Abstract
Myelin basic proteins (MBP) are major constituents of the myelin sheath of oligodendrocytes and Schwann cells in the central nervous system and the peripheral nervous system, respectively. We previously showed that MBP-related transcripts are present in the bone marrow and the immune system. These mRNAs are transcribed from a region called 0′, consisting of three exons, located upstream of the classical MBP exons; these three exons belong to the long MBP gene otherwise called “Golli-MBP.” The most abundant of these mRNAs, now called HMBP (hemopoietic MBP), encompasses the sequence encoded by the region 0′ plus exon 1 and part of intron 1 of the classic MBP gene. Antisera to recombinant HMBP proteins are immunoreactive with proteins of about 26–28 kDa in brain, thymus, and spleen. This report demonstrates that HMBP proteins are present in the vast majority (>95%) of thymic T cells, which express the corresponding transcripts, as do mature T cells from lymph nodes and spleen. HMBP mRNAs and proteins are also manifest in the majority of spleen B lymphocytes and in B cell lines. In addition to lymphoid cells, HMBP proteins are in all types of myeloid lineage cells, i.e., macrophages, dendritic cells, and granulocytes, as well as in megakaryocytes and erythroblasts. Finally, HMBP proteins are present in CD34+ bone marrow cells, and, furthermore, in highly proliferative cultures, these CD34+ cells express HMBP RNAs and proteins. Thus, MBP gene products are present both in the nervous system and in the entire hemopoietic system.
The myelin basic protein (MBP) gene was initially described as coding for proteins that are major constituents of the myelin sheath of oligodendrocytes in the central nervous system (CNS) and Schwann cells in the peripheral nervous system (PNS). MBP-related transcripts are also present in the bone marrow and the immune system (1–3). These mRNAs are transcribed from a region called 0′, consisting of three exons located upstream of the classical MBP exons (exon 1b through 7). The long MBP gene, which contains three new exons, has been called “Golli-MBP” (4). The MBP transcripts containing the region 0′ were subdivided into two distinct families, HMBP-R (hemopoietic MBP-related) and MBP2 according to their 3′ structure. The most abundant mRNA type, HMBP-R, now called HMBP, includes exon 1b of the classical MBP gene followed by a part of the intron 1. In contrast, the MBP2 transcripts contain, in addition, all of the downstream classical MBP gene exons 1b to 7, some of which have been found to be alternatively spliced, as in the classical forms of MBP mRNAs.
Antisera were generated against a recombinant peptide, specific for the deduced mouse HMBP proteins (ref. 5; R.F., unpublished data). These antisera are immunoreactive with multiple proteins and, in particular, with a family of proteins of about 26–28 kDa in brain, thymus, and spleen, tissues in which the new MBP mRNAs are expressed (5).
It has been known for a long time that experimental allergic encephalitis (EAE), considered an experimental model for multiple sclerosis (MS), can be transferred by T lymphocytes specific for myelin proteins such as MBP (6–8). Spontaneous immune conditions in mice can be transferred by hemopoietic stem cells, which differentiate into T and B lymphocytes before the disease is manifest, leading to the hypothesis that the cause of immune diseases may lie in the hemopoietic stem cells, and that syngeneic bone marrow transplantation (in conjunction with an immunosuppressive treatment) induces a state of long-term unresponsiveness to myelin antigens presumably by a mechanism of tolerance that is antigen specific (9–14). Among bone marrow cells, CD34+ cells may be responsible for this effect directly or through their progeny, as exemplified by MS patients who received autologous CD34+ cells (15).
The goal of this study was, therefore, to determine the cell types in which the new MBPs are expressed in the hemopoietic and immune systems. We report that HMBP proteins are present in the vast majority of T and B lymphocytes in thymus, lymph nodes, and spleen. HMBPs are also in the myeloid lineage cells—in macrophages, dendritic cells, and granulocytes—as well as in erythroblasts and megakaryocytes. Finally, CD34+ bone marrow cells express HMBP proteins, and, furthermore, in highly proliferative cultures, these CD34+ cells express HMBP and MBP2 mRNAs, as well as HMBP proteins, suggesting that the therapeutic effect of CD34+ cells may be related to products of the MBP gene.
Materials and Methods
Preparation of Affinity-Purified Antibodies.
Two groups of antisera were used in this study. The first antisera, called 1130 and 1131, were raised against a recombinant GST–HMBP fusion protein encompassing the region 0′ and exon 1 (5). The second group of antisera, named 025 and 026, was obtained independently as follows. HMBP cDNA 5′-coding exons were cloned by PCR by using the specific following sequences: forward TCCGAGCAGCAGCCAGCAC and reverse AAGCTCGTCGGACTCTGAG from total thymus cDNAs. The amplified fragment was cloned, sequenced, and then inserted into a modified pET vector (Stratagene). The protein has an N-terminal 45-aa leader including the first 11 residues from phage T7 (epitope tag), 10 consecutive histidine residues for affinity chromatography on immobilized nickel, and a cleavage site for Xa protease. The protein was purified on a nickel column and assessed for purity on SDS/PAGE gel. Two rabbits were immunized with 50 μg of purified protein in complete Freund's adjuvant (CFA) and received a booster immunization 21 days later. Sera were tested by Western blots (R.F., unpublished data).
Antibody Purification by Affigel.
The GST–HMBP fusion protein was coupled with Affigel 10 (activated immunoaffinity support, Bio-Rad) according to the standard protocol. Serum 1130 was diluted 1/50 in PBS, and 500 μl settled on top of the Affigel-GST HMBP column and left for 2 h. The column was washed with PBS, and the flowthrough was retained. Anti-GST HMBP-specific antibodies were eluted with 0.2 M glycine·HCl at pH 2.2 and immediately neutralized with 1M Tris (pH 8.0). Eluates then were concentrated and dialyzed on Centricon 30 (Amicon) against PBS. Serum 025 was diluted 1/10 and processed as for serum 1130. Purified antibodies were used at a 1/500 dilution throughout this study.
Preparation of Tissues and Cells.
C57BL/6 young mice were perfused intracardially with PBS (pH 7.2), and organs (brain, thymus, and spleen) were excised. To obtain cell suspensions, thymus and spleen were cut into small fragments from which single cells extruded spontaneously. The remaining tissue fragments were eliminated by sedimentation. Peritoneal cells were obtained as previously reported (1). The pre-B cell line P388B (16) and the plasma cell lines T1165 (17) and X-24 (18) were cultured in DMEM with 10% FBS, 200 mM l-glutamine, and a 1/1,000 dilution of β-mercaptoethanol (10 μl in 2.9 ml H2O). IL-6 (4–10 ng/ml) was added to T1165 cells. Bone marrow was extruded from the medullary cavity of femurs, and cells were dispersed by repeated pipetting and used immediately or cultured at 37°C in 10% CO2 in DMEM with 10% FBS, 5 ng/ml IL-3, 10 ng/ml IL-6, 10 ng/ml stem cell factor, and 1/1,000 dilution of mercaptoethanol as above. Bone marrow cells were fed with fresh medium twice weekly, and the floating cells were subcultured 1/20 once per week. They were assayed for HMBP and hemopoietic markers at 5 and 10 wk of culture.
Immunochemistry.
Protein extraction was performed at 4°C on ice; tissues were homogenized in 5 vol of a pH 7.4 solution containing 50 mM Tris, 2 mM MgCl2, 1 mM EDTA, 5 mM mercaptoethanol, 1 mM PMSF, protease inhibitor cocktails, and 1% Nonidet P-40, by using 20 strokes with a tight-fitting glass dounce homogenizer. Tissue fragments were removed by centrifugation at 1,200 × g for 10 min. Cells were washed with PBS, and proteins extracted the same way as for tissues.
Mono- and Two-Dimensional Immunoblots.
Mono-dimensional immunoblots were processed as reported (5). Two-dimensional gels (isoelectric focusing followed by SDS/PAGE) were carried out according to O'Farrel (19), by using Biolyte 3/10 ampholytes (2%) or ampholytes comprising 0.4% Biolyte 3/10; 0.8% Biolyte 4/6; 0.8% Biolyte 5/7 (Bio-Rad), after enrichment for proteins between 20 and 30 kDa as follows: 2 mg of proteins were deposited on a 12.5% SDS/PAGE gel (160 × 160 × 1.5 mm); after migration, proteins between 20 and 30 kDa were isolated, electroeluted with the Bio-Rad electroeluter, concentrated, and dialyzed on Centricon 10 (Amicon) before gels and immunoblots.
Cell Identification.
The following primary antibodies were used: rat anti-mouse CD4 (Harlan Seralab code MAS 110ps); rat anti-mouse CD19 [Caltag (South San Francisco, CA) RM 77004]; rat anti-mouse F4/80 (Caltag RM 2900); rat anti-mouse CD34 (PharMingen cat no. 09431D); rat anti-mouse CD45 (PharMingen cat no. 553076); rat anti-mouse GR1 (PharMingen cat no. 01211A); rat anti-mouse CD41 (PharMingen cat no. 09911D); and rat anti-mouse TER 119 (PharMingen cat no. 09081A). The secondary antibodies were: the goat fluorescein-conjugated F(ab′)2 fragment of anti-rabbit IgG [Rockland (Gilbertsville, PA) code 711 1222] or the goat F(ab′)2 anti-rat IgG rhodamine-conjugated (Cappel cat. no. 55767). For immunocytochemistry, the cell suspensions were centrifuged in Hanks' solution for 10 min at 800 × g, and the cell pellet was fixed in 4% paraformaldehyde (PFA) for 5 min. The fixed cell suspension was then cytocentrifuged on slides at 200 × g for 5 min.
Fluorescence-Activated Cell Sorter (FACS) Staining.
CD3-PECy5, CD4-PE, and CD8-APC (PharMingen) were used to label live cells from thymus and lymph nodes. CD19 (PharMingen) labeling was used on splenocytes and F4/80 on peritoneal macrophages. Staining was done in PBS containing 0.1% NaN3 and 1% BSA at 4°C. The cells then were washed, and three-color analysis was performed by using a FACS Vantage cytometer (Becton Dickinson) equipped with an argon laser (Coherent Radiation, Palo Alto, CA) and a 100-μm nozzle. The cell fractions—i.e., CD4+/CD8+, CD4+ thymus T cells, spleen B cells, and peritoneal macrophages—were over 98% pure as shown after reanalysis.
Reverse Transcription (RT)-PCR.
Total RNA from purified populations was prepared by using the SV total RNA isolation system (Promega) followed by a DNase I treatment. Reverse transcription was performed with Superscript II (CLONTECH) using random hexamers from 1–5 μg total RNA. From a total volume of 50 μl per cDNA, 5 μl were used in the PCR reactions. PCR was performed by using specific HMBP and MBP2 primers: HMBP forward (nucleotides 225–244) 5′-AGAGATTCACCGAGGAGAGG-3′, HMBP reverse (nucleotides 385–365) 5′-GTCTGCTGTGTGCTTGGAGT-3′ (1); MBP2 forward (nucleotides 471–490) 5′-AGAAGCCAGGATTTGGCTAC-3′, MBP2 reverse (nucleotides 749–731) 5′-GCTCCACGGGATTAAGAGA-3′ (from clone M78 in ref. 20). A three-step PCR procedure of 30 sec at 95°C, 40 sec at 56°C, and 60 sec at 72°C was applied for 32 cycles (HMBP) or 36 cycles (MBP2).
Electron Microscopy.
Suspensions of isolated thymocytes were seeded on polylysine-coated coverslips. After adhesion, cells were fixed in 4% PFA, incubated in a 1/400 dilution of the immunopurified 1130 antiserum (see below) and processed as described (21).
Results
T and B Lymphocytes Produce MBP-Related Proteins.
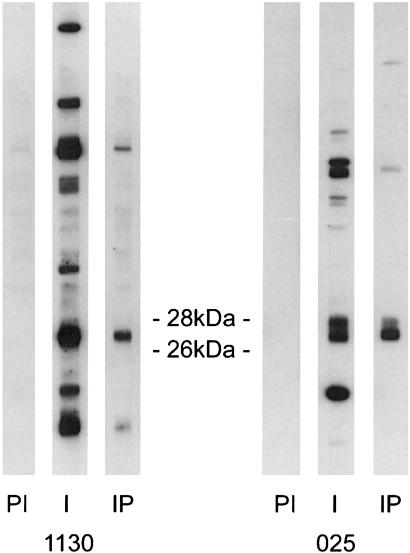
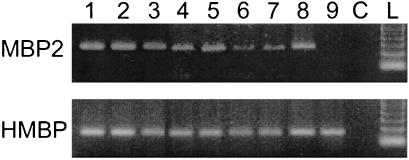
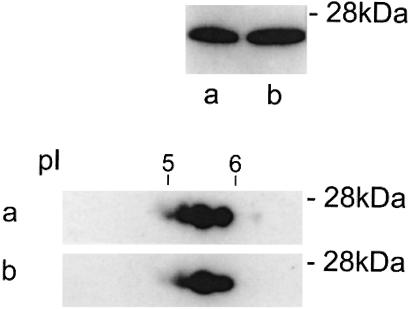
The first goal of this study was to identify the cell type(s) that produce MBP-related proteins in the thymus. To approach this question, adult mouse thymuses were mechanically dissociated into a single cell population made of about 96% T cells. Western blots were performed on the purified T cell population with the crude and corresponding immunopurified antisera. Fig. 1. shows the results of such experiments with antisera 1130 and 025. Although the preimmune sera did not give detectable signals, both crude antisera recognized multiple bands. However, with the corresponding immunopurified antisera, most bands could no longer be detected or decreased drastically, and the only strong remaining bands were located in the 26- to 28-kDa region. All experiments reported here were carried out with immunopurified antisera. These results strongly indicated that thymic T cells produce MBP-related proteins. It was thus of interest to determine whether these proteins were HMBP or MBP2 isoforms. Indeed, RT-PCR experiments showed that FACS-purified thymic T cells express both HMBP and MBP2 at a high level (Fig. 2, lanes 1 and 2). Therefore, experiments were carried out on Shiverer mice, in which the distal part of the MBP gene from exon 2 to 7 is deleted and thus cannot express MBP2 (22). As expected, purified thymus T cells from these mice express only HMBP and not MBP2 transcripts (Fig. 2, lane 9). The immunoreactivity pattern of protein extracts from purified T cells of Shiverer mice is shown in Fig. 3b, in comparison with that of wild-type mice (Fig. 3a). Strikingly, both patterns were equivalent, demonstrating that, indeed, the 26- to 28-kDa MBP proteins from thymus cells are encoded by the HMBP transcripts.
Figure 1.
Thymic T cells produce HMBP proteins. Western blots of proteins from thymic T cells extracts with anti-HMBP antisera 1130 and 025. PI, Preimmune sera; I, crude antisera to HMBP; IP, immunopurified antisera.
Figure 2.
HMBP AND MBP2 mRNA expression in hemopoietic cell lineages. Lane 1, thymus CD4+CD8− cells; lane 2, thymus CD4−CD8+ cells; lane 3, lymph node CD4+ cells; lane 4, lymph node CD8+ cells; lane 5, spleen CD4+ cells; lane 6, spleen CD8+ cells; lane 7, spleen CD19+ cells; lane 8, peritoneal macrophage F4/80+ cells; lane 9, Shiverer thymus; lane C, control, no DNA; lane L, 100-bp DNA ladder.
Figure 3.
T cells from wild-type mice and from Shiverer produce HMBP proteins. Western blots of 20- to 30-kDa proteins of thymus T cells extracts from wild-type mice (a) as compared with Shiverer mice (b) immunodetected with an immunopurifed antiserum. (Upper) Monodimensional gels; (Lower) bidimensional IEF gels with 2% Biolyte 3/10.
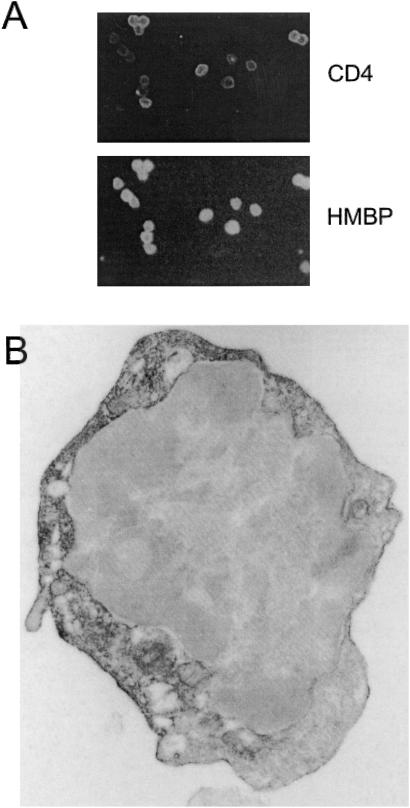
The next question was to determine the proportion of T cells that are immunoreactive to HMBP antisera. Double immunofluorescence experiments were conducted with a CD4 mAb and HMBP antisera on cytocentrifuged thymic T cells. All CD4+ cells were immunolabeled with the HMBP antisera (Fig. 4A). Although the level of intensity of CD4 labeling varied among individual cells, staining with the HMBP antisera appeared strong in most cells. It was thus clear that CD4+ thymocytes produce HMBP proteins. Because the vast majority (>85%) of CD4+ cells in the thymus are immature CD4/CD8 double positive T cells, these results indicated that expression of MBP proteins is an early event in T cell differentiation.
Figure 4.
Thymic CD4+ T cells are immunoreactive to HMBP antisera. (A) Thymic cell suspensions were immunolabeled with an anti-CD4 mAb and an HMBP antiserum. (B) Electron micrograph of a thymic T cell immunolabeled with an HMBP antiserum.
The expression of MBP-related transcripts was studied further in mature T cells isolated from lymph nodes and spleen. RT-PCR experiments, carried out on purified CD4+ and CD8+ T cell populations, show that, indeed, mature cells express both HMBP and MBP2 transcripts (Fig. 2, lanes 3 to 6).
The subcellular distribution of HMBP proteins then was analyzed. No reactivity could be observed on unfixed, live T cells, indicating that HMBP epitopes are not expressed at the cell surface. The localization of HMBP was further investigated at the ultrastructural level. As shown in Fig. 4B, patches of plasma membranes, as well as some intracellular membranes, clearly were immunoreactive. In addition, the cytoplasm, but not the nucleus, showed some diffuse labeling. Thus, the distribution of HMBP in T cells resembles that of the “classic” MBP in oligodendrocytes and Schwann cells.
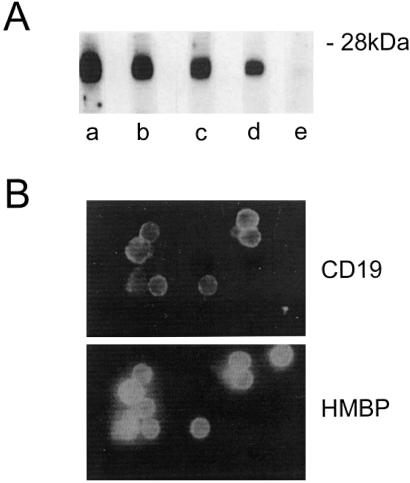
Whether production of HMBP proteins is restricted to the T cell lineage or common to other lymphoid cells, and in particular B cells, was then investigated. RT-PCR experiments carried out on a purified population of CD19+ spleen B cells clearly demonstrated expression of HMBP (Fig. 2, lane 7). Correspondingly, the vast majority of, but not all, the CD19+ B cells were immunolabeled by the HMBP antisera (Fig. 5B). In parallel, B cell lines also were investigated for the production of HMBP. Three cell lines were tested. As shown in Fig. 5A (lanes c and d), two of these lines (X-24 and P388B) exhibited a Western blot pattern equivalent to that of thymic T cells, whereas no immunoreactivity to HMBP antisera could be detected in T-1165 cells (Fig. 5A, lane e).
Figure 5.
HMBP proteins are present in B cells. (A) Western blots of 20- to 30-kDa proteins with an HMBP antiserum. (Aa) T cells; (Ab) spleen cells; (Ac) X-24 B cell line; (Ad) P388 B cell line; (Ae) T 1165 B cell line. (B) Spleen B cells were double labeled with an anti-CD19 and an HMBP antiserum.
Myeloid Cells Express HMBP Proteins.
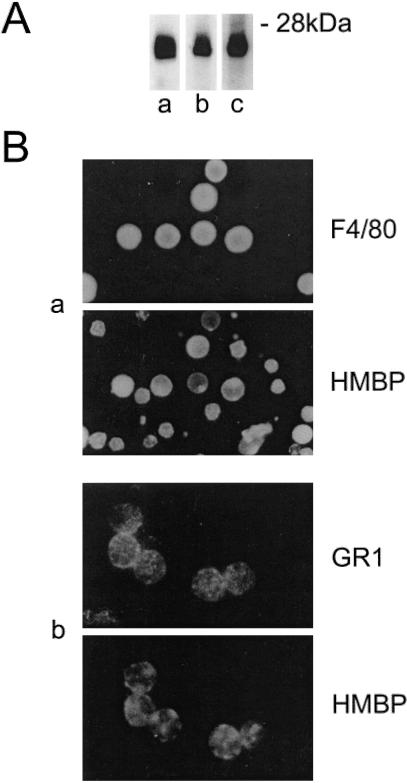
Previous reports had suggested that thymic antigen-presenting cells (APC) produce MBP proteins (23–25). Therefore, immunocytochemistry experiments were performed on macrophages (F4/80) or dendritic cells (NLDC 145) isolated from various tissues. Although the percentage of F4/80 and NLDC 145+ cells isolated from spontaneously extruding thymic cells was very low, most of these were clearly immunolabeled by the HMBP antisera, and similar results were obtained on spleen cell suspensions (data not shown). In addition, immunocytochemistry showed that the vast majority of peritoneal macrophages—but not all—were labeled by HMBP antisera (Fig. 6Ba). Western blot experiments showed that the expected proteins with the molecular masses of 26–28 kDa were abundantly expressed in the vast majority of peritoneal macrophages (Fig. 6Aa), as well as in the P388 macrophagic cell line (Fig. 6Ab). HMBP mRNA expression in peritoneal macrophages was confirmed by RT-PCR performed on highly purified F4/80+ peritoneal cells, showing that both HMBP and MBP2 mRNAs are expressed at a high level (Fig. 2, lane 8).
Figure 6.
Myeloid cells produce HMBP proteins. (A) Western blots of 20- to 30-kDa proteins with an HMBP antiserum. (Aa) Peritoneal cells; (Ab) P388 macrophage cell line; (Ac) bone marrow. (B) Double labeling of (Ba) peritoneal cells (F4/80) and (Bb) granulocytes (Gr1) with an HMBP antiserum.
The detection of HMBP protein expression in macrophages and dendritic cells led to the further investigation of their presence in other cell types of the myeloid lineage. The expression of HMBP proteins therefore was analyzed in bone marrow because HMBP transcripts were initially characterized in this tissue. Western blots performed on bone marrow extracts also exhibit 26- to 28-kDa proteins immunoreactive to the HMBP antisera (Fig. 6Ac). Bone marrow contains a large proportion of granulocytes. To determine whether they produce HMBP proteins, double immunofluorescence experiments were carried out with anti-Gr-1 antibody, whose level of expression is directly related to the granulocytic maturation and which labeled a large proportion of the bone marrow cells. Most of the Gr-1 positive cells were clearly immunoreactive to the HMBP antisera (Fig. 6Bb). Comparable results were obtained with blood granulocytes (data not shown).
Then, HMBP expression was studied in the erythroid and megakaryocytic lineages. Fetal liver cells (ED14) and adult bone marrow cells were double-labeled with TER119, expressed on all cells of the erythroid lineage, and the HMBP antiserum. The majority of erythroblasts, i.e., large TER119+ cells, clearly were immunolabeled by the HMBP antiserum both in fetal liver and bone marrow cells; in contrast, no immunoreactivity was detected in the mature erythrocytes, i.e., small (6 μm in diameter) TER119+ cells. In parallel, a large proportion of megakaryocytes from fetal liver cultures and ex vivo adult bone marrow, labeled by CD41, were immunoreactive to the HMBP antisera (data not shown).
Bone Marrow CD34+ Cells also Express HMBP Proteins.
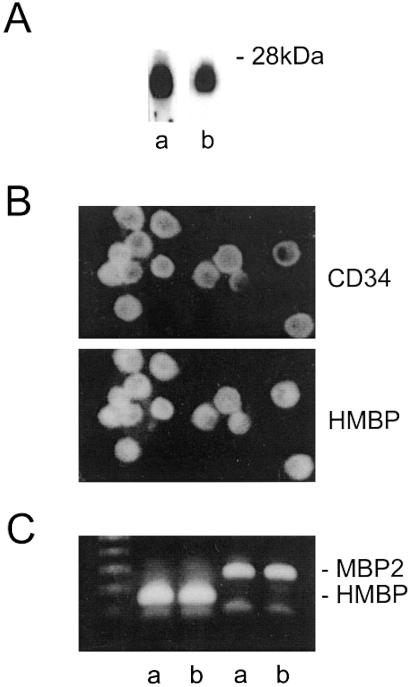
Taken together, these results demonstrated that cells from all lineages of the hemopoietic system can produce HMBP proteins. It was thus of interest to determine at which stage of differentiation these proteins first appear. The hemopoietic system derives from progenitors in the bone marrow that can express CD34 (26–29). As expected, 7–10% of bone marrow cells were found to be CD34+. Because CD34 can be expressed in endothelial cells (30), it was of interest that only a very small proportion of CD34+ cells was immunolabeled by an anti-von Willebrand antiserum, indicating that the vast majority of the bone marrow CD34+ cells are not endothelial cells, but hemopoietic progenitors. The majority of these cells also were immunoreactive to the HMBP antisera (not shown). To further investigate the properties of the CD34+ cells, in vitro cultures of bone marrow cells were performed, in the presence of various cytokines and in the absence of stroma, that were passaged each week (see Materials and Methods). All nonadherent cells expressed CD34 but not the von Willebrand factor. Western blots performed on these nonadherent cells showed that they contain 26- to 28-kDa proteins immunolabeled by the HMBP antisera (Fig. 7Ab) and immunocytochemistry that all CD34+ cells were HMBP+ (Fig. 7B). Furthermore, RT-PCR showed a strong expression of HMBP transcripts and to a lesser extent of MBP2 transcripts (Fig. 7C). These data indicate that primitive hemopoietic progenitors express HMBP proteins.
Figure 7.
Bone marrow CD34+ cells express HMBP proteins. (A) Western blots of protein extracts of CD34+ cells from bone marrow cultures (Ab) as compared with T cell extracts (Aa). (B) All CD34+ cells from bone marrow cultures are HMBP+ as shown by double labeling with a CD34 mAb and an HMBP antiserum. (C) RT-PCR of bone marrow (Ca) and CD34+ cell cultures (Cb).
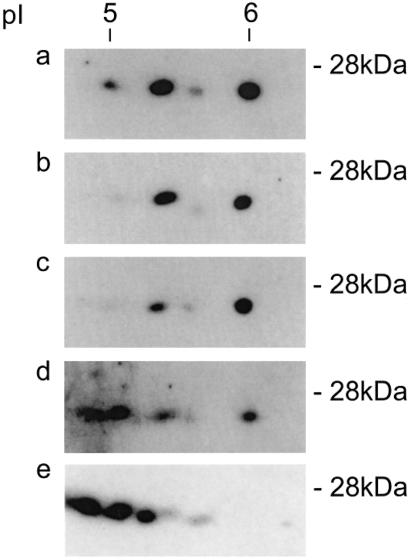
Thus, MBP-related proteins appear to be expressed in all hemopoietic lineages. Therefore, it is of interest that the two-dimensional IEF patterns of 26- to 28-kDa HMBP proteins in all cell types investigated display related patterns, although the intensity of the signal of the different spots, and therefore the ratio of HMBP isoforms, is not identical, a fact that may relate to distinct posttranslational modifications (Fig. 8).
Figure 8.
Western blots of two-dimensional IEF gels of 20- to 30-kDa protein extracts were prepared as in Materials and Methods and probed with an immunopurified HMBP antiserum. (a) T cells; (b) X-24 B cells; (c) peritoneal macrophages; (d) bone marrow; (e) in vitro CD34+ cells.
Discussion
The most striking result of this study is the presence of HMBP proteins in ex vivo bone marrow CD34+ cells with high proliferative activity, in addition to all cell types of the blood and immune systems. This result is consistent with our initial report of the expression of the MBP gene in bone marrow cells from adult mice (1). Two “hemopoietic” MBP mRNA types, called HMBP and MBP2, were originally characterized; they differed in their structure from the classical MBP transcripts by the presence of additional exons, called the 0′ region, upstream of classical MBP. The long MBP gene has been called “Golli-MBP” (4). HMBP and MBP2 mRNAs also were found in tissues and organs colonized by cells of the hemopoietic lineage, such as thymus, spleen, and peritoneal macrophages, in addition to brain. Antisera raised against a recombinant peptide deduced only from the common coding sequences to HMBP and MBP2 isoforms recognize, among others, proteins of about 26–28 kDa that are restricted to brain, thymus, and spleen, in agreement with the mRNA's expression (5). These proteins, visualized by Western blot, comprise unequivocally HMBP isoforms because they also are seen in Shiverer mice, which express HMBP but not the MBP2 transcripts.
Therefore, the identity of cell types of the hemopoietic lineage that express HMBP proteins was investigated. Unexpectedly, HMBP proteins were found in all lineages derived from the hemopoietic progenitors. In regard to the lymphoid lineage, it is significant that all types of T cells—immature T cells, which represent the vast majority of the thymic population, as well as mature T lymphocytes from spleen and lymph nodes—expressed HMBP proteins. In parallel, spleen B lymphocytes, as well as CD19+ plasma cell lines, produce HMBP, raising the possibility that these proteins may be released and autoantibodies produced in vivo. In addition to the lymphoid lineage, all myeloid cells, i.e., granulocytes, macrophages, erythroblasts, and megakaryocytes, express HMBP, whether in the bone marrow, circulating blood, or peritoneal fluid. The only notable exception is erythrocytes, in which HMBP was not detected, probably in relation to the fact that these cells no longer have nuclei, suggesting that HMBP proteins are continuously turning over.
HMBP proteins possess a common sequence (exons 1a and b) with “classical” MBP, so a crossreaction with commercial antisera to MBP could have been expected. However, in our experimental conditions, a sizeable signal on hemopoietic cells with these antisera was not detected although they reacted vigorously with brain extracts. This result may be due to posttranslational modifications or conformational specificities of HMBP proteins. The absence of signal with antisera to classical MBP may account for the fact that they could not detect MBP2 proteins despite the large degree of similarity between MBP2 and classical MBP. Furthermore, MBP2 mRNAs are in low abundance and also may be translated at a low efficiency.
We had observed, years ago, the presence, in T cells, of mRNA signals compatible with those of HMBP and MBP2 (unpublished data) and their immunoreactivity to antisera to HMBP. The report by J.-M. Feng et al. (31) of HMBP RNAs in thymus cells showed that these quite unexpected data were reproducible and that indeed T cells do produce HMBP proteins. However, we did not observe “altered levels of expression in developing thymocytes and lack of significant expression in thymic stromal cells,” and furthermore, HMBP was not detected within T cell nuclei, as J.-M. Feng et al. reported, even at the ultrastructural level. Also, in contrast to Liu et al. (32), we could not detect the presence of “classic” MBPs in immune tissues.
The putative role of MBPs in the immune system in inducing tolerance has been the subject of intense speculation. However, the results reported here demonstrate that the presence of proteins coded by the MBP gene outside the nervous system is not restricted to the immune system, but is a property of the whole hemopoietic lineage, of which the immune system is a branch. Whether the HMBP proteins perform a common function among the diverse cell types of the hemopoietic system, as well as in certain cells of the nervous system, must be determined. One still may ask what the relationship is, if any, between expression of HMBP proteins in the hemopoietic lineage, starting with proliferating bone marrow CD34+ cells, and natural peripheral tolerance of MBP-specific T cells. Hemopoietic cells undergo a rapid turnover, and therefore it is reasonable to assume that HMBP proteins—which are contained within cells—are liberated and can interact with cross-reactive MBP epitopes on T cell antigen receptors (TCRs), thereby leading to tolerization to classical MBP (33). This may provide a mechanism for the therapeutic effect of bone marrow transplantation in MS patients, after immunosuppression. Thus, it might be of interest to study the effects of HMBP proteins or peptides on the experimental allergic encephalitis model and in MS as a substitute for transplantation of CD34+ cells.
Finally, it may be noticed that the name, hemopoietic myelin basic protein related (1), initially given to the new family of products expressed from the long MBP gene, best described its distribution throughout the blood system from stem cells to their differentiated progeny in both the myeloid and lymphoid lineages.
Acknowledgments
We thank Diana Zelenika for the data of Fig. 2, Fawzia Louache, Isabelle Godin, and William Vainchenker for their contributions, and Kenneth Johnson, Michael Potter, and Michael Sela for critical review of the manuscript. We are indebted to Dr. Kent Wilcox for his help in construction of the HMBP-R fusion protein used for immunization at the Medical College of Wisconsin. We also thank S. Barrey for secretarial assistance and M. Louette for the micrographs. B.P. was supported by ARSEP, CANAM, Fondation de France, and Fondation pour la Recherche Medicale. R.B.F. was supported by research Grant AI 30605 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. D.T. was supported by a Veterans Affairs Merit Review Grant.
Abbreviations
- MBP
myelin basic protein
- HMBP
hemopoietic MBP
- MS
multiple sclerosis
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Grima B, Zelenika D, Pessac B. J Neurochem. 1992;59:2318–2323. doi: 10.1111/j.1471-4159.1992.tb10126.x. [DOI] [PubMed] [Google Scholar]
- 2.Zelenika D, Grima B, Pessac B. J Neurochem. 1993;60:1574–1577. doi: 10.1111/j.1471-4159.1993.tb03325.x. [DOI] [PubMed] [Google Scholar]
- 3.Grima B, Zelenika D, Pessac B. Neurobiol Dis. 1994;1:61–66. doi: 10.1006/nbdi.1994.0008. [DOI] [PubMed] [Google Scholar]
- 4.Campagnoni A T, Pribyl T M, Campagnoni C W, Kampf K, Amur-Umarjee S, Landry C F, Handley V W, Newman S L, Garbay B, Kitamura K. J Biol Chem. 1993;268:4930–4938. [PubMed] [Google Scholar]
- 5.Kalwy S, Marty M C, Bausero P, Pessac B. J Neurochem. 1998;70:435–438. doi: 10.1046/j.1471-4159.1998.70010435.x. [DOI] [PubMed] [Google Scholar]
- 6.Patterson P Y. J Exp Med. 1960;111:119–133. doi: 10.1084/jem.111.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Nun A, Wekerle H, Cohen I R. Eur J Immunol. 1981;11:195–199. doi: 10.1002/eji.1830110307. [DOI] [PubMed] [Google Scholar]
- 8.Trotter J, Sriram S, Rassenti L, Chou C-H J, Fritz R B, Steinman L. J Immunol. 1985;134:2322–2327. [PubMed] [Google Scholar]
- 9.Tyndall A, Fassas A, Passweg J. Bone Marrow Transplant. 1992;24:729–734. doi: 10.1038/sj.bmt.1701987. [DOI] [PubMed] [Google Scholar]
- 10.Fassas A, Anagnostopoulos A, Kazis A, Kapinas K, Sakellari I, Kimiskidis V, Tsompanakou A. Bone Marrow Transplant. 1997;20:631–638. doi: 10.1038/sj.bmt.1700944. [DOI] [PubMed] [Google Scholar]
- 11.Karussis D, Vourka-Karussis U, Mizrachi-Koll R, Abramsky O. Mult Scler. 1999;5:17–21. doi: 10.1177/135245859900500104. [DOI] [PubMed] [Google Scholar]
- 12.van Bekkum D W. Stem Cells. 1999;17:172–178. doi: 10.1002/stem.170172. [DOI] [PubMed] [Google Scholar]
- 13.Marmont A M. Annu Rev Med. 2001;51:115–134. doi: 10.1146/annurev.med.51.1.115. [DOI] [PubMed] [Google Scholar]
- 14.Mancardi G L, Saccardi R, Filippi M, Gualandi F, Murialdo A, Inglese M, Marrosu M G, Meucci G, Massacesi L, Lugaresi A, et al. Neurology. 2001;57:62–68. doi: 10.1212/wnl.57.1.62. [DOI] [PubMed] [Google Scholar]
- 15.Burt R K, Traynor A E, Pope R. Blood. 1998;92:3505–3514. [PubMed] [Google Scholar]
- 16.Bauer S R, Holmes K L, Morse H C, III, Potter M. J Immunol. 1986;136:4695–4699. [PubMed] [Google Scholar]
- 17.Nordan P, Potter M. Science. 1986;233:566–569. doi: 10.1126/science.3726549. [DOI] [PubMed] [Google Scholar]
- 18.Mushiski E B, Potter M. Proc Natl Acad Sci USA. 1976;73:932–936. doi: 10.1073/pnas.73.3.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Farrell P H. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 20.Newman S, Kitamura K, Campagnoni A T. Proc Natl Acad Sci USA. 1987;84:886–890. doi: 10.1073/pnas.84.3.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cambier D, Rutin J, Alliot F, Pessac B. Brain Res. 2000;852:191–197. doi: 10.1016/s0006-8993(99)02175-7. [DOI] [PubMed] [Google Scholar]
- 22.Roach A N, Takahashi D, Pravtcheva F, Ruddle F, Hood L. Cell. 1985;42:149–155. doi: 10.1016/s0092-8674(85)80110-0. [DOI] [PubMed] [Google Scholar]
- 23.Fritz R B, Kalvakolanu I. J Neuroimmunol. 1995;57:93–99. doi: 10.1016/0165-5728(94)00167-m. [DOI] [PubMed] [Google Scholar]
- 24.Fritz R B, Zhao M L. J Immunol. 1996;157:5249–5253. [PubMed] [Google Scholar]
- 25.Voskuhl R R. Immunol Rev. 1998;164:81–92. doi: 10.1111/j.1600-065x.1998.tb01210.x. [DOI] [PubMed] [Google Scholar]
- 26.Krause D S, Ito T, Fackler M J. Blood. 1994;84:691–701. [PubMed] [Google Scholar]
- 27.Morel F, Szilvassy S J, Travis M, Chen B, Galy A. Blood. 1996;88:3774–3784. [PubMed] [Google Scholar]
- 28.Donnelly D S, Zelleman D, Sharkia S, Krause D S. Exp Hematol. 1999;27:788–796. doi: 10.1016/s0301-472x(99)00032-6. [DOI] [PubMed] [Google Scholar]
- 29.Matsuoka S, Ebihara Y, Xu M-J. Blood. 2001;97:419–425. doi: 10.1182/blood.v97.2.419. [DOI] [PubMed] [Google Scholar]
- 30.Baumhueter S, Singer M S, Henzel W, Hemmerich S, Renz M, Rosen S D, Lasky L A. Science. 1993;262:436–438. doi: 10.1126/science.7692600. [DOI] [PubMed] [Google Scholar]
- 31.Feng J-M, Givorgri I M, Bongarzone E R, Campagnoni C, Jacobs E, Handley V W, Schonmann V, Campagnoni A T. J Immunol. 2000;165:5443–5450. doi: 10.4049/jimmunol.165.10.5443. [DOI] [PubMed] [Google Scholar]
- 32.Liu H-B, MacKenzie-Graham A J, Palaszynski K, Liva S, Voskuhl R R. J Neuroimmunol. 2001;116:83–93. doi: 10.1016/s0165-5728(01)00284-3. [DOI] [PubMed] [Google Scholar]
- 33.Targoni O S, Lehmann P V. J Exp Med. 1998;187:2055–2063. doi: 10.1084/jem.187.12.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]