Abstract
Peutz–Jeghers syndrome (PJS) is a dominantly inherited human disorder characterized by gastrointestinal hamartomatous polyposis and mucocutaneous melanin pigmentation. LKB1 (STK11) serine/threonine kinase is the product of the causative gene of PJS, which has been mapped to chromosome 19p13.3. However, several studies have produced results that are not consistent with a link between LKB1 gene mutation and PJS. We constructed a knockout gene mutation of Lkb1 to determine whether it is the causative gene of PJS and to examine the biological role of the Lkb1 gene. Lkb1−/− mice died in utero between 8.5 and 9.5 days postcoitum. At 9.0 days postcoitum, Lkb1−/− embryos were generally smaller than their age-matched littermates, showed developmental retardation, and did not undergo embryonic turning. Multiple gastric adenomatous polyps were observed in 10- to 14-month-old Lkb1+/− mice. Our results indicate that functional Lkb1 is required for normal embryogenesis and that it is related to tumor development. The Lkb1+/− mouse is suitable for studying molecular mechanism underlying the development of inherited gastric tumors in PJS.
Keywords: tumor‖embryo development
Peutz–Jeghers syndrome (PJS) is an autosomal dominant disease characterized by the presence of melanin spots on the lips and buccal mucosa and multiple gastrointestinal hamartomatous polyps (1, 2). The incidence of PJS is estimated to be 1 in 120,000 births (3). PJS patients demonstrated the following frequencies of polyps: small bowel, 96% of patients; colon, 27%; rectum, 24%; and stomach, 24% (4). The PJS locus has been mapped to chromosome 19p13.3, and the LKB1 serine/threonine kinase gene (GenBank accession no. U63333) at chromosome 19p13.3 had been identified as the causative gene of PJS (5, 6). However, several studies have reported finding that PJS is not linked to LKB1 gene mutation (7–9). Recently, it has been reported that Lkb1 plays an important role in the vascular endothelial growth factor signaling pathway and embryonic vascular formation (10). In the present study, to determine the causative gene of PJS and analyze the biological role of Lkb1, we created mice lacking Lkb1 by targeted mutagenesis. Our mouse model of human PJS patients appears to be suitable for use in further studies of the onset of PJS.
Materials and Methods
Cloning of Mouse Lkb1 Gene.
Mouse Lkb1 genomic cosmid clones were purchased from the Resource Center of the German Human Genome Project (Berlin), after screening of a prespotted library (strain 129/Ola) by hybridization with human LKB1 cDNA as the probe. Two positive clones (P2436Q3 and L07209Q3) were identified. Mouse Lkb1 gene fragments were amplified from the two positive cosmid clones by PCR, using primers based on the mouse Lkb1 cDNA sequence (GenBank accession no. AB015801). These amplified fragments were sequenced directly. Comparison of the mouse sequence with that of the human gene revealed that both cosmid clones lacked exon 1 and part of intron 1. Consequently, using primers for intron 1 (5′-ACTGCAGCTGACCCAAGCCAGGAT-3′) and the 5′ untranslated region of mouse Lkb1 cDNA (5′-CGAAGGACAGAGGACAAAGAGTGG-3′), we amplified a region including intron 1 and exon 1 from normal mouse genomic DNA. The sequence of the complete mouse Lkb1 gene was determined by combining these sequences (the sequence of the mouse Lkb1 gene was deposited into GenBank with accession no. AB026255).
Generation of Lkb1 Knockout Mice.
Mouse Lkb1 gene fragments were subcloned from a cosmid clone containing intron 1 (≈7 kb) to a DT-A cassette B plasmid vector (11), which contains a diphtheria toxin A gene fragment as a negative selection marker. A fragment of PGK-neo cassette flanked by two loxP sequences (5′-ATAACTTCGTATAGCATACATTATACGAAGTTAT-3′) was generated and inserted into the HindIII site of intron 1. The targeting vector was then constructed by inserting a loxP fragment into a HindIII site immediately downstream from exon 8 (Fig. 1a). AB2.2-Prime embryonic stem (ES) cells (Lexicon Genetics, The Woodlands, TX) were transfected by electroporation with a linearized targeting vector, and then cultured in a medium supplemented with 300 μg/ml G418 (GIBCO/BRL). G418-resistant clones were screened by PCR using the primers LOXP3 S2 (5′-CCGGTGTTCCACATAACTTC-3′) and MPJ37 (5′-GTTTCCCAAGCTTTATTTATTGCC-3′). The presence of the 3-loxP allele in KpnI-cut genomic DNA from ES cell clones was previously confirmed by Southern blot analysis. ES cells were injected into C57BL/6J (CLEA Japan, Tokyo) blastocyst (12). To obtain Lkb1 3-loxP heterozygous mutants, male chimeras were mated with C57BL/6J females. To produce null mutant mice in which the deleted region of Lkb1 encompasses exons 2–8, the circular expression vector pCre-pac (13) was microinjected into pronuclei of fertilized C57BL/6J eggs that had been inseminated with Lkb1 3-loxP heterozygous mutant spermatozoa. These eggs were cultured, and then transferred into the oviducts of ICR (CLEA Japan) pseudopregnant recipients. Screening for deletion of exons 2–8 was accomplished by performing PCR on DNA from cells at the tip of the tail. Genotyping by PCR, with Takara Ex Taq (Takara, Kyoto), was performed with the following primers: MPJ37 and MPJ69 (5′-CCTTTGGCTGCTGGGTGACTTCTG-3′) (Fig. 1a). After an initial hot start at 94°C for 2 min, 36 cycles (94°C for 30 sec, 68°C for 3 min) were run.
Figure 1.
Mouse Lkb1 gene targeting. (a) Structures of mouse Lkb1 gene (exons 1–10), targeting vector, and the predicted structures of targeted and null alleles, which can be generated from targeted allele by Cre-loxP-mediated recombination. In the targeting vector, the neomycin gene (neor) flanked by two loxP sequences was inserted in the HindIII site at intron 1, and one more loxP sequence was inserted in another HindIII site just after exon 8. Localization of each PCR primer and probes for Southern blots used to detect each allele are indicated. The positions of restriction sites of BamHI (B) and KpnI (K) are also indicated. (b) PCR analysis (PCR, Left) and Southern blot analysis (STN, Right) of ES cell clones. In Southern analysis, DNAs from each ES cell clone (whose clone number is indicated above the gel) and normal mouse tissue (Normal) were digested with KpnI and hybridized with probe A. To detect targeted alleles by PCR, LOXP3 S2 and MPJ37 primers were used. The bands derived from wild-type allele (W) and targeted allele (T) are indicated by arrows. (c) Southern blot analysis of Lkb1 3-loxP heterozygous mice, which were generated from two independent ES cell clones (clones 256 and 358). Southern blot analysis was performed as in b. (d) PCR analysis of Lkb1+/− offspring generated by pCre-pac plasmid injection into pronuclei of fertilized C57BL/6J eggs that were inseminated with Lkb1 3-loxP heterozygous mutant spermatozoa. MPJ69 and MPJ37 primers were used to detect the null allele (N), which lacks exons 2–8, flanked by loxP sequences.
Analysis of Embryos.
At 8.5 to 17.5 days postcoitum (dpc) (plug date, 0.5 dpc), the embryos were dissected free of the decidua and yolk sac, the amnion was removed, and the embryos were then photographed under stereomicroscopic observation. Tissue from embryo tail was used for DNA analysis. This tissue was suspended in 50 μl of cell lysis buffer (0.5% Nonidet P-40/800 ng/ml proteinase K/1× PCR buffer), and incubated at 55°C for 2 h. It was boiled at 95°C for 15 min, and then chilled on ice. PCR analysis was then performed by using 1-μl samples of this suspension. Genotyping by PCR, with Takara Ex Taq, was performed with the following primers: MPJ54 (5′-TGAGACAGCCACCTGAGGGA-3′), MPJ56 (5′-ACAGAGCTCTCCAAGGGAGA-3′), MPJ69, MPJ105 (5′-GCCGAGGCAGGGCTTAGCGA-3′), MPJ107 (5′-AGTACAGGCACAAGGAGATG-3′), and MPJ109 (5′-TGCCTGGGTATCAGACCTGA-3′) (Fig. 1a). To detect null and wild-type alleles, the PCR products generated by MPJ69/MPJ56 primers and MPJ54/MPJ56 primers were subjected to nested PCR with MPJ109/MPJ 107 primers (null allele) and MPJ105/MPJ107 primers (wild-type allele), respectively. After an initial hot start at 94°C for 2 min, 36 cycles (94°C for 30 sec, 68°C for 3 min) were run.
Immunohistochemistry of Lkb1 in Embryo.
Wild-type pregnant mice at 8.5 and 9.0 dpc were killed and perfused with periodate-lysine-paraformaldehyde (PLP) fixative containing 4% paraformaldehyde. Wild-type embryos were removed and fixed with PLP fixative for another 6–8 h at 4°C. The specimens were then dehydrated in acetone for 16 h at 4°C. Acetone was then removed, and the specimens were embedded in paraffin and sectioned (14). The sections were deparaffinized by immersion in xylene and acetone, and then subjected to immunohistochemical staining. After treatment with blocking reagent (Block Ace, Snow Brand Milk Production, Tokyo), the sections were incubated with one of the following antibodies for 16 h at 4°C: rabbit anti-Lkb1 P6 antibody (1 μg/ml) (15), raised against a peptide mapping near the amino terminus (amino acids 27–45) of mouse Lkb1; goat anti-Lkb1 D19 (Santa Cruz Biotechnology), raised against a peptide near the amino terminus (amino acid positions unknown) of mouse Lkb1; or normal rabbit IgG (1 μg/ml, Dako). Bound antibody was visualized by incubating sections with biotin-labeled goat anti-rabbit IgG antibody (Vector Laboratories) or rabbit anti-goat IgG antibody (Vector Laboratories), followed by incubation with horseradish peroxidase-conjugated streptavidin (Vector Laboratories) and reaction with 0.05% diaminobenzidine (Wako Pure Chemical, Osaka) in 0.05 M Tris⋅Cl (pH 7.4), 0.05% sodium azide, and 0.036% H2O2. The sections were counterstained with hematoxylin.
Fecal Occult Blood Tests.
Kits using Shionogi A (orthotolidine) and Shionogi B (guajac) slides (Shionogi, Osaka) were used for fecal occult blood tests. Fecal samples from Lkb1+/− and wild-type mice were spread on the slides and examined by using the protocol recommended by the manufacturer. In this test, samples were considered positive if both Shionogi A and B kits gave positive results.
Histological Analysis of Polyps.
Tissues from the glandular stomach and intestine were fixed with neutralized 10% formalin solution, embedded in paraffin, cut into 4- to 5-μm sections, stained with hematoxylin/eosin or azan, and examined microscopically.
Analysis of Lkb1 Expression in the Polyps by Northern Blotting and Reverse Transcriptase–PCR.
Total RNA was extracted from the glandular stomach of wild-type and Lkb1+/− mice by using ISOGEN (Nippon Gene, Toyama, Japan). Next, 20 μg of total RNA was electrophoresed on 1% agarose gel and transferred to a Hybond N+ membrane. These membranes were hybridized overnight at 42°C in a buffer containing 50% formamide, 5× saline-sodium phosphate EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 0.5% SDS, 5× Denhardt's solution, 10% dextran sulfate, 250 μg/ml denatured salmon sperm DNA, and a 32P-labeled mouse Lkb1 cDNA probe. The membranes were washed for 30 min in 2× SSC and 0.2% SDS at 65°C, and then for 30 min in 0.5× SSC and 0.2% SDS at 65°C. cDNA was synthesized from total RNA of the glandular stomach of wild-type and Lkb1+/− mice by using the Thermoscript reverse transcriptase–PCR system (GIBCO/BRL). RNA was primed with 2.5 μM of the primer MPJ23 (5′-AGAAACCAACCCAAAGACAG-3′), which is specific for the exon 10 noncoding region of Lkb1. Amplification was performed by using Takara Ex Taq (Takara), with the following primer pairs: LKS1 (5′-ATGGACGTGGCGGACCCGA-3′), specific for exon 1, and MPJ18 (5′-GGTGAAGTCTCCTCTCCCAATGTT-3′), specific for exon 6; MPJ4 (5′-TTCAAGGTGGACATCTGGT-3′), specific for exon 5, and LKAS1 (5′-CACTGCTGCTTGCAGGCCGA-3′), specific for exon 9. After an initial hot start at 94°C for 2 min, 40 cycles (94°C for 30 sec, 60°C for 30 sec, 72°C for 2 min) were run. The PCR products were cloned into the plasmid by using the pGEM-T Easy Vector system (Promega). They were then sequenced by using an Applied Biosystems 377 automated DNA sequencer and the BigDye terminator cycle sequencing version 2.0 ready reaction (Perkin–Elmer), with T7 and SP6 primers. For each polyp, the sequences of more than 20 plasmid clones were examined.
All experiments described in this article were conducted in accordance with the Guiding Principles for the Care and Use of Research Animals promulgated by Chugai Pharmaceutical.
Results
Generation of Lkb1 Mutant Mice.
The homologous recombinant ES cell clones were injected into blastocysts to generate the chimeras. The chimeras of four ES cell clones (Fig. 1b) were bred with C57BL/6J to produce Lkb1 3-loxP heterozygous mice. We obtained two independent mutant mouse lines, which had 3-loxP sequences (Fig. 1c). Mice of the 256 and 358 lines were bred and used for further analysis. To establish the Lkb1+/− mice, in which the region of Lkb1 encompassing exons 2–8 is deleted, pCre-pac was injected into Lkb1 3-loxP heterozygous fertilized eggs. Seventy-five injected eggs were transferred into recipients. Three of 12 resultants were identified as Lkb1+/− mice with deleted exons 2–8 resulting from Cre-loxP recombination (Fig. 1d).
Lack of Functional Lkb1 Gene Results in Death in Utero.
At 9.5 dpc, most Lkb1−/− embryos had been resorbed (genotyping by PCR; data not shown). At 8.5 dpc, live Lkb1−/− embryos could still be recovered (Table 1). The Lkb1−/− embryos recovered at 9.0 dpc were significantly smaller and exhibited developmental retardation, compared with wild type (Fig. 2). The Lkb1−/− embryos did not undergo embryonic turning, formed only about 10 pairs of somites, and resembled wild-type embryos at 8.0–8.5 dpc (Fig. 2 b and c). The body and head sizes of Lkb1−/− embryos at 9.0 dpc were equivalent to those of wild-type embryos at 8.5 dpc. However, the sizes of somites in Lkb1−/− embryos at 9.0 dpc were less than half of those of wild-type embryos at 8.5 dpc.
Table 1.
Genotypes of embryos and live young from intercrosses of LKB-1 heterozygous mice
| dpc | No. of embryos
|
||||
|---|---|---|---|---|---|
| Genotype
|
Resorbed | Total | |||
| Wild type | Hetero | Homo | |||
| 3.5 | 3 | 13 | 3 | — | 19 |
| 8.5 | 6 | 19 | 5 | 4* | 34 |
| 9.5 | 8 | 10 | 0 | 8† | 26 |
| 10.5 | 3 | 12 | 0 | 2 | 17 |
| 16.5–17.5 | 4 | 17 | 0 | 7 | 28 |
| Postnatal | 55 | 91 | 0 | — | 146 |
Included four hetero.
Included four homo, one hetero, and three of unknown genotype.
Figure 2.
Morphology of Lkb1−/− embryo and expression of Lkb1 in wild-type embryo. (a–d) Morphology of Lkb1−/− embryos. Wild-type (a) and Lkb1−/− (c and d) at 9.0 dpc and wild-type (b) at 8.5 dpc. The size of Lkb1−/− embryo was similar to that of normal embryos of 8.0–8.5 dpc. Note the Lkb1−/− embryos did not undergo embryonic turning and had very small somite (s). (e–g) Immunostaining of Lkb1 in wild-type embryo at 8.5 dpc (e) and 9.0 dpc (f and g) using anti-Lkb1 antibody. The heart (g) and nucleated embryonic blood cells (f) of wild-type embryo at 9.0 dpc immunostained for Lkb1. S: somite. Only one nucleated embryonic blood cell (e) of wild-type embryo at 8.5 dpc was immunostained for Lkb1. (Magnifications: a–d, ×40; e and f, ×400; g, ×100.)
Expression Patterns of Lkb1 Protein in Embryos.
Immunohistochemical analysis was performed to identify the cells that expressed Lkb1. In wild-type embryos at 9.0 dpc, nucleated embryonic blood cells bound anti-Lkb1 P6 antibody and anti-Lkb1 D19 antibody (Fig. 2f), and heart tissue bound anti-Lkb1 D19 antibody (Fig. 2g). However, few embryonic blood cells in wild-type embryos at 8.5 dpc bound anti-Lkb1 P6 antibody (Fig. 2e). When normal IgG was used instead of these antibodies, no cells in wild-type embryos at 9.0 dpc were stained (data not shown).
Lkb1+/− Mice Developed Gastrointestinal Polyps.
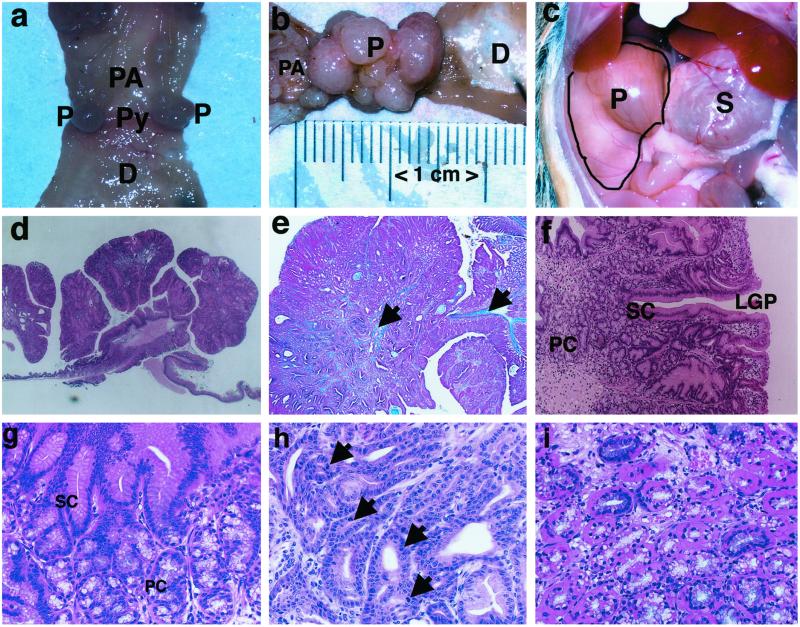
No occult blood was detected in wild-type mice aged 4–14 months. However, 36% and 82% of Lkb1+/− mice aged 4 and 14 months, respectively, were positive for occult blood (Table 2). Macroscopic examination revealed aggregates of polyps in 10-month-old (but not in 4-month-old) mice positive for occult blood (Fig. 3a). The size of these polyp aggregates was almost similar to that of the stomach in 14-month-old Lkb1 mutant mice (Fig. 3 b and c). Most polyps localized at the junction of pyloric antrum and duodenum in the glandular stomach. Found moribund or immediately after death, Lkb1+/− mice were found to have food contents in the intestine, in contrast to the glandular stomach filled with food (Fig. 3c). Most Lkb1+/− mice only had polyps in the glandular stomach; only one of the 12 Lkb1+/− mice had polyps in both the small intestine and glandular stomach. No polyps were found in other organs including large intestine examined (Table 3). The spleens of 14-month-old Lkb1+/− mice were enlarged after bleeding from polyps. These mice also exhibited, at the pylorus, polypoid tumorous growth into the gastric lumen (Fig. 3d). The core of the polyp was formed by a tree-like branching of the muscularis mucosa with connective tissue (Fig. 3e). Polyps consisted of two types of cells: cells that resembled surface mucous cells and cells that resembled those of a pyloric gland. The relative proportions of these cells varied, but all polyps exhibited differentiation into a structure resembling a pyloric gland. The surface of the polyps was covered with cells that resembled mucous cells. These cells extended deep into the mucosa, forming glandular tubules that resembled gastric pits (Fig. 3f); all of these cells were columnar, and some were hypertrophic with a high columnar appearance (Fig. 3g). In a relatively deep region of each polyp, cells that resembled those of pyloric glands, with large nuclei and pale cytoplasm, formed a glandular lumen. Continuities of surface cells that resembled mucous cells and cells that resembled those of pyloric glands were seen in some areas of the polyps (Fig. 3f). The cells that formed glandular structures exhibited less nuclear density and less nuclear atypia than other cells and exhibited a single-layer arrangement (Fig. 3g). There was no multilayered growth, but nuclear density was high in some parts of the polyps, and nuclear atypia and increased mitosis were also observed in some polyp cells (Fig. 3h). Few of the cells that resembled those of pyloric glands contained eosinophilic material in the cytoplasm near the basement membrane (Fig. 3i). Cystic dilatations mainly lined with cells that resembled those of pyloric glands were also observed. Edema was observed in the superficial region of the polyps. Fibrosis and infiltration by mononuclear cells was occasionally seen in the interstitium. There was no extensive necrosis or invasion by proliferating cells into the submucosa or the muscularis layer. The polypoid tumors found in the gastric pylorus were diagnosed as adenomatous polyps, based primarily on the following features: proliferation of well-differentiated glandular stomach cells in a polypoid pattern, very little atypia, and no definite invasion into the submucosa or the muscularis layer.
Table 2.
Results of occult blood tests in LKB1 mutant mice
| Age | Genotype | No. of mice
|
||
|---|---|---|---|---|
| Total no. of mice tested | Positive | Negative | ||
| 4 months | Hetero | 11 | 4 | 7 |
| Wild type | 6 | 0 | 6 | |
| 14 months | Hetero | 10 | 9 | 2 |
| Wild type | 10 | 0 | 10 | |
Figure 3.
Macroscopic and histological appearance of Lkb1+/− mice. (a–c) Macroscopic appearance of polyps in Lkb1+/− mice. Gross appearance of the polyps (P) at junction of pyloric antrum (PA) and duodenum (D). Pylorus (Py) (a) 10 months, (b) 14 months. (c) The size of the polyp is almost equivalent to that of the glandular stomach at 14 months of age. Note that the duodenum and small intestine contained no food. S: glandular stomach; P: polyps. (d–i) Histological examination of polyps at 14 months. (d and f–i) Hematoxylin and eosin staining. (e) Azan stain. (d) Low-power section of the polyp protruding into the gastric lumen. (e) The core of the polyp is formed by a tree-like branching of the muscularis mucosa with connective tissue (arrow). (f) The polyps consist of two types of cells: surface mucous epithelium-like cells (SC) and pyloric gland-like cells (PC) and form glandular tubules like gastric pits (LGP). (g) Surface mucous epithelium-like cells of columnar or high columnar type (SC). Pyloric gland-like cells have large nuclei and pale cytoplasm (PC). (h) High nuclear density, nuclear atypia, and mitosis (arrowhead) are observed in a proportion of proliferating cells. (i) Pyloric gland-like cells also contain eosinophilic material in the cytoplasm. (Magnifications: a–c, ×2; d, ×5; e, ×20; f, ×100; g–i, ×200).
Table 3.
Location of polyps in LKB1 mutant mice
| Genotype | No. of mice
|
|||
|---|---|---|---|---|
| Total | Location of polyps
|
|||
| Stomach | Intestine | Other organs | ||
| Wild type | 6 | 0 | 0 | 0 |
| Hetero | 12 | 12 | 1 | 0 |
These mice were killed and observed at 11–20 months old.
Lkb1 Expression in Gastric Polyps.
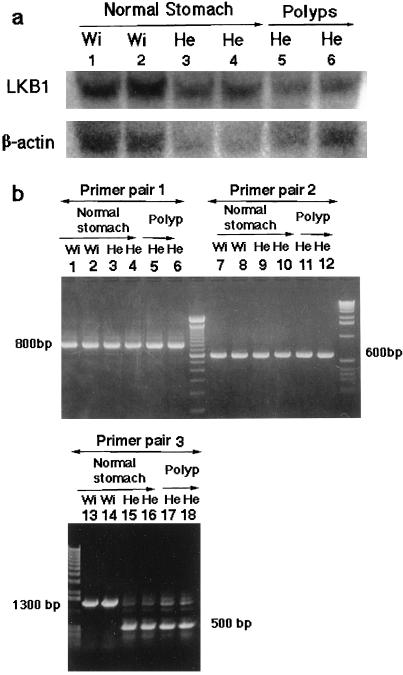
The wild-type Lkb1 gene was expressed in gastric polyps in Lkb1+/− mice, in a pattern similar to that of Lkb1 expression in glandular stomach cells of control mice, but the signaling intensity was approximately half the control value (Fig. 4a). Both mutant and wild-type Lkb1 mRNAs were detected in the polyps of Lkb1+/− mice (Fig. 4b). The sequencing of reverse transcriptase–PCR products cloned into plasmids showed that wild-type mRNAs in the polyps from two Lkb1+/− mice did not have any mutations (data not shown).
Figure 4.
Northern blot and reverse transcriptase–PCR analysis of Lkb1 in polyps. (a) Lanes 1–6: 20 μg total RNA was isolated from 18-month-old Lkb1+/− and wild-type mice. Lanes 1 and 2: glandular stomach of wild-type (wi). Lanes 3 and 4: normal parts of the glandular stomachs of Lkb1+/− (He). Lanes 5 and 6: gastric polyps of Lkb1+/−. Lanes 3 and 5, and 4 and 6: RNAs from the same mouse. The mouse Lkb1 cDNA was used for probe. Blots were reprobed for β-actin, which was used as the loading control. (b) Lanes 1–6: amplification by primer pair 1, LKS1 located in exon 1 and MPJ18 located in exon 6. Lanes 7–12: amplification by primer pair 2, MPJ4 located in exon 5 and LKAS1 located in exon 9. Lanes 13–18: amplification by primer pair 3 LKS1 located in exon 1 and LKAS1 located in exon 9. Lanes 1, 2, 7, 8, 13, and 14: glandular stomach of wild-type (wi); lanes 3, 4, 9, 10, 15, and 16: normal part of the glandular stomach of Lkb1+/− (He); lanes 5, 6, 11, 12, 17, and 18: gastric polyps of Lkb1+/−. Note that for lanes 15–18 the mutant mRNAs (about 500 bp) from targeted allele, deleted exons 2–8, were expressed in heterozygous mice.
Discussion
In the present study, we demonstrated LKB1 to be the causative gene of PJS, as shown by the fact that polyps were found in all Lkb1+/− mice. Previous findings that argue against the involvement of LKB1 gene mutation in PJS may be the result of differences in methods of mutational analysis, locus heterogeneity, epigenetic inactivation of LKB1 (16), or the involvement of a second gene in induction of PJS.
The histological characteristics of polyps in the Lkb1+/− mice were similar to those of polyps found in human PJS. These include epithelium misplacement, the presence of many cells of normal appearance, rare or nonexistent nuclear atypia, and lack of lymphatic invasion, suggesting early-stage adenomatous origin (17, 18). In human PJS, the most common site of polyps is the small intestine, followed by the large intestine and stomach (19, 20). However, in the present Lkb1+/− mice, the polyps were predominantly located in the glandular stomach. Differences in preferential sites for tumors were observed in mice and humans with regard to adenomatous polyposis coli (APC) mutation (21). The most polyps in patients with familial adenomatous polyposis exhibiting APC mutation and polyps in Apc mutant mouse are found in the large and small intestines, respectively. Such differences are probably caused by differences in diet and/or effects of modifier genes. In the present study, histological evidence of progression of gastric polyps from hamartomatous to adenomatous to carcinomatous lesions was not found in Lkb1+/− mice until they were at least 14 months old. In a case study of a human twin with PJS, one of the twins died from intestinal obstruction at the age of 20 years, and the other died from cancer at the age of 52 years (1, 22). Early manifestations in PJS patients include multiple hamartomatous polyps (1, 2). Later in life, PJS patients have an increased incidence of cancer (23, 24). Lkb1+/− mice may die before the onset of carcinomatous changes and metastasis, as a result of intestinal obstruction and/or bleeding from polyps.
Previous studies have shown negative immunostaining for LKB1 of polyps from PJS patients (25), and we assessed loss of heterozygosity (LOH) by using mRNA isolated from whole polyps of Lkb1+/− mice. However, our results do not suggest that LOH was responsible for polyp formation in the Lkb1+/− mice. Our findings regarding expression of mRNA from the mutant Lkb1 allele indicate that it is possible that a dominant negative mechanism directly participates in polyp formation. Because the mutant protein might have at least a 96-aa protein (exon 1). Or, because the expression levels of Lkb1 mRNA in the Lkb1+/− mice were approximately half of the wild type, haplo-insufficiency (26) may influence polyp formation. However, contamination of the polyps with normal cells containing a wild-type allele could interfere with LOH assessment, as reported previously (27, 28).
Lkb1−/− embryos died between 8.5 and 9.5 dpc. It is possible that developmental arrest of Lkb1−/− embryos occurred at around the 10-paired-somites stage because the size and development of dead mutant embryos corresponded to normal embryos at 8.0–8.5 dpc. Lkb1−/− embryos exhibited malformed somites and did not undergo embryonic turning. These findings indicate that the Lkb1 gene plays an important role in embryonic development, as recently reported (10). Another study has found that Lkb1 mRNA is ubiquitously expressed during the early stages (7–11 dpc) of mouse embryogenesis (29). However, in the present study, only nucleated embryonic blood cells and heart tissue bound Lkb1 antibody at 9.0 dpc. This difference may be caused by variation in expression levels of Lkb1 protein. The marked change in the expression of Lkb1 protein in nucleated embryonic blood cells between 8.5 dpc and 9.0 dpc may be associated with the death of Lkb1−/− embryos at around 9.0 dpc. Interestingly, recent studies have reported that multipotent hemopoietic cells start to emerge in the yolk sac and para-aortic splanchnopleura of mouse embryos at around 9.0 dpc (30). Furthermore, Lkb1-deficient mice exhibit vascular abnormalities and deregulation of vascular endothelial growth factor and die at midgestation (10). Together, these findings suggest that the expression of Lkb1 in nucleated embryonic blood cells at 9.0 dpc is essential for multipotent hemopoietic growth and/or differentiation.
Our Lkb1+/− mouse model appears to be a useful tool for the study of PJS and may aid in the design of new therapies for this disease.
Recently, Miyoshi et al. (31) reported that gastrointestinal hemartomatous polyps developed in Lkb1 heterozygous knockout mice. The results presented in Miyoshi et al.'s paper support our results for the involvement of Lkb1 deficiency in gastrointestinal tumor formation.
Acknowledgments
We thank S. Uchida for providing excellent technical assistance, M. Masutani, K. Ueno, and T. Watanabe for their helpful comments, and T. Yagi for providing the pCre-pac plasmid.
Abbreviations
- PJS
Peutz–Jeghers syndrome
- ES
embryonic stem
- dpc
days postcoitum
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB026255).
References
- 1.Jeghers H, McKusick V A, Karz K H. N Engl J Med. 1949;241:992–1005. doi: 10.1056/NEJM194912292412601. [DOI] [PubMed] [Google Scholar]
- 2.Peutz J L. Ned Tijdschr Geneeskd. 1921;10:134–146. [Google Scholar]
- 3.Linder N M, Greene M H. J Natl Cancer Inst. 1998;90:1039–1071. doi: 10.1093/jnci/90.14.1039. [DOI] [PubMed] [Google Scholar]
- 4.McGarrity T J, Kulin H E, Zaino R J. Am J Gastroenterol. 2000;95:596–604. doi: 10.1111/j.1572-0241.2000.01831.x. [DOI] [PubMed] [Google Scholar]
- 5.Jenne D E, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, Muller O, Back W, Zimmer M. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 6.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, et al. Nature (London) 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 7.Olschwang S, Boissonm C, Thomas G. J Med Genet. 2001;38:356–360. doi: 10.1136/jmg.38.6.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olschwang S, Markie D, Seal S, Neale K, Phillips R, Cottrell S, Ellis I, Hodgson S, Zauber P, Spigelman A, et al. J Med Genet. 1998;35:42–44. doi: 10.1136/jmg.35.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Launonen V, Avizienyte E, Loukola A, Laiho P, Salovaara R, Jarvinen H, Mecklin J P, Oku A, Shimane M, Kim H C, et al. Cancer Res. 2000;60:546–548. [PubMed] [Google Scholar]
- 10.Ylikorkala A, Rossi D J, Korsisaari N, Luukko K, Alitalo K, Henkemeyer M, Makela T P. Science. 2001;293:1323–1326. doi: 10.1126/science.1062074. [DOI] [PubMed] [Google Scholar]
- 11.Yagi T, Nada S, Watanabe N, Tamemoto H, Kohmura N, Ikawa Y, Aizawa S. Anal Biochem. 1993;214:77–86. doi: 10.1006/abio.1993.1459. [DOI] [PubMed] [Google Scholar]
- 12.Kawase Y, Iwata T, Watanabe M, Kamada N, Ueda O, Suzuki H. Contemp Topics Lab Ani Sci. 2001;40:31–34. [PubMed] [Google Scholar]
- 13.Taniguchi M, Sanbo M, Watanabe S, Naruse I, Mishina M, Yagi T. Nucleic Acids Res. 1998;26:679–680. doi: 10.1093/nar/26.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato Y, Mukai K, Watanabe S, Goto M, Shimosato Y. Am J Pathol. 1986;125:431–435. [PMC free article] [PubMed] [Google Scholar]
- 15.Nezu J, Oku A, Shimane M. Biochem Biophys Res Commun. 1999;261:750–755. doi: 10.1006/bbrc.1999.1047. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M, Avizienyte E, Corn P G, Lothe R A, Baylin S B, Aaltonen L A, Herman J G. Oncogene. 2000;19:164–168. doi: 10.1038/sj.onc.1203227. [DOI] [PubMed] [Google Scholar]
- 17.Shepherd N A, Bussey H J, Jass J R. Am J Surg Pathol. 1987;11:743–749. doi: 10.1097/00000478-198710000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Estrada R, Spjut H J. Am J Surg Pathol. 1983;7:747–754. [PubMed] [Google Scholar]
- 19.Utsunomiya J, Gocho H, Miyanaga T, Hamaguchi E, Kashimure A. Johns Hopkins Med J. 1975;136:71–82. [PubMed] [Google Scholar]
- 20.Burdick D, Prior J T. Cancer. 1982;50:2139–2146. doi: 10.1002/1097-0142(19821115)50:10<2139::aid-cncr2820501028>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 21.Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, Matsumoto H, Takano H, Akiyama T, Toyoshima K, et al. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 22.Weber F P. Q J Med. 1949;12:404–408. [Google Scholar]
- 23.Giardiello F M, Welsh S B, Hamilton S R, Offerhaus G J, Gittelsohn A M, Booker S V, Krush A J, Yardley J H, Luk G D. N Engl J Med. 1987;316:1511–1514. doi: 10.1056/NEJM198706113162404. [DOI] [PubMed] [Google Scholar]
- 24.Spigelman A D, Murday V, Phillip R K. Gut. 1989;30:1588–1590. doi: 10.1136/gut.30.11.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karuman P, Gozani O, Odze R D, Zhou X C, Zhu H, Shaw R, Brien T P, Bozzuto C D, Ooi D, Cantley L C, et al. Mol Cell. 2001;7:1307–1319. doi: 10.1016/s1097-2765(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 26.Quon K C, Berns A. Genes Dev. 2001;15:2917–2921. doi: 10.1101/gad.949001. [DOI] [PubMed] [Google Scholar]
- 27.Hemminki A, Tomlinson I, Markie D, Jarvinen H, Sistonen P, Bjorkqvist A M, Knuutila S, Salovaara R, Bodmer W, Shibata D, et al. Nat Genet. 1997;15:87–90. doi: 10.1038/ng0197-87. [DOI] [PubMed] [Google Scholar]
- 28.Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Proc Natl Acad Sci USA. 1995;92:4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luukko K, Ylikorkala A, Tiainen M, Makela T P. Mech Dev. 1999;83:187–190. doi: 10.1016/s0925-4773(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 30.Godin I, Dieterlen-Lievre F, Cumuno A. Proc Natl Acad Sci USA. 1995;92:773–777. doi: 10.1073/pnas.92.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyoshi H, Nakau M, Ishikawa T, Seldin M F, Oshima M, Taketo M M. Cancer Res. 2002;62:2261–2266. [PubMed] [Google Scholar]