m144 has primary sequence homology to MHC class I detectable by blast (25). Surprisingly, however, we found that MCMV contains at least 11 other ORFs encoding molecules with potential MHC-like folds, as determined by structural homology analysis [3d-pssm (29)], even though they show little sequence relationship to known MHC class I family members (Table 1). Many of these ORFs were in the m145 family and became prime candidates for the putative Ly49H ligand.
Table 1.
MCMV ORFs encoding proteins with predicted MHC-like folds
| MCMV ORF | Top hit* (MHC-fold) | E-value of top hit (% confidence) | Sequence comparison to top hit†
|
No. of hits to MHC-folds | |
|---|---|---|---|---|---|
| Aligned/total | Identical/similar | ||||
| m17 | HLA-A2 | 0.35 (70) | 151/314 | 31/68 | 3 |
| m37 | ZAG | 0.118 (80) | 170/256 | 29/62 | 6 |
| m90 | ZAG | 0.027 (95) | 153/191 | 13/45 | 4 |
| m144 | HLA-B8 | 0.00013 (95) | 229/252 | 59/209 | 6 |
| m145 | mCD1d1 | 0.0287 (95) | 245/411 | 30/96 | 7 |
| m150 | MIC-A | 0.0146 (95) | 246/298 | 45/98 | 11 |
| m151 | T22 | 0.0299 (90) | 235/316 | 39/118 | 6 |
| m152 | MIC-A | 0.03 (95) | 286/317 | 31/118 | 7 |
| m153 | T22 | 0.0534 (90) | 235/318 | 36/67 | 5 |
| m155 | mCD1d1 | 0.056 (90) | 244/313 | 39/87 | 3 |
| m157 | H2-M3 | 0.089 (90) | 225/289 | 50/74 | 7 |
| m159 | ZAG | 0.0094 (95) | 240/343 | 44/91 | 7 |
All predicted ORFs of the MCMV genome were analyzed with the 3d-pssm search algorithm (29), which recognizes remote protein sequence homologies based on three-dimensional position-specific scoring matrices. The predicted mature ectodomains of 12 MCMV ORFs had top alignment hits with profiles of MHC class I or class I-related crystal structures with statistical E-values less than unity. The E-value is the number of database hits expected by chance. Lower E-values represent more significant scores to which 3d-pssm assigns % confidence. The protein database (PDB) ID codes for the top hit classical and non-classical MHC class I molecules are: HLA-A2 (1duz), HLA-B8(1 age), ZAG (1zag; zinc-α2-glycoprotein), mCD1d1 (1cd1; murine CD1d), MIC-A (1hyr), T22 (1c16), H2-M3 (1mhc). Reported in the rightmost column is the total number of hits with E-values less than unity that are MHC-related structures for the given ORF.
Shown is the number of viral ORF residues that were sequence aligned to the top 3d-pssm top hit, as is the total number of residues in the mature ectodomains that were queried. Also detailed is the number of identical residues, as well as the total number of identical and conservatively substituted residues in the alignments. Analysis of these MCMV ORFs by blast produced significant MHC-fold similarities (E-values smaller than 1) only for m144, a previously identified class I-related molecule that associates with β2m (44). All 12 of these MHC class I-like MCMV ORFs, except m157, are predicted type I transmembrane proteins, with a canonical N-terminal leader peptide signal sequence and short cytoplasmic tail. (Analyzed by the signal peptide and transmembrane region prediction servers at the Center for Biological Sequence Analysis, Technical University of Denmark, www.cbs.dtu.dk.) m157, on the other hand, has a predicted glycosylphosphatidylinositol (GPI) membrane linkage site. [Prediction from the DGPI server (available via Proteomics Tools on Expasy Molecular Biology server).]
Identification of m157 as the Ligand for Ly49H.
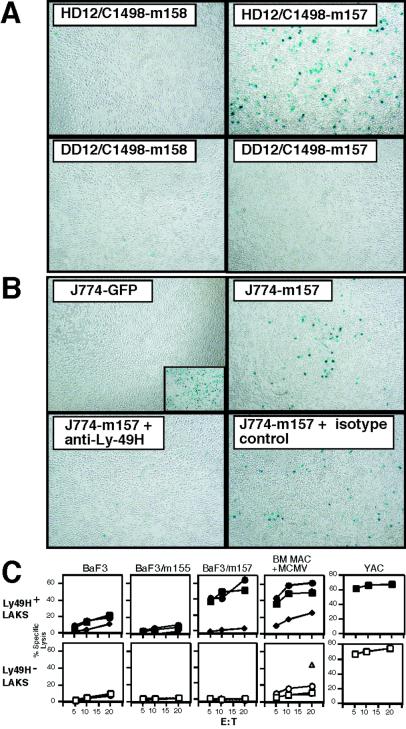
PCR primers for the m145 family and other candidate ORFs were used to amplify cDNAs from an MCMV-infected IC-21 library. Twenty nine readily produced amplicons (m17, m37, m42, m73, m74, m90, m92, m100, m117.1, m124, m136, m138, m144, m145, m146, m150, m151, m152, m153, m154, m155, m157, m158, m159, m160, m163, m164, m165, and m167) were expressed in cell lines with highly efficient retroviral vectors. Only transfectants expressing m157, one of the predicted MHC class I folded proteins, stimulated HD12 cells (Fig. 2A and data not shown), and this stimulation was blocked specifically by anti-Ly49H mAb (Fig. 2B). None of the other transduced ORFs stimulated HD12 cells (data not shown). This list included m144, the MCMV MHC class I-like molecule (6), previously speculated to be the ligand for Ly49H (12). Furthermore, expression of m157 in resistant targets conferred markedly enhanced susceptibility to lysis by bulk NK cells (data not shown), and in particular to Ly49H+ but not Ly49H− NK cells (Fig. 2C). Susceptibility was specifically reversed by the anti-Ly49H mAb (Fig. 2C). Finally, MCMV-infected bone marrow (BM) macrophages retained susceptibility to Ly49H+ NK cells whereas they became more resistant to Ly49H− NK cells, an effect also reversed by the anti-Ly49H mAb (Fig. 2C). Taken together, these data indicated that Ly49H on NK cells specifically recognizes the product of the m157 ORF.
Figure 2.
Recognition of m157 by Ly49H. (A) Specific activation of HD12 cells by m157 transfectants. HD12 or DD12 cells were incubated with C1498 cells transfected with m157, or control m158, as indicated. (B) Specific blockade of m157 activation of HD12 by anti-Ly49H mAb. HD12 cells were incubated with J774 cells transfected with GFP or m157, in the presence of anti-Ly49H or isotype control mAb, as indicated. (Inset) Maximal β-gal induction with PMA + ionomycin. (C) Ly49H+ NK cells kill MCMV-infected C57BL/6 BM macrophages and BaF3-m157 transfectants. A standard 4-h 51Cr-release assay was performed with Ly49H sorted LAK cells from C57BL/6 mice. Ly49H+ (filled symbols) or Ly49H− (open symbols) LAKs were incubated with indicated targets at various effector-to-target (E:T) ratios in media alone (squares) or with the anti-Ly49H mAb 3D10 (diamonds) or isotype control (circles). In the BM macrophage experiments, F(ab′)2 fragments were used to avoid redirected lysis whereas whole antibodies were used for other panels. The isotype control for BM macrophage experiments was mAb B8-24-3, and mAb 9E10 was used as isotype control in the remaining experiments. The lysis of uninfected BM macrophages is represented by a triangle.
Stimulation of Fresh NK Cells by m157.
The m157 ORF encodes a deduced 329-aa polypeptide with no sequence homology to any other molecule in GenBank except for r157, its apparent homolog in the rat CMV genome (data not shown). m157 contains a potential N-terminal 21-aa signal peptide, and an ectodomain with five potential sites for N-linked glycosylation. However, a putative hydrophobic transmembrane domain is interrupted by a stop codon. Because the related domain in r157 does not contain a stop codon, we verified the sequence of m157 with multiple reverse transcription–PCR primers (data not shown). All cDNA clones contained m157 sequence identical to that in the published annotated MCMV genome (25) and m157 has a putative glycosylphosphatidylinositol linkage (Table 1).
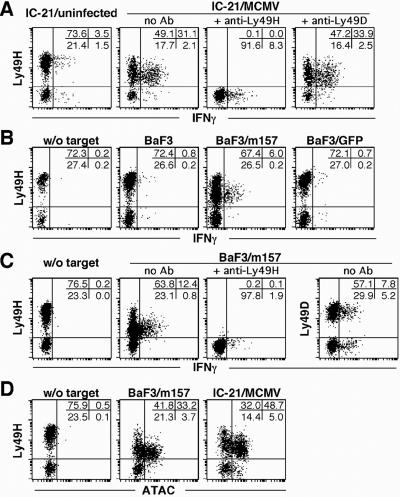
Importantly, the identification of the Ly49H ligand also permits advances in understanding of the in vivo NK cell response to MCMV because it provided a means to examine whether Ly49H+ NK cells are selectively activated soon after exposure to MCMV-infected cells. After only a brief coincubation of freshly isolated C57BL/6 RAG-1−/− splenocytes (devoid of mature B and T cells) with MCMV-infected IC-21 targets in vitro, there was selective activation of Ly49H+ NK cells as indicated by enhanced intracellular IFN-γ staining, primarily in Ly49H+ NK1.1+ cells (Fig. 3A). This effect was blocked by the anti-Ly49H mAb (Fig. 3A). A similar effect was also seen with m157-transfected BaF3 cells that was not observed with untransfected or other transfected BaF3 cells (Fig. 3B) or in costaining with anti-Ly49D (Fig. 3C). The m157 effect was also reversed with anti-Ly49H (Fig. 3C) but not a control Ab (data not shown). Strikingly, we also observed decreased anti-Ly49H binding in response to m157 targets and to a lesser extent with MCMV infection. It is also notable that the IFN-γ-expressing Ly49H+ cells showed lower reactivity with anti-Ly49H, indicating a correlation with NK cell activation. Finally, MCMV infection and m157 also triggered Ly49H+ NK cells to express a chemokine, ATAC/lymphotactin (ref. 30; Fig. 3D), that may serve to recruit or activate other inflammatory cells. Thus, m157 triggers specific activation of primary Ly49H+ NK cells, strongly suggesting that m157 quickly stimulates specific NK cell activation in vivo.
Figure 3.
Interaction of Ly49H with m157 induces production of IFN-γ and ATAC by fresh NK cells. Freshly isolated splenocytes from B6.RAG-1-deficient mice were cocultured with (A) uninfected or MCMV-infected IC-21 cells, (B) parental BaF3 cells or BaF3 cells transfected with either m157 or GFP, (C) BaF3 cells transfected with m157, and (D) BaF3 cells transfected with m157 or MCMV-infected IC-21 cells. All cocultures were performed at effector-to-target ratio of 1:1. Where indicated, F(ab′)2 fragments of anti-Ly49H or anti-Ly49D were added during incubation. Six to 8 h later, cells were stained for intracellular IFN-γ (A, B, and C) or ATAC (D) after cell surface marker staining for NK1.1 and Ly49H or Ly49D. As controls, profiles of cytokine production by NK cells in the absence of target cells are shown (w/o target). Gated NK1.1+ cells are shown.
Discussion
m157 belongs to the m145 family, comprised of related glycoproteins (≈20% amino acid identity) including m152 (25). These proteins are generally expressed early during infection and are dispensable for viral growth and replication in vitro (31). The Ly49H ligand appears to be less well expressed on MCMV-infected 3KO macrophages (see supporting information), suggesting that m157 expression may be enhanced by association with MHC class I or β2-microglobulin molecules. In this regard, m152 can retain MHC class I heavy chains in the endoplasmic reticulum-Golgi intermediate compartment (32) and also has a predicted MHC class I fold. These data suggest that m157 may have a role in altering the MHC class I assembly and presentation pathway, perhaps in an allotype or tissue-specific manner (33, 34). Alternatively, because it is known that individual MHC class I ligands can bind both NK cell activation and inhibitory receptors (3, 23, 35), m157 may also bind an inhibitory receptor in C57BL/6 or other strains. These possible alternative immune evasion functions for m157, the limited sequence availability of MCMV strains, and the restricted expression of Ly49H to a relatively small number of inbred mouse strains (9, 36) may explain why MCMV retains this ORF in its genome. The complexities of interactions between the various ORF products, especially m157 and the putative inhibitory receptor ligand m144, and their effects on NK cells during infection also require further analysis.
Interestingly, using IFN-γ production as a surrogate marker of activation, we were able to detect specific activation of freshly explanted Ly49H+ NK cells after only a brief exposure to m157-expressing targets. When extrapolated to the in vivo setting by accounting for MCMV trafficking from the peritoneum to the spleen, infection and induction of m157 expression, and production of IFN-γ by Ly49H+ NK cells, one would have expected to detect similar Ly49H+ NK cells responses in vivo within the early period after infection. In addition, viral replication is already controlled by Ly49H+ NK cells during this time. However, during this period, there is enhanced IL-12 production (37) that may stimulate most NK cells to produce IFN-γ regardless of whether they express Ly49H or are triggered through this activation receptor. Thus, our current data provide additional evidence that the specific activation of naive Ly49H+ NK cells may be masked by such systemic responses.
The m157 induction of IFN-γ may nonetheless be important in helping to control MCMV because IFN-γ would be delivered in close proximity of infected cells. By contrast, systemic production of IFN-γ by IL-12 or other cytokines may be less effective. In addition, we also showed that a chemokine, ATAC/lymphotactin, was also specifically produced by m157 activation of Ly49H+ NK cells, suggesting that the response to m157 may also provide a means for NK cells to trigger other inflammatory events, such as the attraction or activation of other immune cells. Thus, in addition to cytotoxicity, specific activation of NK cells may help provide local control of virus infection and lead to a cascade of other events in innate immunity.
A notable finding in our studies is the marked decreased reactivity of the anti-Ly49H mAb with Ly49H+ NK cells that appeared to have encountered the m157 ligand. This effect appeared to be specific and could be due to a variety of effects, including receptor down-regulation akin to effects seen with T cell antigen receptor cross-linking. Alternatively, down-regulation of Ly49H expression could be due to receptor shedding, or simply blockade of Ab binding by ligand. Nevertheless, the effect can be viewed as a specific activation event on cells that have engaged ligand and may itself have physiological consequences.
Another unexpected outcome of our studies was the identification of a large number of other MCMV ORFs encoding proteins with potential MHC-like structures. Whereas unequivocal determination of their three dimensional folds will require crystallographic studies, these findings nonetheless provide candidates for further immunological study. The putative MHC class I-like molecules may well be involved in immune evasion or detection because they are dispensable for in vitro growth and replication. Perhaps these molecules also perturb MHC class I assembly and antigen presentation or interact with NK cell receptors, like m152 and m157, respectively. These findings also suggest that other viruses, especially large DNA viruses, may contain previously unrecognized homologues or functional mimics of NK cell receptor ligands. This result may be particularly relevant for other viruses manifesting phenotypic resistance that is genetically linked to the NK gene complex or other mammalian genomic regions that encode NK cell receptors. Murine loci for genetic resistance to mousepox, and herpes simplex, for example, map to the NKC (38, 39), suggesting that they may encode NK cell receptors that interact with viral encoded ligands. In addition, perhaps a similar bioinformatics approach to mammalian genomes will reveal other putative MHC-like molecules that may serve as endogenous ligands for Ly49H and other NK cell receptors. Thus, further analysis of these putative MHC class I-like molecules may be revealing.
Finally, it is intriguing to note that the m145 family and m157 itself are present in the genome of rat CMV but absent in human CMV (40). Likewise, the Ly49 gene cluster is also present in rats (41), but only a remnant is present in the human genome (42, 43), perhaps reflecting the coordinated evolution of pathogen immune evasion and innate host response genes, a virus-host counterpart to an international arms race.
Supplementary Material
Acknowledgments
We thank Nilabh Shastri, Toshio Kitamura, Ted Hansen, Eva-Marie Wormstall, Pamela Stanley, David Leib, Tom Shenk, and Skip Virgin for reagents; Marco Colonna, Leon Carayannopoulos, and Skip Virgin for encouragement and helpful discussions; Jennifer Laurent, Kim Marlotte, and Darryl Higuchi for technical assistance; and Emil Unanue, Tony French, and Emily Ho for critical reading of the manuscript. This work was supported by the Barnes-Jewish Hospital Foundation and grants from the National Institutes of Health to W.M.Y., who is an investigator of the Howard Hughes Medical Institute. A.A.S. is supported by Project Grant 990646 from the National Health and Medical Research Council of Australia. Work in the Fremont laboratory is supported by the “Midwest Center for Structural Genomics” (National Institutes of Health/National Institute of General Medical Sciences P50 GM62414).
Abbreviations
- NK
natural killer
- MCMV
murine cytomegalovirus
- NKC
NK gene complex
- ITAM
immunoreceptor tyrosine-based activation motif
- KARAP
killer cell-activating receptor-associated protein
- β-gal
β-galactosidase
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- GFP
green fluorescent protein
- CHO
Chinese hamster ovary
- ATAC
activation-induced T cell-derived and chemokine-related cytokine
- BM
bone marrow
References
- 1.Tortorella D, Gewurz B E, Furman M H, Schust D J, Ploegh H L. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 2.Ljunggren H G, Karre K. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Karlhofer F M, Ribaudo R K, Yokoyama W M. Nature (London) 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 4.Lanier L L, Phillips J H. Immunol Today. 1996;17:86–91. doi: 10.1016/0167-5699(96)80585-8. [DOI] [PubMed] [Google Scholar]
- 5.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu M-L. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 6.Farrell H E, Vally H, Lynch D M, Fleming P, Shellam G R, Scalzo A A, Davis-Poynter N J. Nature (London) 1997;386:510–514. doi: 10.1038/386510a0. [DOI] [PubMed] [Google Scholar]
- 7.Tomasec P, Braud V M, Rickards C, Powell M B, McSharry B P, Gadola S, Cerundolo V, Borysiewicz L K, McMichael A J, Wilkinson G W. Science. 2000;287:1031–1033. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 8.Scalzo A A, Fitzgerald N A, Simmons A, La Vista A B, Shellam G R. J Exp Med. 1990;171:1469–1483. doi: 10.1084/jem.171.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scalzo A A, Lyons P A, Fitzgerald N A, Forbes C A, Yokoyama W M, Shellam G R. Genomics. 1995;27:435–441. doi: 10.1006/geno.1995.1074. [DOI] [PubMed] [Google Scholar]
- 10.Scalzo A A, Fitzgerald N A, Wallace C R, Gibbons A E, Smart Y C, Burton R C, Shellam G R. J Immunol. 1992;149:581–589. [PubMed] [Google Scholar]
- 11.Brown M G, Dokun A O, Heusel J W, Smith H R, Beckman D L, Blattenberger E A, Dubbelde C E, Stone L R, Scalzo A A, Yokoyama W M. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 12.Lee S H, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal S M. Nat Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 13.Daniels K A, Devora G, Lai W C, O'Donnell C L, Bennett M, Welsh R M. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith K M, Wu J, Bakker A B, Phillips J H, Lanier L L. J Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- 15.Smith H R, Chuang H H, Wang L L, Salcedo M, Heusel J W, Yokoyama W M. J Exp Med. 2000;191:1341–1354. doi: 10.1084/jem.191.8.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasello E, Olcese L, Vely F, Geourgeon C, Blery M, Moqrich A, Gautheret D, Djabali M, Mattei M G, Vivier E. J Biol Chem. 1998;273:34115–34119. doi: 10.1074/jbc.273.51.34115. [DOI] [PubMed] [Google Scholar]
- 17.Dokun A O, Kim S, Smith H R, Kang H S, Chu D T, Yokoyama W M. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 18.Biron C A, Nguyen K B, Pien G C, Cousens L P, Salazar-Mather T P. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 19.Bauer S, Groh V, Wu J, Steinle A, Phillips J H, Lanier L L, Spies T. Science. 1999;285:727–729. [PubMed] [Google Scholar]
- 20.Sanderson S, Shastri N. Int Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura T. Int J Hematol. 1998;67:351–359. doi: 10.1016/s0925-5710(98)00025-5. [DOI] [PubMed] [Google Scholar]
- 22.Dorner B G, Scheffold A, Rolph M S, Huser M B, Kaufmann S H, Radbruch A, Flesch I E, Kroczek R A. Proc Natl Acad Sci USA. 2002;99:6181–6186. doi: 10.1073/pnas.092141999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehta I K, Smith H R C, Wang J, Margulies D H, Yokoyama W M. Cell Immunol. 2000;209:29–41. doi: 10.1006/cimm.2001.1786. [DOI] [PubMed] [Google Scholar]
- 24.Idris A H, Smith H R C, Mason L H, Ortaldo J H, Scalzo A A, Yokoyama W M. Proc Natl Acad Sci USA. 1999;96:6330–6335. doi: 10.1073/pnas.96.11.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawlinson W D, Farrell H E, Barrell B G. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hengel H, Brune W, Koszinowski U H. Trends Microbiol. 1998;6:190–197. doi: 10.1016/s0966-842x(98)01255-4. [DOI] [PubMed] [Google Scholar]
- 27.Virgin H W, 4th, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira S A, Park S H, Lee P, Bendelac A, Shenk T E. J Virol. 2002;76:885–894. doi: 10.1128/JVI.76.2.885-894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley L A, MacCallum R M, Sternberg M J. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 30.Dorner B, Muller S, Entschladen F, Schroder J M, Franke P, Kraft R, Friedl P, Clark-Lewis I, Kroczek R A. J Biol Chem. 1997;272:8817–8823. doi: 10.1074/jbc.272.13.8817. [DOI] [PubMed] [Google Scholar]
- 31.Thale R, Szepan U, Hengel H, Geginat G, Lucin P, Koszinowski U H. J Virol. 1995;69:6098–6105. doi: 10.1128/jvi.69.10.6098-6105.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziegler H, Thale R, Lucin P, Muranyi W, Flohr T, Hengel H, Farrell H, Rawlinson W, Koszinowski U H. Immunity. 1997;6:57–66. doi: 10.1016/s1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]
- 33.Kavanagh D G, Gold M C, Wagner M, Koszinowski U H, Hill A B. J Exp Med. 2001;194:967–978. doi: 10.1084/jem.194.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hengel H, Reusch U, Geginat G, Holtappels R, Ruppert T, Hellebrand E, Koszinowski U H. J Virol. 2000;74:7861–7868. doi: 10.1128/jvi.74.17.7861-7868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura M C, Linnemeyer P A, Niemi E C, Mason L H, Ortaldo J R, Ryan J C, Seaman W E. J Exp Med. 1999;189:493–500. doi: 10.1084/jem.189.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown M G, Scalzo A A, Stone L R, Clark P Y, Du Y, Palanca B, Yokoyama W M. Immunogenetics. 2001;53:584–591. doi: 10.1007/s002510100365. [DOI] [PubMed] [Google Scholar]
- 37.Orange J S, Biron C A. J Immunol. 1996;156:4746–4756. [PubMed] [Google Scholar]
- 38.Brownstein D G, Gras L. Am J Pathol. 1997;150:1407–1420. [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira R A, Scalzo A, Simmons A. J Immunol. 2001;166:5869–5873. doi: 10.4049/jimmunol.166.10.5869. [DOI] [PubMed] [Google Scholar]
- 40.Vink C, Beuken E, Bruggeman C A. J Virol. 2000;74:7656–7665. doi: 10.1128/jvi.74.16.7656-7665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dissen E, Ryan J C, Seaman W E, Fossum S. J Exp Med. 1996;183:2197–2207. doi: 10.1084/jem.183.5.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westgaard I H, Berg S F, Orstavik S, Fossum S, Dissen E. Eur J Immunol. 1998;28:1839–1846. doi: 10.1002/(SICI)1521-4141(199806)28:06<1839::AID-IMMU1839>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 43.Trowsdale J, Barten R, Haude A, Stewart C A, Beck S, Wilson M J. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 44.Chapman T L, Bjorkman P J. J Virol. 1998;72:460–466. doi: 10.1128/jvi.72.1.460-466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.