Abstract
The natural history of follicular lymphoma (FL) is frequently characterized by transformation to a more aggressive diffuse large B cell lymphoma (DLBCL). We compared the gene-expression profiles between transformed DLBCL and their antecedent FL. No genes were observed to increase or decrease their expression in all of the cases of histological transformation. However, two different gene-expression profiles associated with the transformation process were defined, one in which c-myc and genes regulated by c-myc showed increased expression and one in which these same genes showed decreased expression. Further, there was a striking difference in gene-expression profiles between transformed DLBCL and de novo DLBCL, because the gene-expression profile of transformed DLBCL was more similar to their antecedent FL than to de novo DLBCL. This study demonstrates that transformation from FL to DLBCL can occur by alternative pathways and that transformed DLBCL and de novo DLBCL have very different gene-expression profiles that may underlie the different clinical behaviors of these two types of morphologically similar lymphomas.
Histological transformation is a pivotal event in the natural history of cancers, typically coincident with more aggressive clinical behavior. Follicular lymphoma (FL) is often a precursor to a more aggressive lymphoma. FL accounts for 25–40% of all non-Hodgkin's lymphomas (NHL) (1, 2). These tumors apparently derive from follicular center B cells and in the majority of cases they harbor the t(14;18)(q32;q21) chromosomal translocation, resulting in deregulated expression of the antiapoptotic protein BCL-2. Although initially an indolent disease, sensitive to a variety of chemotherapeutic agents, FL is incurable, with a continuous pattern of relapse associated with decreasing sensitivity to chemotherapy (3). Transformation to more aggressive large cell lymphoma occurs in 25–60% of patients with FL (3, 4). In this process a more virulent subclone of cells emerges, typically associated with the loss of follicular histological architecture, a rapidly progressive clinical course refractory to treatment, and short survival. Several secondary genetic abnormalities associated with histological transformation of FL have been described, including a number of nonrandom chromosomal changes. These include gains on 2q, 6p, 7p, 12q, and 17q and losses on 5p and 8q (5–8), c-myc gene rearrangements (9), p53 mutations (10, 11), accumulation of mutations in the 5′ untranslated regulatory region of BCL-6 gene (12), somatic mutations of the translocated BCL-2 gene (13), and inactivation of p16 and p15 by deletions, mutations, and hypermethylation (14, 15). The marked heterogeneity of these secondary aberrations—each observed only in a subset of transformed lymphomas—suggests that no single genetic mechanism is responsible for all of the transformation events.
We hypothesized that the genomic alterations that lead to higher-grade transformation of FL would be accompanied by alterations in the gene-expression program and that systematic analysis of these alterations in the genomic expression program might provide new insights into molecular pathogenesis. We therefore used DNA microarrays to characterize the genome-wide gene-expression patterns associated with morphological transformation of FL, and their relationships to the expression patterns in the antecedent FL and de novo diffuse large B cell lymphoma (DLBCL).
Materials and Methods
Tumor Specimens and Cell Lines.
Sequential biopsy specimens from 12 patients with FL diagnosed and treated with chemotherapy at Stanford University Hospital were selected for this study. Overall, 24 biopsy specimens, 12 obtained at the time of FL diagnosis and 12 subsequent biopsy specimens obtained at the time of morphologic transformation to DLBCL were evaluated. All lymphoma specimens were re-reviewed and classified according to the Revised European-American Lymphoma (REAL) Classification (1). All of the specimens, except tumors from cases IL122 and IL124, in which insufficient number of cells was available, were analyzed by flow cytometry for expression of Ig heavy and light chains and B and T cell markers. Lymph node histology at the time of transformation was classified as DLBCL in all of the 12 specimens (T cell-rich B cell diffuse large-cell lymphoma in specimen IL119B). In five cases (IL105B, IL114C, IL119B, IL122B, and IL125B) no remaining follicles were observed, whereas in the other seven transformed cases scattered residual follicles were seen in addition to the dominant DLBCL component. The clonal relationship between the members of each pair of tumors was confirmed in all cases by examination of Ig and/or BCL-6 genes mutations and translocations. Samples from 11 patients were tested for the presence of BCL-2 rearrangements by PCR. Ten of the tested samples harbored Ig-BCL-2 rearrangements, which were identical between the original FL tumors and the post-transformation tumors.
In addition, we analyzed purified germinal center B (GCB) lymphocytes, 3 DLBCL cell lines (SUDHL6, OCI Ly3, OCI Ly10; ref. 16), and 11 tumor tissues frozen from patients with primary DLBCL. These cell lines and specimens were previously classified as Germinal Center B cell (GCB) type DLBCL (six DLBCL specimens and SUDHL6 cell line) or Activated B cell (ABC) type DLBCL (five DLBCL specimens and OCI Ly3, OCI Ly10 cell lines) (16).
RNA Isolation and Amplification.
All tumor specimens were retrieved from a tissue bank where they were stored either as tissue fragments in Tissue-Tek Optimal Cutting Temperature (OCT) compound 4583 (Miles) or as single-cell suspension in liquid nitrogen. Total cellular RNA or mRNA was isolated from the tumor specimens and cell lines by using the RNeasy kit (Qiagen) or the FAST TRACK 2.0 kit (Invitrogen), respectively, according to the manufacturers' instructions. RNA from all of the specimens and cell lines was subjected to one round of linear amplification as described (17). RNA from specimens IL124 A and B was amplified twice each on two separate days and each amplified sample was analyzed separately to control for amplification reproducibility.
cDNA Microarray Procedures.
The custom DNA microarray used in this study included all of the Lymphochip genes used in our previous studies (16, 18), supplemented with cDNA clones representing additional named UniGene clusters. The microarrays comprised a total of 37,632 spots, representing 32,876 unique cDNA clones, corresponding to ≈17,622 unique UniGene clusters, including at least 10,250 unique named genes. Microarray analysis of gene expression was performed as described (16). In each experiment, fluorescent cDNA probes were prepared from an experimental amplified RNA (aRNA) sample (Cy5-labeled) and a control aRNA sample (Cy3-labeled) isolated from a pool of 11 cell lines (MCF7, Hs578T, NTERA2, Colo205, OVCAR-3, UACC-62, MOLT-4, RPMI-8226, NB4+ATRA, SW872, and HepG2). The use of a common reference cDNA pool allows the determination of the relative expression of each gene across all of the samples. In separate experiments, gene expression was compared directly for each patient between the post-transformation (Cy5-labeled) and the specimen obtained at the time of FL diagnosis (Cy3-labeled). Agglomerative hierarchical clustering was applied either to the gene axis alone or sequentially to both gene axis and to the tumor axis by using the cluster program (M. Eisen, http://rana.lbl.gov/EisenSoftware.htm; ref. 19). The results were analyzed with tree view (M. Eisen; http://rana.lbl.gov/EisenSoftware.htm). The databases, as well as tools for analysis and visualization of the data, are available at http://genome-www.stanford.edu/transformation/index.shtml.
Analysis of the c-myc Gene Mutations.
Genomic DNA was extracted from 5.0 × 106 cells by using a commercially available kit as described by the manufacturer (QIAmp Tissue Kit, Qiagen). The c-myc gene was amplified from the genomic DNA by using primers designed from GenBank accession no. X00364. Exons were amplified using primers complementary to adjacent intron sequence, covering 1,888 base pairs of exon and 827 base pairs of intron sequence. Intron 1 was amplified in two overlapping segments, covering the first 1,010 base pairs. Primer sequences were synthesized as follows: Exon 1 (forward), 5′-GAT CCT CTC TCG CTA ATC TCC G-3′; Exon 1 (reverse), 5′-CAG GAA TGG GAG AAA AGA CAC C-3′; Intron1-part 1 (forward), 5′-5′-ACT TTG CAC TGG AAC TTA CAA CA-3′; Intron1-part 1 (reverse), 5′-AAG CCA AAT GCC AAC TTC TT-3′; Intron1-part 2 (forward), 5′-CGG ACA TTC CTG CTT TAT TGT G-3′; Intron1-part 2 (reverse), 5′-TGC CAG CTT TTC TTC TTT CTC T-3′; Exon 2 (forward), 5′-TCC GCA CCA AGA CCC CTT TAA C-3′; Exon 2 (reverse), 5′-AAG AGT GGC CCG TTA AAT AAG CTG-3′; Exon 3 (forward), 5′-GCT CTT TGG GGA GAT AAT TTT GT-3′; Exon 3 (reverse), 5′-TGA TTG CTC AGG ACA TTT CTG T-3′ (Amersham Pharmacia Life Technologies).
PCR amplification was performed with AmpliTaq Gold (Applied Biosystems) in 30-μl reactions by using a Touchdown PCR protocol: 95°C, 2 min; (95°C, 20 s; 63°C, 1 min, decrease 0.5°C per cycle; 72°C, 1 min) repeat 14 times; (95°C, 20 s; 56°C, 45 s; 72°C, 45 s) repeat 19 times; 72°C, 7 min. The PCR products were then sequenced in both forward and reverse directions by using ABI Big dye terminator chemistry (Applied Biosystems).
Results and Discussion
Gene-Expression Comparison Between FL and Post-Transformation DLBCL.
We profiled gene expression by using DNA microarrays in 12 biopsy specimens obtained at the time of FL diagnosis and 12 biopsy specimens obtained at the time of morphologic transformation to DLBCL. Two different experimental approaches were used: type 1, gene expression was compared directly between pairs of pre- and post-transformation cDNA preparations labeled with different fluorochromes and cohybridized on the same array; or type 2, relative gene expression in each tumor specimen was determined by comparison to a common reference cDNA derived from a pool of 11 cell lines. The type 1 experiments have the advantage of direct measurement of gene-expression changes between the two evaluated specimens and thus were used for evaluation of gene-expression changes associated with the transformation process, whereas in type 2 experiments the use of a common reference allows the relative expression of each gene to be compared across all of the samples. The latter approach was used to compare gene-expression profiles between FL, post-transformation, and de novo DLBCL.
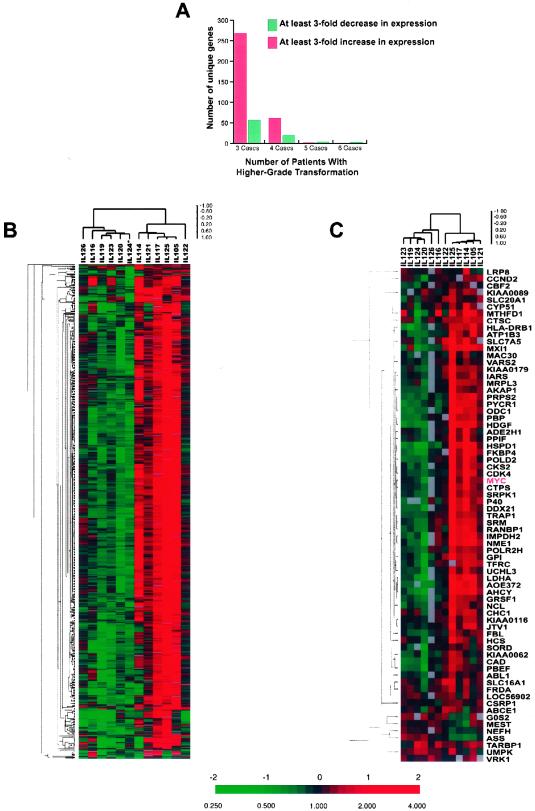
Initially we examined whether the morphological and clinical evolution in higher-grade transformation of follicular lymphomas is associated with prominent gene-expression changes. We searched for consistent features in the changes in gene-expression patterns associated with FL transformation to DLBCL. From flow cytometric data we know that the pre- and post-transformation specimens can contain variable numbers of normal T cells, varying in relative abundance by up to a factor of 1.5 [except in case IL119B, which, by standard criteria, is a T cell-rich B cell lymphoma (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org)]. Therefore, we limited our analysis to genes that exhibited at least a 3-fold variation in expression to reduce the contribution of variation in the number of T cells in the pre- and post-transformation biopsies. In three or more pairs of specimens, 671 unique genes (415 with names and 256 ESTs) met this criterion. None of these 671 genes showed a consistent increase or decrease in expression in all of the cases. In fact, levels of expression of most of these genes changed with transformation in only a subset of cases. Fig. 1A demonstrates in how many cases the 415 unique named genes demonstrated a consistent change. We performed hierarchical clustering of these 671 unique genes and then clustered the 12 cases based on similarity in their pattern of expression of these genes. Distinct clones representing the same gene typically clustered in adjacent rows in this gene map, indicating that each gene has a distinct pattern of expression and that our expression measurements are sufficiently precise to distinguish them. Similarly, expression patterns determined using independent RNA amplifications of tumor pair IL124 were closely matched, clustering together (data not shown). The cases could be divided into two groups based on transformation-associated changes in their patterns of expression of these 671 genes (Fig. 1B). In these two groups of cases expression of the same cluster of genes either increased or decreased on transformation. However, within each of the two subgroups significant residual heterogeneity was apparent, because the changes occurring with transformation in each individual case had distinctive characteristics. The post-transformation survival of patients in these two groups was similar (but the power of this small study to detect differences in survival was very limited).
Figure 1.
Heterogeneous gene-expression changes accompany histological and clinical transformation of FL. (A) Number of transformed FL cases demonstrating at least 3-fold up- or down-regulation of identical unique named genes. The majority of genes demonstrated similar changes in four or less transformed specimens, thus further emphasizing the marked genetic heterogeneity of the transformation process. (B) Hierarchical clustering of the 671 unique genes and 12 transformed FL specimen pairs (type 1 experiments). Two expression profiles each accounting for half of the transformed pairs are disclosed. Expression of these 671 genes was either up- or down-regulated on transformation and none of the genes demonstrated a uniform change in the same direction in the majority of tested tumor pairs. The results of the repeated analyses of case IL124 were averaged and presented as a single value and are marked by *. The ratios are a measure of relative gene expression in each experimental sample and were depicted according to the color scale shown at the bottom. As indicated, the scale extends from fluorescence ratios of 0.25–4 (−2 to + 2 in log base 2 units). Gray indicates missing or excluded data. (C) Hierarchical clustering of the 91 human genes reported as regulated by c-myc and 12 transformed FL specimen pairs. Stratification of the specimen pairs into two major divisions similar to those observed in B.
Closer examination of the genes whose expression changed significantly with transformation suggested possible functional interpretations (see Table 2, which is published as supporting information on the PNAS web site). Some of the alterations in gene expression could be related to the known change in the cell size observed on transformation, including increase in expression of genes involved in the cytoskeleton (e.g., actins, tubulins) and in ribosomal genes (e.g., ribosomal proteins and eukaryotic translation initiation and elongation factors). The 415 named genes with the largest alterations in expression included only one oncogene previously implicated in transformation of FL: the c-myc gene. Many regulatory target genes of the human c-myc gene have recently been identified (20, 21). Fifteen of these c-myc-regulated genes were among the 671 selected genes. The covariation in expression of these 15 c-myc-regulated genes in the samples we analyzed was highly significant (P < 10−7, by hypergeometric distribution). We therefore performed hierarchical clustering of the 12 specimen pairs, based only on the transformation-associated changes in the expression of the 91 reported c-myc-regulated genes that were present on the array irrespective of their levels of expression. Remarkably, this resulted in segregation of the cases into the same two groups (Fig. 1C) as observed in Fig. 1B. This observation suggests that c-myc and its regulated genes may play an important role in histological transformation in one or both of the groups. One group of cases, comprising specimens from five patients (IL121, IL105, IL114, IL117, and IL125), was characterized by transformation-associated increases in expression of c-myc and its target genes. A second group of cases, comprising specimens from four patients (IL123, IL119, IL124, and IL120) was characterized by decreased expression of c-myc and its target genes on transformation. A third, smaller group of cases (IL126, IL116, and IL122), although not distinguished by hierarchical clustering, was characterized by minimal or no change in the expression of c-myc and its target genes.
The c-myc gene is known to be target of chromosomal translocation, amplification, or mutations, all of which may affect the expression of c-myc and its target genes. We therefore performed a thorough search for c-myc gene mutations in each of our cases by sequencing of all of the 3 exons and the first intron of the c-myc gene, regions previously reported to be affected by mutations (22). We found an acquisition of a new mutation on transformation in one case—IL125. In this case a new mutation at position 4698 (C → T), leading to P60S amino acid substitution on transformation was observed. This case exhibited marked increase in c-myc and its target gene expression on transformation (Fig. 1C). This mutation is located in the c-myc box I mutation hot spot region of the gene was previously reported in NHL specimens (23, 24). It is located in the vicinity of the Thr-58 and Ser-62, which are the major sites of c-myc phosphorylation. Mutations in this region were previously reported to deregulate c-myc function by interfering with c-myc protein ubiquitination leading to decreased proteasome-mediated c-myc turnover (24). In addition, mutations in this region were reported to impart to the c-myc protein increased transforming activity (25, 26). We also performed an analysis of c-myc gene amplification in our cases by a microarray based comparative genomic hybridization. No evidence for c-myc gene amplification on transformation was found in any of our cases although a number of amplifications and deletions in other genes could be detected (Martinez-Climent et al., unpublished data). Unfortunately, the amount of genomic DNA available from these cases was insufficient for Southern blot analysis to search for c-myc gene rearrangements; however, from previous reports we would expect no more than one of the tumor cases to have had c-myc gene rearrangement on transformation (9). Proliferation is often accompanied by increased expression of c-myc and its target genes. The observed increase in c-myc and its target gene expression in this group of transformed cases might therefore be a consequence, rather than a cause, of the loss of growth control and accelerated cellular proliferation response in the transformed lymphomas.
In the second group of the transformation cases that exhibited a decrease in expression of c-myc and its target genes, it is likely that the transformation was accompanied by alterations in programmed cell death pathways, because we observed no increase in expression of the genes characteristically associated with loss of growth control and accelerated proliferation. c-myc has been reported to induce apoptosis (27) as well as growth arrest (28). It is therefore possible that the diminished expression of c-myc and/or its target genes may contribute to transformation by impairing these processes.
Comparison of de Novo DLBCL and DLBCL Arising from FL.
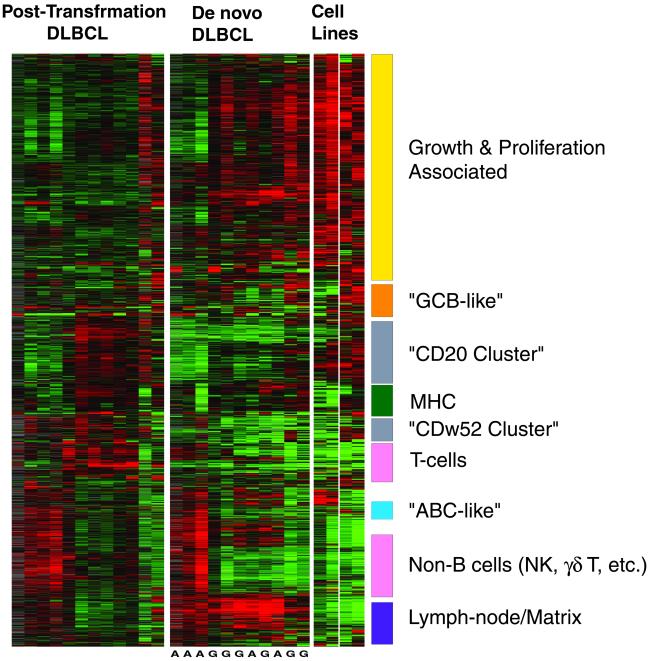
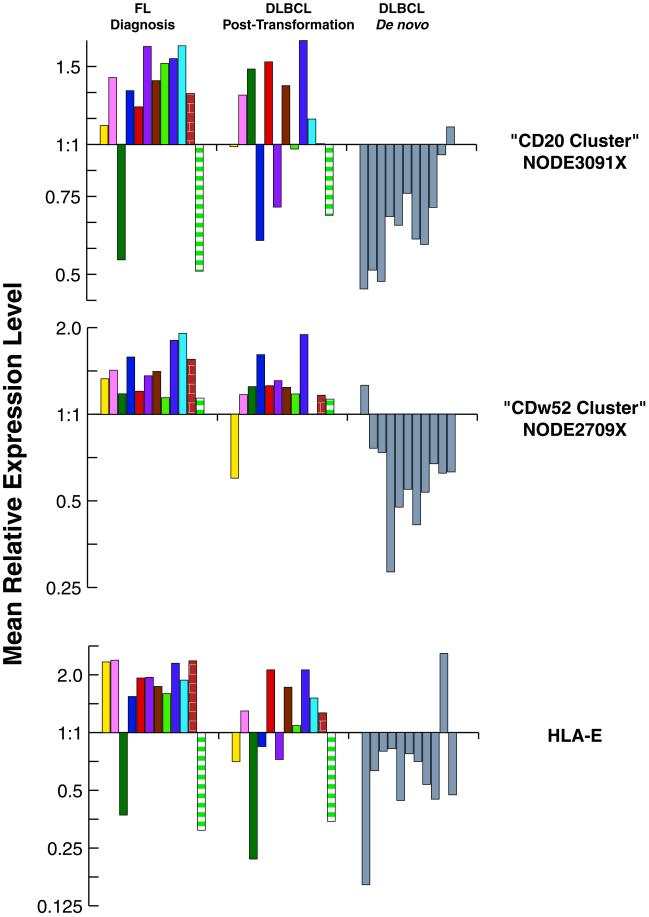
Because post-transformation and de novo DLBCL are clinically distinct, we hypothesized that differences in gene-expression profiles could be identified between these two groups of NHL. We compared gene-expression patterns of the 12 post-transformation DLBCL with those of 11 de novo DLBCL specimens. We first identified a set of 2,205 genes that exhibited at least 3-fold variation from the mean in 2 or more of these 23 cases. Hierarchical clustering of the 23 tumors based on this group of genes could not perfectly separate the post-transformation from the de novo DLBCL, although a trend was observed (data not shown). We therefore searched to identify gene groups with distinct expression patterns by using a different approach. Fig. 2 depicts the expression profiles of the same gene set, with the gene axis hierarchically clustered across both tumor types, whereas on the specimen axis, the tumors have been clustered separately by type (i.e., de novo and transformed DLBCL). As depicted, it is apparent that gene-expression patterns in some of the gene clusters are similar between the post-transformation and the de novo DLBCL, whereas the expression profiles of several gene clusters are quite distinct (Fig. 2). Genes associated with cell proliferation, cell cycle genes, and c-myc target genes were more highly expressed in most of the de novo DLBCL than in most of the post-transformation DLBCL. Genes in a cluster that contains CD20 (“CD20 Cluster”) and a cluster that contains CD52w (“CDw52 Cluster”) (Figs. 2 and 3) were expressed at higher levels in most post-transformation cases than in de novo DLBCL. These two clusters consisted of a heterogeneous group of genes including genes encoding components of B lymphocyte signaling pathways (e.g., CD79A, CD79B, and PAG genes in the “CD52w cluster” and SYK, LYN, BLK, WAS, and WASPIP genes in the “CD20 cluster”), antiproliferative genes (e.g., WEE1 and BTG1) and several surface marker genes. Expression of MME (CD10), MS4A2 (CD20), and CDw52 (CAMPATH1) were all higher in post-transformation DLBCL compared with de novo DLBCL. Most of the major histocompatibility (MHC) cluster of genes (Fig. 2), containing HLA genes and HLA-related genes (e.g., β2 microglobulin, PSMB8, PSMB9), were expressed at similar levels in the post-transformation DLBCL and the de novo DLBCL cases. However, the HLA-E gene exhibited significantly higher expression in most of the post-transformation DLBCL than in the majority of the de novo DLBCL cases (Fig. 3). The difference in the expression of HLA-E is particularly interesting in view of the known function of HLA-E as a major ligand for the natural killer inhibitory receptor (29).
Figure 2.
Gene-expression clusters in post-transformation and de novo DLBCL. Depicted are 2,205 gene-expression measurements made on 27 microarray analyses of 12 post-transformation DLBCL, 11 de novo DLBCL, cell lines, and purified germinal center B (GCB) cells. Each row represents a separate cDNA clone on the microarray and each column a separate mRNA sample. The results presented represent the ratio of hybridization of fluorescent cDNA probes prepared from each experimental mRNA sample to a reference mRNA sample, relative gene expression, depicted according to the color scale shown in Fig. 1. A, ABC type DLBCL; G, GCB type DLBCL [as determined previously for these cases (16)].
Figure 3.
Mean relative expression of genes comprising the “CD20” and the “CD52W” clusters and of the HLA-E gene in FL diagnosis, transformed, and de novo DLBCL specimens. Mean relative expression was calculated from the sum of expression of each gene in the cluster divided by the number of genes in the cluster. Results from different specimens from the same patient are marked by the same color. All of the results in de novo DLBCL are in gray.
To evaluate the expression signatures of genes in these clusters at different stages of the natural history of FL and to compare them to the de novo DLBCL, we have evaluated mean relative expression levels of genes in CD20 and CDw52 clusters and of the HLA-E gene in specimens obtained from FL at diagnosis, at time of transformation to DLBCL, and from de novo DLBCL at diagnosis (Fig. 3). Post-transformation DLBCL exhibited gene-expression signatures more similar to those observed in FL specimens than to those observed in de novo DLBCL specimens (Fig. 3). Some of the transformed DLBCL biopsy specimens contained residual untransformed FL components that could have contributed to a distinct gene-expression signature. However, even the five “pure” specimens, in which FL remnants were not observed, showed the characteristic expression patterns that distinguished the post-transformation from the de novo DLBCL cases. Therefore, these observations suggest that on transformation some of the gene-expression signatures characteristic of FL are conserved.
Another confounding factor that must be considered is the effect of chemotherapy. While all of the de novo DLBCL were evaluated before exposure to chemotherapy, all of the evaluated post-transformation DLBCL had received prior chemotherapy. The similarity of gene-expression signatures in chemotherapy-treated post-transformation DLBCL to those observed in untreated FL specimens suggests that the observed differences between the post-transformation and de novo DLBCL specimens are not related to the effects of chemotherapy but rather represent primary differences in the gene-expression profiles between these two types of DLBCL.
In summary, our results may provide useful molecular markers distinguishing the de novo from the transformed forms of DLBCL, suggest that either loss of proliferation control associated with increased c-myc expression or loss of an apoptotic mechanism associated with a decreased c-myc expression may underlie the histological transformation, and perhaps even guide choice of therapeutic interventions (anti-CDw52 [CAMPATH1] therapy) for a subset of these tumors.
Supplementary Material
Acknowledgments
This work was supported by Grants CA33399, CA34233, and HG01932 from the U.S. Public Health Service–National Institutes of Health and by the Howard Hughes Medical Institute. P.O.B. is an Associate Investigator of the Howard Hughes Medical Institute. R.L. is an American Cancer Society Clinical Research Professor.
Abbreviations
- FL
follicular lymphoma
- DLCBL
diffuse large B cell lymphoma
References
- 1.Harris N L, Jaffe E S, Stein H, Banks P M, Chan J K, Cleary M L, Delsol G, De Wolf-Peeters C, Falini B, Gatter K C. Blood. 1994;84:1361–1392. [PubMed] [Google Scholar]
- 2.Harris N L, Jaffe E S, Diebold J, Flandrin G, Muller-Hermelink H K, Vardiman J, Lister T A, Bloomfield C D. J Clin Oncol. 1999;17:3835–3849. doi: 10.1200/JCO.1999.17.12.3835. [DOI] [PubMed] [Google Scholar]
- 3.Horning S J, Rosenberg S A. N Engl J Med. 1984;311:1471–1475. doi: 10.1056/NEJM198412063112303. [DOI] [PubMed] [Google Scholar]
- 4.Acker B, Hoppe R T, Colby T V, Cox R S, Kaplan H S, Rosenberg S A. J Clin Oncol. 1983;1:11–16. doi: 10.1200/JCO.1983.1.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Armitage J O, Sanger W G, Weisenburger D D, Harrington D S, Linder J, Bierman P J, Vose J M, Purtilo D T. J Natl Cancer Inst. 1988;80:576–580. doi: 10.1093/jnci/80.8.576. [DOI] [PubMed] [Google Scholar]
- 6.Richardson M E, Chen Q G, Filippa D A, Offit K, Hampton A, Koduru P R, Jhanwar S C, Lieberman P H, Clarkson B D, Chaganti R S. Blood. 1987;70:444–447. [PubMed] [Google Scholar]
- 7.Yunis J J, Frizzera G, Oken M M, McKenna J, Theologides A, Arnesen M. N Engl J Med. 1987;316:79–84. doi: 10.1056/NEJM198701083160204. [DOI] [PubMed] [Google Scholar]
- 8.Hough R E, Goepel J R, Alcock H E, Hancock B W, Lorigan P C, Hammond D W. Br J Cancer. 2001;84:499–503. doi: 10.1054/bjoc.2000.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yano T, Jaffe E S, Longo D L, Raffeld M. Blood. 1992;80:758–767. [PubMed] [Google Scholar]
- 10.Sander C A, Yano T, Clark H M, Harris C, Longo D L, Jaffe E S, Raffeld M. Blood. 1993;82:1994–2004. [PubMed] [Google Scholar]
- 11.Lo Coco F, Gaidano G, Louie D C, Offit K, Chaganti R S, Dalla-Favera R. Blood. 1993;82:2289–2295. [PubMed] [Google Scholar]
- 12.Lossos I S, Levy R. Blood. 2000;96:635–639. [PubMed] [Google Scholar]
- 13.Matolcsy A, Casali P, Warnke R A, Knowles D M. Blood. 1996;88:3937–3944. [PubMed] [Google Scholar]
- 14.Elenitoba-Johnson K S, Gascoyne R D, Lim M S, Chhanabai M, Jaffe E S, Raffeld M. Blood. 1998;91:4677–4685. [PubMed] [Google Scholar]
- 15.Pinyol M, Cobo F, Bea S, Jares P, Nayach I, Fernandez P L, Montserrat E, Cardesa A, Campo E. Blood. 1998;91:2977–2984. [PubMed] [Google Scholar]
- 16.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 17.Wang E, Miller L D, Ohnmacht G A, Liu E T, Marincola F M. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 18.Alizadeh A, Eisen M, Davis R, Ma C, Sabet H, Tran T, Powell J, Yang L, Marti G, Moore D, et al. Cold Spring Harbor Symp Quant Biol. 1999;64:71–78. doi: 10.1101/sqb.1999.64.71. [DOI] [PubMed] [Google Scholar]
- 19.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coller H A, Grandori C, Tamayo P, Colbert T, Lander E S, Eisenman R N, Golub T R. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm G W, Laux G, Polack A, Weidle U H, Eick D. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti R S, Kuppers R, Dalla-Favera R. Nature (London) 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 23.Raffeld M, Yano T, Hoang A T, Lewis B, Clark H M, Otsuki T, Dang C V. Curr Top Microbiol Immunol. 1995;194:265–272. doi: 10.1007/978-3-642-79275-5_31. [DOI] [PubMed] [Google Scholar]
- 24.Bahram F, von der Lehr N, Cetinkaya C, Larsson L G. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- 25.Hoang A T, Lutterbach B, Lewis B C, Yano T, Chou T Y, Barrett J F, Raffeld M, Hann S R, Dang C V. Mol Cell Biol. 1995;15:4031–4042. doi: 10.1128/mcb.15.8.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pulverer B J, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett J R. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 27.Prendergast G C. Oncogene. 1999;18:2967–2987. doi: 10.1038/sj.onc.1202727. [DOI] [PubMed] [Google Scholar]
- 28.Felsher D W, Zetterberg A, Zhu J, Tlsty T, Bishop J M. Proc Natl Acad Sci USA. 2000;97:10544–10548. doi: 10.1073/pnas.190327097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty D E. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.