Abstract
Limb-girdle muscular dystrophy, type 2A (LGMD 2A), is an autosomal recessive disorder that causes late-onset muscle-wasting, and is due to mutations in the muscle-specific protease calpain 3 (C3). Although LGMD 2A would be a feasible candidate for gene therapy, the reported instability of C3 in vitro raised questions about the potential of obtaining a stable, high-level expression of C3 from a transgene in vivo. We have generated transgenic (Tg) mice with muscle-specific overexpression of full-length C3 or C3 isoforms, which arise from alternative splicing, to test whether stable expression of C3 transgenes could occur in vivo. Unexpectedly, we found that full-length C3 can be overexpressed at high levels in vivo, without toxicity. In addition, we found that Tg expressing C3 lacking exon 6, an isoform expressed embryonically, have muscles that resemble regenerating or developing muscle. Tg expressing C3 lacking exon 15 shared this morphology in the soleus, but not other muscles. Assays of inflammation or muscle membrane damage indicated that the Tg muscles were not degenerative, suggesting that the immature muscle resulted from a developmental block rather than degeneration and regeneration. These studies show that C3 can be expressed stably in vivo from a transgene, and indicate that alternatively spliced C3 isoforms should not be used in gene-therapy applications because they impair proper muscle development.
Mutations in calpain 3 (C3) cause the disease limb-girdle muscular dystrophy (LGMD), type2A, the most prevalent form of LGMD (1). The calpains are a family of calcium-dependent, cytosolic proteases of unknown function (2) with calpain 3 (C3) being the only muscle-specific member (3). C3 is an extremely unstable protein that has not been successfully purified because of its ability to autolyze rapidly (4). Expression of the protein from constructs in vitro has also been extremely inefficient, unless the active site is mutated or truncations of certain exons (ex) are performed (3, 5). In addition, purification in a baculovirus system has only succeeded in producing a truncated C3 (6). Because of the high instability of C3, most of the biochemical information has been inferred from comparisons with the ubiquitous calpains, through analysis by using yeast two-hybrid screens and through expression of constructs in COS-7 cells (5, 7). C3 has three insertion sequences that differentiate it from all other calpain isoforms and may contribute to the muscle-specific function of C3. These regions, called NS, IS1, and IS2, have no known function or homologue; however, autolytic cleavage occurs at three sites in IS1 (4), and IS2 has been implicated in both protein stability (3) and interaction with the cytoskeletal protein titin (7).
Several observations have suggested that calpains may play a role in normal muscle development. Muscle expresses at least four different isoforms of calpain (2). Two of the isoforms, m- and μ-calpain, are ubiquitously expressed. Blockade of m-calpain, by using antisense oligos, blocks myoblast fusion in vitro (8). In addition, overexpression of calpastatin, the calpain-specific inhibitor (9, 10), also blocks muscle cell differentiation in vitro. Furthermore, alternatively spliced C3 isoforms lacking portions of either IS1and/or IS2 have been observed in developing human and mouse skeletal muscle by using reverse transcription–PCR (5). After birth, expression levels of the deletion isoforms decline and the full-length, mature isoform predominates. Although it is unknown whether alternatively spliced isoforms of C3 are translated in muscle or whether they have a function in vivo, the precise embryonic regulation of their expression suggests that they perform a developmental role.
Because more than 100 gene mutations have been identified for LGMD 2A and the mutation is recessive, patients could potentially benefit from gene therapy with a C3 transgene. Because previous studies have found that C3 is unstable, it was unclear whether C3 could be stably expressed from transgenes in vivo. Therefore, we developed transgenic (Tg) mice to test the feasibility of overexpressing C3 in skeletal muscle. Alternatively spliced developmental isoforms of C3, which are more stable than full-length C3, were also examined. We have shown that overexpression of full-length C3 in Tg mice is not toxic to muscle, nor does it produce a phenotype; however, overexpression of C3 isoforms lacking ex 6 or ex 15 produces skeletal muscle that does not seems to be fully differentiated. Although these Tg mice are not disease models for LGMD, they do provide insight into the potential for C3 to be used in gene therapy trials and can be used to identify in vivo substrates of C3.
Methods
Construct Synthesis and Creation of Tg Mice.
Tg constructs were made with the human skeletal α-actin promoter upstream of three different C3 cDNAs. The three constructs were (i) C3 wild-type (wt) cDNA, (ii) C3 ex 6− (C3 lacking exon 6) and (iii) ex 15− (C3 lacking exon 15) (Fig. 1). The human skeletal α-actin construct is described in detail (11). Tg mice were generated at the University of California Irvine Tg Mouse Facility by microinjection of linearized, purified plasmid into F2 hybrid zygotes from C57BL/6J × Balb c parents, as described (12). N1 mice were crossed with C57/BL10, and the N1 or N2 generations were analyzed. All comparisons were made with age-matched, non-Tg littermates. Mice were genotyped by PCR by using primers in the human skeletal α-actin promoter (5′-CCC GAG CCG AGA GTA GCA GT-3′) and the vp1 intron (5′-CCC TTC CCT GTT GGC TAC T-3′).
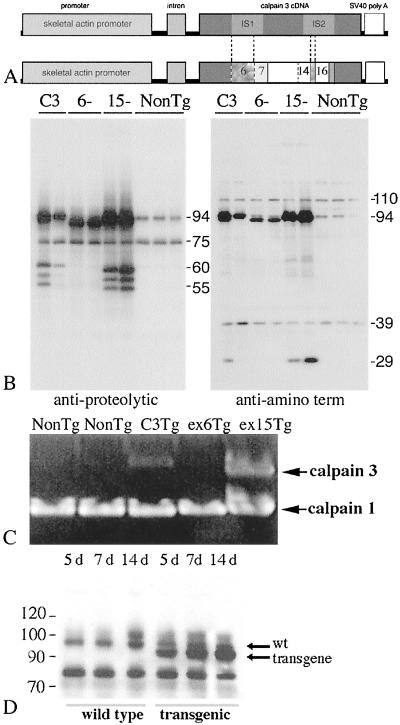
Figure 1.
Tg constructs, protein expression, and analysis of transgene activity. (A) Constructs used to make Tg mice. Ex deletions are indicated by dotted lines. Numbers are ex numbers. (B) Immunoblot of muscle extracts from Tgs probed with 12A2 (left blot) or pNS (right blot). Wild-type C3 is 94 kDa. Deletion isoforms produce 89.5-kDa (ex 6) and 93-kDa (ex 15) proteins. (C) Zymogram of Tg muscle extracts. (D) Time course of transgene expression. Whole-muscle extracts from 5-, 7-, and 14-day (d) non-Tg and ex 6− Tg were immunoblotted for C3. Arrows indicate wt and transgene products.
Tissue Preparation and Immunohistochemistry.
Tissue preparation and immunohistochemistry were performed as described (13). Antibodies used for staining were rat anti-NCAM (Chemicon, mAB 310), rat anti-Mac 1 (PharMingen), anti-AchRα (Transduction Laboratories, Lexington, KY), and mouse anti-fast MHC (NovoCastra, Newcastle, U.K.).
Immunoblots.
Immunoblotting was performed as described (14). Analysis of C3 was performed by using anti-12A2 (15) and anti-IS2 (3).
Casein Zymography.
Zymograms were performed according to the protocol of Raser et al. (16). Gels were overrun for 1 h at 80 V. Five hundred micrograms of whole-muscle lysate was loaded per lane of the gel. Incubation in activation buffer was performed overnight in 5 mM CaCl2.
RNA Isolation and Gene Array Analysis.
Total RNA was isolated according to Chomczynski and Sacchi (17) and sent to the University of California San Diego Gene Chip Facility for labeling and hybridization to Affymetrix gene chips. Data were analyzed with Gene Spring software. Each sample hybridized contained RNA from two to three animals combined.
Apoptosis Assays.
Tg muscle (gastrocnemius and triceps) was assayed for apoptosis by terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling (TUNEL) assay (18). Thymus was used as a positive control. Thymus and muscle without TUNEL reaction were used as negative controls.
Results
Characterization of C3 Transgenics.
Three types of Tg mice were generated that overexpress wt and alternatively spliced isoforms of C3 in muscle. At least two lines were analyzed per Tg. These lines were as follows: two lines of wt C3 Tg (lines 7.2 and 7.9), six lines of Tg with a deletion in the IS1 domain (lines 41.1, 37.9, 39.1, 37.3, 27.10, and 42), and two lines of Tg with a deletion in the IS2 domain (lines 10.2 and 10.3). The ex 6 deletion removes 48 amino acids, and the ex 15 deletion removes 6 amino acids from C3. The six amino acids in ex 15 (KKKKTK) encode most of a sequence that is reported to be a nuclear localization signal (7).
C3 Transgenes Can Be Stably Expressed in Muscle.
Transgene expression was examined by immunoblotting with anti-C3 (Fig. 1). High levels of muscle-specific expression of C3 were observed in all Tg mice analyzed. On the basis of gene expression profiling (data not shown), the mRNA for C3 is elevated in all Tg mice 25–60 times relative to endogenous levels of C3 in non-Tg mice.
C3 Transgenes Have Autolytic Activity.
Previous studies have demonstrated that C3 has three autolytic sites within the IS1 domain (4). The predicted proteolytic fragments of 60, 58, and 55 kDa that result from autolysis in IS1 are observed when tissue extracts from C3 Tg and ex 15− Tg mice are probed with an antibody against the proteolytic domain (12A2; Fig. 1B, left blot). These autolytic products are not observed in non-Tg lanes, probably because they are below the limit of detection. Two of the three autolytic sites that produce these fragments are removed in the ex 6− transgene. Although the third autolytic site remains in this transgene, the expected 55-kDa autolytic product is not observed. Blots probed with an antibody against the N terminus (pNS; ref. 3) show the ≈30-kDa N-terminal fragment resulting from autolysis in IS1 in the mature C3 and ex 15− transgenes. These immunoblots show that autolytic activity is preserved in full length C3 and C3 ex 15− transgenes.
Activity of C3 Transgenes in Zymograms.
To assess activity of the transgenes in Tg muscle extracts, we performed zymograms with casein as a substrate (16). This assay was not sensitive enough to detect endogenous C3 from non-Tg mice; however, extracts from transgenics expressing C3 and ex 15− transgenes had detectable activity in a zymogram (Fig. 1C). The ex 6− Tg showed no activity above non-Tg control muscle, even though expression levels of this transgene were approximately 60-fold higher than endogenous C3. Because it has been demonstrated in vitro that ex 6− constructs retain some proteolytic activity, it cannot be concluded that all C3 activity is abolished when this transgene is overexpressed (5). However, these data suggest that extracts from ex 6− Tg may have reduced C3 proteolytic activity compared with non-Tg and show that extracts from mice expressing full-length C3 and ex 15− Tg produce higher C3 activity than extracts from non-Tg mice.
Time Course of Transgene Expression and Examination of Endogenous C3 Expression.
Previous studies have shown that transgenes expressed from the human skeletal α-actin promoter are not expressed in utero or 1 day after birth (19). To examine the time course of transgene expression in neonatal mice, transgene expression was examined 5, 7, and 14 days after birth in one line of ex 6− Tg (line 37.3) mouse muscles by immunoblotting for C3 protein. The size of this transgene is smaller than the wt protein (94 kDa), which allows for specific study of the transgene without interference from endogenous C3. This analysis shows that the transgene is turned on as early as 5 days of age (Fig. 1D). Endogenous C3 does not seem to be down-regulated in response to transgene expression.
Analysis of C3 Tg (Full-Length, Mature C3 Transgene).
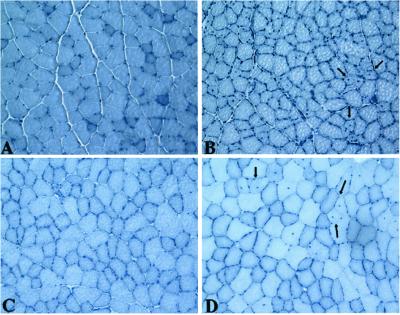
Two different lines of C3 Tg mice were analyzed at 6 weeks and 8–9 months of age. Both lines looked normal on a gross level. Triceps, quadriceps, hamstring, gastrocnemius, and soleus muscles were all examined morphologically (Fig. 2). These muscles looked normal except for the infrequently observed centrally nucleated fibers in the quadriceps, tibialis anterior, and soleus muscle (Fig. 2). These abnormal fibers were of normal or slightly reduced diameter and were observed in small patches in some mice. Although some muscles had 10–50 centrally nucleated fibers in a cross section, other mice did not have them at all. C3 Tg mice had normal muscle strength, by grip-strength analysis (data not shown). These data show that C3 can be expressed at high levels from a transgene, without toxicity and without disruption of normal muscle function.
Figure 2.
Morphology of C3 Tg Muscles. (A–D) Cross sections of muscles stained with hematoxylin. (A) Non-Tg 6-week quad. (B) Line 7.9, 6-week quad. (C) Line 7.2, 16-week quad. (D) Line 7.2, 16-week tibialis anterior. Arrows point to centrally located nuclei. All micrographs were photographed at the same magnification.
C3 Tg Lacking Ex 6 Have Abnormal Muscle.
All ex 6− Tg lines showed reduced musculature, kyphosis, and a waddling gait. Muscles of ex 6− Tg mice were also overtly smaller than wt muscles (Fig. 3 A–C). The phenotype was observed in nine different lines. Two of these were founders that were not able to breed (lines 37.8 and 41.7), and a third was also unhealthy but was mistakenly killed by the Tg facility and was not analyzed. Founder 37.9 was very affected but was able to breed for a period and also produced offspring with strong phenotype. Founder 41.1 was healthy but produced several sick mice that showed phenotype at 3 weeks of age. Lines 39.1 and 37.3 showed a milder phenotype, and lines 42 and 27.10 showed the mildest phenotypes. Severely affected mice died by 4–6 weeks of age. Less severely affected mice could survive at least 7–8 months. C3 protein analysis by immunoblotting did not show any correlations between the severity of the phenotype and the level of transgene expression.
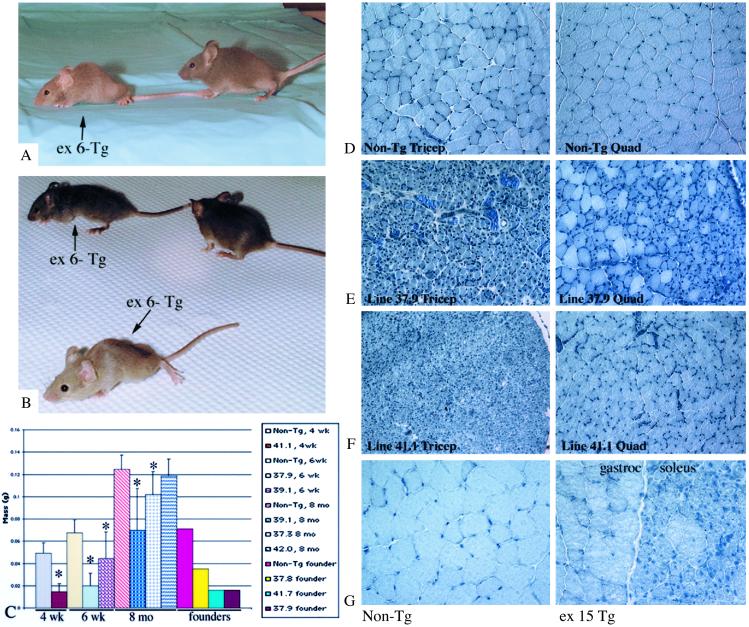
Figure 3.
Phenotype of ex 6− and ex 15− Tg. (A and B) Photographs of ex 6− Tg and non-Tg littermates. Arrows indicate ex 6− Tg mice. Non-Tg littermates do not have arrows. (C) Graph represents average muscle masses of 4-week, 6-week, and 8-month triceps muscles of different ex 6− Tg lines. *, P < 0.05 is significantly different from age-matched controls. Non-Tg controls are the left-most bar for each age group. Weights of founder triceps also are shown on the graph. For each of these founder bars, n = 1. All others are n > 5, except for 37.9, which is n = 2. Vertical bars represent SD. (D–F) Sections of 4- to 6-week non-Tg and ex 6− Tg muscles stained with hematoxylin. All ex 6− micrographs were photographed at the same magnification. (G) Sections of non-Tg gastrocnemius and ex 15− Tg gastrocnemius/soleus. Both micrographs were taken at the same magnification.
Morphological examination of the ex 6− Tg muscles showed that they had many small-diameter, centrally nucleated fibers, resembling regenerating or developing muscle cells. The most severely affected mice were lines 41.1, 37.9, and 39.1. In addition, two founders' muscles (lines 37.8 and 41.7) had the same morphology as those other lines (Fig. 3 D–F). Not all small-diameter fibers in ex 6− Tg had central nuclei. By light microscopy, the triceps, hamstrings, quadriceps, soleus, psoas, and gastrocnemius were more affected than diaphragm, deltoid, tibialis anterior, and biceps brachii. The level of transgene expression did not differ in any of these muscles by immunoblotting (data not shown). For the more affected muscles, the entire cross section was homogeneously affected (e.g., triceps), whereas some muscles only showed patchy areas with small-diameter or centrally nucleated fibers (e.g., tibialis anterior). Triceps mass was reduced in all but one ex 6− Tg line (Fig. 3C).
Ex 15− Tg Have Abnormal Solei.
Two lines of ex 15− Tg were examined at 6 weeks and 8 months of age. Ex 15− Tg had a normal gross phenotype. Morphological examination of triceps, hamstrings, psoas, diaphragm, gastrocnemius, tibialis anterior, and soleus was performed and only the soleus muscle and a few areas of the gastrocnemius (Fig. 3G) displayed the morphology observed in the ex 6− Tg mice. This abnormal morphology was observed in one of the lines studied (10.3) but not the other (10.2). The soleus muscle showed a wider variation in fiber diameter than the muscles of ex 6− Tg (see Fig. 5C).
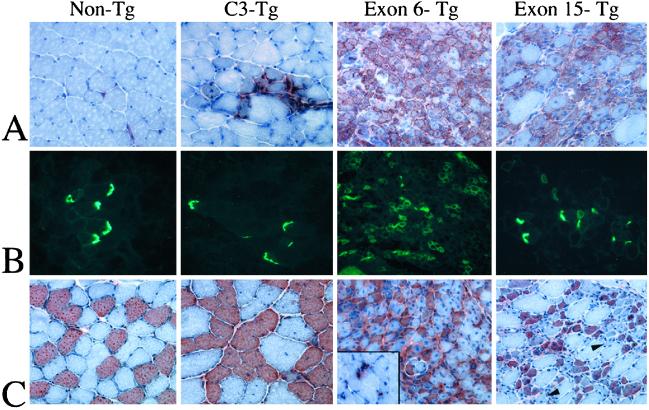
Figure 5.
C3 Tg lacking ex 6 and 15 are developmentally immature. (A) Cross sections of Tg and non-Tg soleus muscles stained for NCAM (red) and counterstained with hematoxylin (blue). NCAM is normally concentrated at the neuromuscular junction in adult tissue. In the ex 6− Tg, and in the ex 15− Tg soleus, NCAM is distributed around the sarcolemma in many of the fibers. (B) Cross sections of Tg and non-Tg gastrocnemius and soleus stained for AchRα. Note the distribution of AchRα in normal vs. ex 6− and ex 15− Tg is similar to that of NCAM. (C) Cross sections of non-Tg and Tg soleus muscles stained for slow MHC (red) and counterstained with hematoxylin (blue). Inset of ex 6− Tg shows an area of gastrocnemius muscle that is positive for slow MHC (red). This area probably represents new myofiber growth. Arrowheads show fibers with strong phenotype that do not stain for slow MHC.
C3 Tg, Ex 6− Tg, or Ex 15− Tg Do Not Show Evidence of Muscle Degeneration.
The high percentage of centrally nucleated fibers observed in ex 6− Tg is reminiscent of developing or regenerating muscle. Because muscle regeneration recapitulates muscle development, it was essential to determine whether these fibers were a result of degeneration or, alternatively, a defect in muscle maturation. Tg mice were assayed for degeneration because it is usually followed by regeneration. Muscles were analyzed for the presence of cells expressing Mac 1, which stains macrophages and neutrophils, because muscle degeneration is usually accompanied by inflammatory cell invasion (Fig. 4 A–D; ref. 20). No areas of the gastrocnemius, soleus, or triceps were observed to contain Mac 1 cells at the concentration or distribution that is characteristic of degenerative muscle.
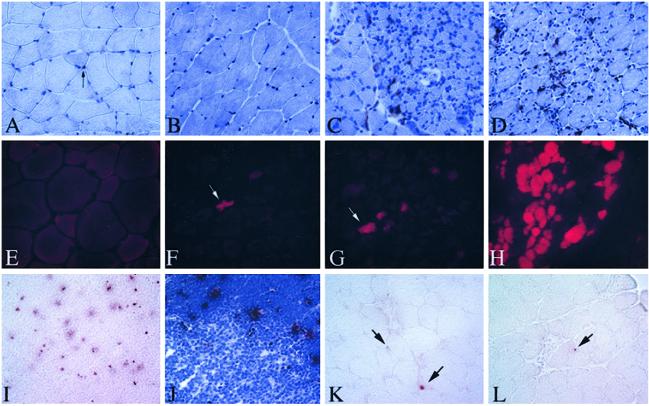
Figure 4.
Absence of indicators of necrosis/degeneration in Tg mice. (A–D) Sections of Tg soleus stained with mac 1 (red) and counterstained with hematoxylin (blue). (A) Non-Tg. (B) C3-Tg. (C) Ex 6− Tg. (D) Ex 15− Tg. (E–H) Triceps muscles from EB-injected mice (E) non-Tg mouse, (F and G) ex 6− Tg, (H) quadriceps from an mdx mouse. EB shows red fluorescence under UV light. (I–L) TUNEL-stained sections. (I and J) TUNEL-stained thymus, positive control. Red dots are positive nuclei. (J) Counterstained with hematoxylin showing the specificity of the TUNEL reaction in the cortical regions where the majority of apoptosis occurs in thymus. (K and L) Gastrocnemius from 6-week, ex 15− Tg. In K, one myonucleus is stained positively. Another positive cell can be seen in the endomysium (upper arrow). In L, an endomysial-positive nucleus is shown (arrow). All micrographs were photographed at the same magnification.
Muscles from Tg lines were next analyzed for the presence of muscle membrane lesions, because membrane damage is a common feature of dystrophic and injured muscle (21). These lesions can be visualized by the presence of the extracellular marker dyes Evans blue (EB), within the muscle fibers. We examined at least two lines from each Tg triceps for EB+ fibers at 6 weeks of age. Few EB+ fibers were observed in any Tg line (Fig. 4 E–G). Therefore, EB dye analysis did not reveal any degenerating areas or compromise of the plasma membrane.
Finally, analysis of serum creatine kinase (CK) was performed because elevated serum CK is a common feature of myopathies and muscle injuries. Increased serum CK was not observed in any of the transgenic lines (data not shown). Taken together, these assays of muscle degeneration support the hypothesis that the small-diameter fibers in ex 6− Tg do not result from degeneration/regeneration.
C3 Tg Mice Do Not Show Prominent Apoptosis.
A feature common to many mouse models and human muscular dystrophies is the presence of apoptotic myonuclei (18, 22). We examined all isoforms of C3 Tg mice for apoptotic nuclei in the triceps, gastrocnemius, and soleus. Although occasional apoptotic nuclei were observed in the endomysial space, few myonuclei were TUNEL-positive. Positive myonuclei, when observed, were found in some small fibers (Fig. 4 K and L).
Evidence for Developmental Immaturity in Ex 6− Tg Muscles.
The data presented above indicate that muscle degeneration/regeneration is not the cause of the small-diameter, centrally nucleated fibers observed in ex 6− Tg. An alternative explanation for the phenotype is that the muscles of these Tg mice are developmentally immature. Support for this hypothesis first came from gene array analysis (data not shown). By using gene chips, many markers of early myogenesis, that are not normally expressed highly in mature muscle, were shown to be up-regulated in the ex 6− Tg including RNA encoding neural cell adhesion molecule (NCAM), embryonic myosin heavy chain (MHC), myf 5, and myf 6.
Immunoblotting (not shown) and immunohistochemistry (Fig. 5 A and B) confirmed the elevated level of NCAM as well as acetylcholine receptor α (AchRα), another gene expressed highly in development, in ex 6− Tg muscles (Fig. 5 A and B). Although many ex 6− Tg muscles showed uniformly elevated NCAM and AchRα staining, the soleus muscle of ex 15− Tg and a few small patches in the C3 Tg also had positive fibers. Both NCAM and AchRα showed a dispersed, sarcolemmal membrane distribution rather than a restricted distribution to the neuromuscular junction that would usually be expected in adult muscle. This distribution is consistent with the immature phenotype of the tissue and lends further support for the idea that these muscles are developmentally immature.
Previous studies have shown that muscle fibers of the “fast” type express higher levels of C3 than fibers of the “slow” type (23). Staining for slow MHC was performed to determine whether the predominantly affected fibers were of the fast or slow type. No correlation was observed between slow MHC expression and developmental phenotype in the ex 6− Tg; however, in the ex 15− Tg, a higher proportion of fibers expressing slow MHC seemed abnormal (Fig. 5C).
Discussion
Previous studies showing rapid autolysis of C3 in vitro suggested that it might be similarly autolyzed in vivo. The inability to express C3 efficiently in vitro without significant degradation (3, 6) and the inability to purify the intact protein by traditional methods (4) has led to questions of whether C3 could be stably expressed from a transgene for use in gene therapy treatments. The results of this study show that C3 can be expressed stably by a transgene, and that expression of mature C3 is not toxic to muscle.
The results of the present study also suggest that the ex 6− isoform of C3 should not be used for gene therapy because its overexpression results in muscle that seems developmentally abnormal. The abnormal muscle does not reflect degeneration/regeneration because ex 6− Tg mice show no observable degenerative areas, they have normal serum CK and do not display evidence of muscle membrane damage. To ensure that the ex 6− Tg phenotype could not have resulted from integration of the transgene into another gene, multiple lines were examined and the phenotype was observed repeatedly. The abnormal morphology of slow fibers in the soleus in the ex 15− Tg also suggests that this transgene might also be deleterious for gene therapy of slow muscle fibers.
The abnormal muscle observed in ex 6− Tg must have resulted from defects in myoblast fusion, or defects in maturation after fusion. Defects in fusion would likely produce few myotubes with many satellite cells in the surrounding endomysium. Because many myotubes and small fibers were identified that were not surrounded by large numbers of mononucleated cells, the likelihood of a fusion defect is minimal. Furthermore, because fusion of primary myotubes occurs before birth, and this transgene is not expressed highly in utero (19), this hypothesis is not supported. Therefore, a more likely scenario is that a postfusion defect in maturation caused the immature muscles.
The in vivo role of C3 has not yet been explored, although mice lacking exons 2 and 3 and a dominant negative Tg have a mild dystrophic phenotype (24, 25). Both of these models express C3 with reduced activity and are models of LGMD2A. In this study, the ex 6− Tg is not a model for LGMD 2A, although these mice also express an isoform of C3 with reduced proteolytic activity. Previous studies in vitro have shown that some LGMD 2A mutations result from loss of C3 activity and not from altered binding to titin (26). Although the common feature between these former studies and the current investigation is altered C3 activity, the lack of a common phenotype between this Tg and those other mouse models suggests that the ex 6− Tg phenotype is not simply explained by the expression of a protease with reduced activity. In this investigation, the constitutive expression of a normally regulated isoform affected the maturation process. The overexpression of a developmental isoform in mature muscle interfered with the function of wt calpain, which may have prevented cleavage of substrate in a developmentally appropriate manner.
Although findings of this study suggest that C3 may have a role in muscle maturation, the late onset of muscle pathology in LGMD 2A indicates that C3 is not essential for production of functional muscle. Although these two observations may seem contradictory, it is possible that the atrophy observed in LGMD 2A humans during adulthood results from a reduced myogenic capacity that is not apparent until it is accompanied by the impaired regenerative capacity of aging muscle. Although mice overexpressing C3 lacking exons 6 and 15 are not developmental models, the embryonic phenotype lends support for a role for C3 in muscle maturation. These findings also suggest that alternatively spliced isoforms of C3 serve specific roles in regulating muscle differentiation and provide evidence that the IS1 and IS2 regions may be important in defining these roles. Perhaps IS1 and IS2 regions help define substrate specificity or their specific splicing may help regulate where and when substrate is cleaved and help distinguish it from other calpains in muscle, particularly m-calpain.
Acknowledgments
We thank K. Wen, W. Winckler, J. Moylan, T. Tranh, G. Yip, E. Lerner, S. Wilsterman, L. Suel, and P. Hajdari for technical contributions; T. Fielder from the University of California, Irvine, Transgenic Facility for expertly microinjecting the Tg constructs; and Dr. R. Crosbie for use of her microscope. Funding for this work was provided by National Institutes of Health Grants RO1 AR48177, RO3 AR45838, and the Muscular Dystrophy Association.
Abbreviations
- C3
calpain 3
- ex
exon
- LGMD
limb-girdle muscular dystrophy
- MHC
myosin heavy chain
- EB
Evans blue
- AchRα
acetylcholine receptor α
- NCAM
neural cell adhesion molecule
- Tg
transgenic
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- wt
wild type
References
- 1.Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 2.Sorimachi H, Suzuki K. J Biochem. 2001;129:653–664. doi: 10.1093/oxfordjournals.jbchem.a002903. [DOI] [PubMed] [Google Scholar]
- 3.Sorimachi H, Toyama-Sorimachi N, Saido T C, Kawasaki H, Sugita H, Miyasaka M, Arahata K, Ishiura S, Suzuki K. J Biol Chem. 1993;268:10593–10605. [PubMed] [Google Scholar]
- 4.Kinbara K, Ishiura S, Tomioka S, Sorimachi H, Jeong S Y, Amano S, Kawasaki H, Kolmerer B, Kimura S, Labeit S, et al. Biochem J. 1998;335:589–596. doi: 10.1042/bj3350589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herasse M, Ono Y, Fougerousse F, Kimura E, Stockholm D, Beley C, Montarras D, Pinset C, Sorimachi H, Suzuki K, et al. Mol Cell Biol. 1999;19:4047–4055. doi: 10.1128/mcb.19.6.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branca D, Gugliucci A, Bano D, Brini M, Carafoli E. Eur J Biochem. 1999;265:839–846. doi: 10.1046/j.1432-1327.1999.00817.x. [DOI] [PubMed] [Google Scholar]
- 7.Sorimachi H, Kinbara K, Kimura S, Takahashi M, Ishiura S, Sasagawa N, Sorimachi N, Shimada H, Tagawa K, Maruyama K, et al. J Biol Chem. 1995;270:31158–31162. doi: 10.1074/jbc.270.52.31158. [DOI] [PubMed] [Google Scholar]
- 8.Cottin P, Brustis J J, Poussard S, Elamrani N, Broncard S, Ducastaing A. Biochim Biophys Acta. 1994;1223:170–178. doi: 10.1016/0167-4889(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 9.Barnoy S, Zipser Y, Glaser T, Grimberg Y, Kosower N S. J Cell Biochem. 1999;74:522–531. doi: 10.1002/(sici)1097-4644(19990915)74:4<522::aid-jcb2>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- 10.Temm-Grove C J, Wert D, Thompson V F, Allen R E, Goll D E. Exp Cell Res. 1999;247:293–303. doi: 10.1006/excr.1998.4362. [DOI] [PubMed] [Google Scholar]
- 11.Crawford G E, Faulkner J A, Crosbie R H, Campbell K P, Froehner S C, Chamberlain J S. J Cell Biol. 2000;150:1399–1409. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan B, Constantini F, Lacey E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. p. 332. [Google Scholar]
- 13.Spencer M J, Walsh C M, Dorshkind K A, Montecino-Rodriguez E, Tidball J G. J Clin Invest. 1997;99:2745–2751. doi: 10.1172/JCI119464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer M J, Croall D E, Tidball J G. J Biol Chem. 1995;270:10909–10914. doi: 10.1074/jbc.270.18.10909. [DOI] [PubMed] [Google Scholar]
- 15.Anderson L V, Davison K, Moss J A, Richard I, Fardeau M, Tomé F M, Hübner C, Lasa A, Colomer J, Beckmann J S. Am J Pathol. 1998;153:1169–1179. doi: 10.1016/S0002-9440(10)65661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raser K J, Posner A, Wang K K. Arch Biochem Biophys. 1995;319:211–216. doi: 10.1006/abbi.1995.1284. [DOI] [PubMed] [Google Scholar]
- 17.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 18.Tidball J G, Albrecht D E, Lokensgard B E, Spencer M J. J Cell Sci. 1995;108:2197–2204. doi: 10.1242/jcs.108.6.2197. [DOI] [PubMed] [Google Scholar]
- 19.Brennan K J, Hardeman E C. J Biol Chem. 1993;268:719–725. [PubMed] [Google Scholar]
- 20.Spencer M J, Montecino-Rodriguez E, Dorshkind K, Tidball J G. Clin Immunol. 2001;98:235–243. doi: 10.1006/clim.2000.4966. [DOI] [PubMed] [Google Scholar]
- 21.Straub V, Rafael J A, Chamberlain J S, Campbell K P. J Cell Biol. 1997;139:375–385. doi: 10.1083/jcb.139.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baghdiguian S, Martin M, Richard I, Pons F, Astier C, Bourg N, Hay R T, Chemaly R, Halaby G, Loiselet J, et al. Nat Med. 1999;5:503–511. doi: 10.1038/8385. [DOI] [PubMed] [Google Scholar]
- 23.Jones S W, Parr T, Sensky P L, Scothern G P, Bardsley R G, Buttery P J. J Muscle Res Cell Motil. 1999;20:417–424. doi: 10.1023/a:1005572125827. [DOI] [PubMed] [Google Scholar]
- 24.Richard I, Roudaut C, Marchand S, Baghdiguian S, Herasse M, Stockholm D, Ono Y, Suel L, Bourg N, Sorimachi H, et al. J Cell Biol. 2000;151:1–9. doi: 10.1083/jcb.151.7.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tagawa K, Taya C, Hayashi Y, Nakagawa M, Ono Y, Fukuda R, Karasuyama H, Toyama-Sorimachi N, Katsui Y, Hata S, et al. Hum Mol Genet. 2000;9:1393–1402. doi: 10.1093/hmg/9.9.1393. [DOI] [PubMed] [Google Scholar]
- 26.Ono Y, Shimada H, Sorimachi H, Richard I, Saido T C, Beckmann J S, Ishiura S, Suzuki K. J Biol Chem. 1998;273:17073–17078. doi: 10.1074/jbc.273.27.17073. [DOI] [PubMed] [Google Scholar]