Abstract
Loss of function of the adenomatous polyposis coli (APC)/Apc tumor suppressor gene occurs early in the etiology of intestinal cancer in mammals. In human colonic tumors, genomic instability is proposed to be associated with tumor initiation by inducing loss of APC function. We have used a mouse model of inherited intestinal cancer (ApcMin/+, Min/+) to analyze the earliest stages of tumorigenesis in this organ. We find that tumors from C57BL/6 Min/+ mice have a stable karyotype and stable microsatellites. In contrast to previous claims, we find that homozygosity for the Min allele of Apc in tumors can proceed by homologous somatic recombination. Further, our analysis of early, benign human colorectal adenomas failed to reveal any evidence for generalized chromosomal or microsatellite instability. These results cast doubt on the hypothesis that either of these forms of genomic instability is necessary for the initial development of colorectal adenomas. We contrast our analysis of autochthonous primary tumors to other studies involving xenografts or cultured cells.
Cancer is a disease typified by defects in DNA metabolism and cellular homeostasis, the molecular bases of which are under intense scrutiny. Current models portray the evolution from adenoma to carcinoma as a linear, multistage process, with each stage relying on multiple independent mutations. Indeed, it has been proposed that a typical colorectal cancer contains at least 11,000 genomic alterations (1). Given that the basal spontaneous mutation rate is unlikely to account for the number of mutations found in most cancers, Loeb (2) hypothesized that a mutator phenotype arises early in the tumorigenic process to stimulate the acquisition of the required mutations. This hypothesis has been borne out by the finding that inherited mutations in certain DNA repair genes predispose mammals to cancer (3, 4).
Recent studies have focused on the role of a mutator phenotype/genomic instability in the pathogenesis of human colorectal cancer. Cell lines from human colorectal adenocarcinomas invariably display genomic instability, either karyotypic/chromosomal or in microsatellite length (5). Whereas microsatellite instability (MI) is diagnostic of defects in DNA mismatch repair (6), chromosomal instability (CI) is sometimes found to result from a defect in the mitotic checkpoint pathway (7). These observations, combined with analysis of allelic imbalance in benign colorectal adenomas, have led to the hypothesis that genomic instability arises early in the development of colorectal cancer in the human and that karyotypic instability drives allelic loss at the adenomatous polyposis coli (APC) locus (8).
There are many caveats associated with studies of cancer cell lines, including the fact that most cell lines and xenografts are derived from advanced adenocarcinomas, not early benign adenomas. Thus, it may be dangerous to make generalizations from these cell lines to the events that occur early in the tumorigenic process. To this end, it is crucial to be able to validate in vivo any genomic changes that are commonly found in cell lines and xenografts. Mouse models may act as valuable tools for beginning to understand the earliest stages of human cancer. For example, ApcMin/+(Min/+) mice develop dozens of benign adenomas throughout the intestinal tract, sharing with human colonic adenomas a requirement for inactivation of APC/Apc. In C57BL/6 (B6) mice heterozygous for a mutant allele of Apc, loss of Apc function occurs almost exclusively by loss of heterozygosity (LOH) (9–11).
We initially sought to determine whether CI plays a role in the essential LOH event that initiates tumorigenesis in mice heterozygous for mutant alleles of Apc. The mechanism of this LOH event is uncertain, being reported to occur by nondisjunction or nondisjunction with reduplication (9, 12). Our findings in this regard have led us in turn to analyze the genome of early, benign adenomas from the human colon and to re-evaluate the roles of the CI and MI classes of genomic instability in the very early stages of intestinal tumorigenesis.
Materials and Methods
Mouse Husbandry and Genotyping.
Animals were bred and housed at the McArdle Laboratory for Cancer Research. Animals were maintained on a Purina 5020 diet with 9% fat and 20% protein. Min/+ mice were genotyped as described (13). Tyrc-2J animals were genotyped by PCR with the primers Tyr-AluF (5′-CTTCAAAGGGGTGGATGAGC-3′) and Tyr-AluR (5′-CTAATCAAGACTAGCTTCTCTG-3′). After PCR, products were digested with AluI to generate a 150-bp wild-type (Tyr+) band and a ≈130-bp mutant (Tyrc-2J) band. Rb(7.18)9Lub (Rb9) is carried on the C57BL/6JEi genetic background (14). At the time of breeding, this strain had been separated from C57BL/6J for approximately 60 generations. Rb9 carriers were identified by karyotyping peripheral blood lymphocytes with the protocol of Davisson and Akeson (15).
Preparation of Interphase Fluorescent in Situ Hybridization (FISH) Samples.
Interphase FISH on mouse and human tissues was performed as described by Jacoby et al. (16). For both mouse and human, 5-μm sections from paraffin-embedded tissues were used. For mouse tissues, adjacent normal intestinal epithelium was used as a control. Because many of the human adenomas did not contain visible normal intestinal epithelium, nonepithelial cells within the lamina propria were used as a normal control. Human FITC-labeled centromeric probes were obtained from ID Labs Biotechnology (London, Ontario, Canada). Mouse biotin-labeled, chromosome-specific cosmid probes were obtained from Applied Genetics Laboratories (Melbourne, FL). A probe specific to the Apc locus on mouse chromosome 18 was isolated by screening the RPCI-23 B6 bacterial artificial chromosome (BAC) library (P. DeJong, Children's Hospital, Oakland Research Institute, Oakland, CA) with an Apc PCR product amplified from B6 genomic DNA. BAC DNA was used for FISH after being labeled with digoxygenin by nick translation (Roche Molecular Biochemicals).
Human Cell Lines.
Human colorectal adenocarcinoma cell lines were obtained from the American Type Culture Collection and grown in McCoy's 5A medium supplemented with 10% FBS. Karyotypes from these cells were generated by the same protocol as for peripheral blood lymphocytes, omitting the 42-h culturing step. Interphase nuclei from these preparations were used for interphase FISH.
Karyotyping Multiple Intestinal Neoplasia (Min) Tumors.
To obtain karyotypes from B6 Min tumors, intestines were isolated and flushed with RPMI supplemented with 10% FBS. Individual tumors were then microdissected and diced with a razor blade. The tumor cell suspension was incubated in RPMI + 10% FBS with 5 μg/ml colchicine for 30 min and karyotyped as described above. Each sample was confirmed to contain primarily tumor cells by quantitative PCR analysis (see below).
Immunohistochemistry on Tissues.
Immunohistochemistry was performed on 5-μm paraffin-embedded tissue with either a mouse anti-β-catenin mAb (Transduction Laboratories, Lexington, KY) or an affinity-purified rabbit anti-APC polyclonal antibody (17), as described by Merritt et al. (18). This antibody was detected with biotin-labeled goat α-rabbit secondary antibody and the Vectastain ABC kit (Vector Laboratories). Sections were lightly counterstained with hematoxylin. Anti-β-catenin mAb was detected with goat anti-mouse AlexaFluor 594 (Molecular Probes). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole.
Analysis of LOH and MI.
Quantitative PCR to assess the allelic status at the Apc locus was performed as described (9). Quantitative PCR for the Tyr locus was performed with the same PCR primers as the genotyping reaction, spiking the reaction with [32P] dCTP. Analysis for MI was performed as described by Luongo et al. (9). Primers were obtained from Research Genetics (Huntsville, AL) and end-labeled with [32P] ATP by using T4 polynucleotide kinase.
Results and Discussion
Analysis of Genomic Instability in Intestinal Adenomas from Min/+ Mice.
To study the initiation of tumorigenesis in B6 Min/+ animals, we analyzed adenomas for evidence of CI and/or MI. We focused specifically on these two forms of genomic instability because one or the other is commonly found in human adenocarcinoma cell lines (5). Interphase FISH was used to enumerate specific chromosomes in Min tumors. As a control, we used a probe specific to chromosome X. This probe detected two copies of chromosome X in tumors from female Min/+ animals and one copy in tumors from male Min/+ animals (Table 1). All autosomes examined were found to be present in two copies, including the Apc-bearing chromosome, 18 (Table 1). A bacterial artificial chromosome probe spanning Apc also detected two copies of this locus (Table 1).
Table 1.
Interphase FISH on intestinal adenomas from B6 Min/+ mice
| Adenoma source | Probe | N | FISH signals per nucleus
|
|
|---|---|---|---|---|
| Normal tissue | Tumor tissue | |||
| B6Min/+ ♂ | Chr. X-specific | 4 | 1.04 ± 0.02 | 1.06 ± 0.02* |
| B6Min/+ ♀ | Chr. X-specific | 5 | 1.68 ± 0.03 | 1.61 ± 0.01 |
| B6Min/+ | Chr. 18-specific | 23 | 1.65 ± 0.08 | 1.63 ± 0.07† |
| B6Min/+ | Apc locus | 17 | 1.69 ± 0.06 | 1.67 ± 0.04 |
| B6Min/+ | Chr. 4-specific | 14 | 1.53 ± 0.06 | 1.52 ± 0.04 |
| B6Min/+ | Chr. 7-specific | 5 | 1.59 ± 0.03 | 1.68 ± 0.04 |
Chr., chromosome.
P > 0.15 when compared with corresponding normal tissue (Wilcoxon Rank Sum analysis), and P < 0.01 when compared with tumor or normal tissue from B6 Min/+ ♀ tumors.
All autosomes examined are present in two copies.
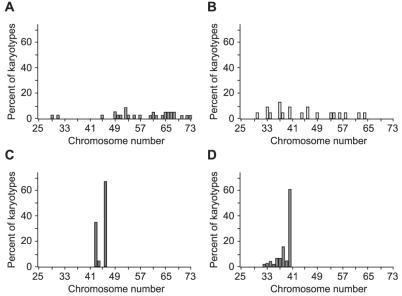
To gain a more complete view of the chromosome complement of tumors from B6 Min/+ animals, we generated karyotypes directly from tumors of the small and large intestine. As a control, we analyzed human colorectal adenocarcinoma cell lines. Two of these adenocarcinoma cell lines, HT29 and SW480, demonstrated karyotypic instability, as manifested by a high variance of the number of chromosomes per cell rather than by alteration of the mean number of chromosomes per cell for a given sample: HT29 = 58 ± 11 chromosomes/cell, SW480 = 45 ± 10 chromosomes/cell (Fig. 1 A and B). Note that although the SW480 line had a near-diploid mean chromosome number, it had a large standard deviation in the number of chromosomes per cell. By contrast, the HCT116 line had a near-diploid mean and a much lower standard deviation (45 ± 1.5 chromosomes/cell, Fig. 1C), indicating that this mismatch repair-deficient cell line has a stable karyotype. We analyzed 77 karyotypes that originated from 18 tumors from two different B6 Min/+ animals. We found the Min tumor karyotypes to be largely normal, having a near-diploid mean and a low standard deviation (39 ± 2 chromosomes/cell, Fig. 1D). Importantly, we never observed a karyotype with more than the diploid number of chromosomes, in contrast to the karyotypically unstable cell lines HT29 and SW480. This lack of hyperdiploid chromosome counts is responsible for the hypodiploid mean chromosome number of 39 in Min tumors. From these observations, we conclude that tumors from B6 Min/+ mice have a stable and apparently normal karyotype.
Figure 1.
Karyotypes from metaphase-arrested colorectal adenocarcinoma cell lines and B6 Min tumors. CI cell lines (A and B) can be either highly aneuploid (HT29; chromosome number = 58 ± 11) or near diploid (SW480; chromosome number = 45 ± 10), but always have a high variance in the number of chromosomes per karyotype. The chromosomally stable cell line (C), by contrast, is nearly diploid and has a low variance (HCT116; chromosome number = 45 ± 1.5). Tumors from B6 Min/+ mice (D) fall into the karyotypically stable class, being nearly diploid with a low variance (chromosome number = 39 ± 2). The number of karyotypes analyzed from each sample is as follows: HT29, 29; SW480, 20; HCT166, 20; B6 Min tumors, 77.
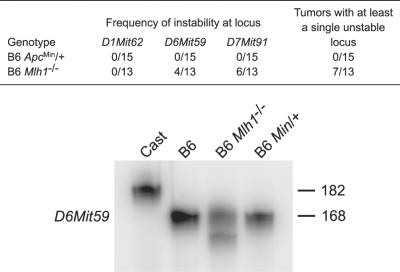
Because adenomas from B6 Min/+ mice were found to have a stable karyotype, we sought to determine whether microsatellites were genetically unstable in the early tumors. Three microsatellite loci were analyzed for each tumor: D6Mit59 and D7Mit91 have been shown to be frequently unstable in tissues from mismatch repair-deficient animals, whereas D1Mit62 is a locus that remains stable in the absence of DNA mismatch repair activity (19). Tumors from B6 Apc+/+; Mlh1−/− animals were used as a positive control for MI. Tumors from B6 Apc+/+; Mlh1−/− animals frequently showed nonparental microsatellite alleles (Fig. 2), indicating that these simple sequence-length repeats were unstable. By contrast, none of the adenomas from B6 Min/+ mice displayed MI (Fig. 2).
Figure 2.
Tumors from B6 Min/+ mice have stable microsatellites. Although approximately 50% of B6 Mlh1−/− tumors display new alleles for at least one microsatellite, no new alleles were detected in B6 Min/+ tumors. Shown is a representative amplification of D6Mit59 from Mus musculus castaneus genomic DNA (Cast), B6 genomic DNA (B6), B6 Mlh1−/− tumor DNA (B6 Mlh1−/−), and B6 Min/+ tumor DNA (B6 Min/+). Novel allele sizes can be seen for B6 Mlh1−/−.
The Mechanism of LOH in Tumors from B6 Min/+ Mice.
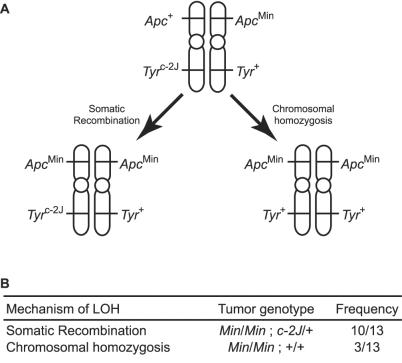
If loss of Apc function in adenomas from Min/+ mice is not caused by a CI phenotype, what is the mechanism of LOH? Min adenomas carry two copies of the Apc locus on chromosome 18 (Table 1), indicating that LOH does not occur by simple nondisjunction or a large interstitial deletion. Further, it is unlikely that LOH involves a small interstitial deletion, given that the spontaneous LOH event is chromosomewide (9). Instead, LOH occurs either by somatic recombination or a process rendering the mutant chromosome homozygous. The acrocentric structure of the mouse chromosomes makes it difficult to resolve this issue. Therefore, we generated a mouse strain that carries the Min mutation on the metacentric chromosome, Rb(7.18)9Lub (Rb9) (14). To determine the mechanism of LOH in Rb9/Rb9 tumors, we marked the long arm of the chromosome, Rb9q, with a spontaneous albino mutation in the Tyr gene (Tyrc-2J), which arose on the B6 genetic background. The experimental animals therefore are homozygous for Rb9 and heterozygous at Apc and Tyr, with the two mutant alleles linked in repulsion phase. LOH analysis revealed that most adenomas with LOH at the Apc locus on Rb9p maintained heterozygosity at Tyr on Rb9q (Fig. 3), indicating that LOH in Rb9/Rb9 Min/+ Tyrc-2J/+ tumors occurs largely, if not entirely, by somatic recombination. Although the somatic behavior of the metacentric Rb9 chromosome may differ from the wild-type acrocentric chromosome, it remains to be seen whether the somatic recombination event that induces LOH occurs at a recombination hotspot on proximal chromosome 18.
Figure 3.
LOH in tumors from B6 Min/+ mice occurs by homologous somatic recombination. (A) LOH at the Apc locus by somatic recombination can leave the Tyr locus intact (heterozygous), whereas chromosomal homozygosis leads to LOH at both loci. (B) Among the tumors from B6 Rb9/Rb9 Min/+ Tyrc-2J/+ that showed LOH at the Apc locus, 10/13 (77%) maintained heterozygosity at the Tyr locus. This result indicates that LOH has occurred by somatic recombination. A minority of tumors (3/13: 23%) showed LOH at both loci (homozygosity for ApcMin and Tyr+). It is important to note that this tumor genotype can result from either chromosomal homozygosis or double somatic recombination.
Analysis of Genomic Instability in Early Adenomas from Human Colon.
Our observations of Min/+ mice apparently contradict the hypothesis that one or the other form of genomic instability, CI or MI, is required for tumorigenesis in the human colon. There are several a priori explanations for this apparent contradiction: (i) the requirement for genomic instability differs between hereditary and sporadic tumors, (ii) there is a difference between intestinal tumorigenesis in the mouse versus the human, (iii) the benign adenoma differs from the adenocarcinoma in its requirement for genomic instability, or (iv) xenografts and cultured cells are released from an in vivo requirement for a balanced diploid karyotype.
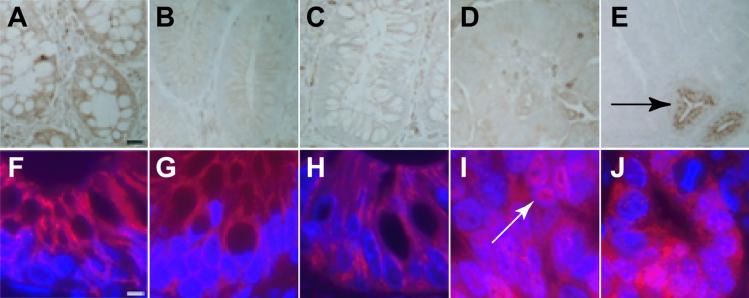
Does the Min adenoma truly reflect the benign adenoma from the human colon, or does it represent a distinct tumorigenic pathway? To address this question, we analyzed very early human colonic adenomas for evidence of CI or MI. These lesions were classified histologically as either benign tubular adenomas or benign villous adenomas with no dysplasia. Further, small tumors (diameter <1 cm) were chosen to enrich for early lesions. To confirm at the molecular level that we were analyzing the very earliest adenomas, we stained tissue sections for both APC and β-catenin. Herter et al. (20) have previously demonstrated that benign adenomas without any histological evidence of dysplasia display normal, lateral localization of β-catenin. All of the sporadic human adenomas, as well as adenomas from patients with familial adenomatous polyposis, displayed lateral β-catenin staining (Fig. 4). By contrast, sections of Min tumors and sporadic human adenocarcinomas displayed clear nuclear β-catenin staining (Fig. 4). This finding is consistent with previous reports (21) and may indicate that Min tumors are more advanced than very early human colonic adenomas. However, the cells in adenomas from both humans and Min/+ mice could be specifically diagnosed by being negative for APC/Apc immunoreactivity.
Figure 4.
APC and β-catenin staining of human adenomas. (A–E) APC staining. (F–J) β-catenin staining. (A and F) Normal human intestinal epithelial cells stain heavily for APC. In addition, β-catenin is found at the lateral edges of cells, but not in the nucleus. (B and G) Sporadic human adenoma cells do not express APC, but display lateral β-catenin localization. Importantly, although the level of cytoplasmic β-catenin may be increased, there is very little β-catenin in the nucleus. (C and H) Adenomas from familial adenomatous polyposis patients look similar to sporadic human adenomas. (D and I) Sporadic human adenocarcinomas fail to express APC. In contrast to normal tissue and benign adenomas, however, adenocarcinomas show bright nuclear β-catenin staining (white arrow). (E and J) Adenomas from Min/+ animals fail to express Apc. An adjacent normal crypt is shown to express Apc (black arrow). Interestingly, β-catenin is often found in the nucleus of Min adenoma cells. (Scale bar: A–E, 25 μm; F–J, 5 μm.)
We examined human adenomas for evidence of CI by interphase FISH analysis. In particular, chromosomes 7 and 18 were examined, as these chromosomes are among the first with detectable cytogenetic abnormalities during the development of colorectal cancer (22, 23). Cytogenetic analysis of adenocarcinoma cell lines revealed that karyotypic instability cannot be determined from the mean number of chromosomes per karyotype, but instead is reflected by the variance in the number of chromosomes per karyotype (Fig. 1). To confirm that CI could be detected by interphase cytogenetics, FISH was performed on interphase cells from adenocarcinoma cell lines. For each cell line, at least 50 interphase nuclei were analyzed. Individual cells from cell lines with a known CI phenotype (LoVo, SW480) frequently displayed more than two copies—up to six copies—of chromosome 7 or 18 (Table 2). Individual cells from the karyotypically stable lines (DLD1, HCT116), by contrast, had only one or two, and occasionally three, copies of each of these chromosomes per cell (Table 2). The quantitative difference between the two classes lies in the variance in the number of signals per nucleus. The CI cell lines had a standard deviation of ≈1 signal per nucleus, whereas the stable lines had a standard deviation of only 0.5 signal per nucleus (Table 2). Qualitatively, the stable cell lines display a complete absence of cells with more than three copies of a given chromosome, whereas karyotypically unstable cell lines frequently have four, five, or six copies per cell. Using the bootstrap method, we generated confidence intervals for the variance in the number of chromosomes per cell (24). For the karyotypically stable cell lines, the 50% confidence interval for standard deviations is 0.41 to 0.46. For the lines with CI, the 99.9% confidence interval is 0.92 to 1.3.
Table 2.
Interphase FISH on human adenomas and adenocarcinoma cell lines
| Sample* | Chromosomal instability phenotype | Avg. counts per nucleus ± SD
|
|
|---|---|---|---|
| Normal | Tumor | ||
| DLD1 | Stable | n.d. | 2.10 ± 0.54 |
| HCT116 | Stable | n.d. | 2.14 ± 0.40 |
| LoVo | Unstable | n.d. | 2.92 ± 0.92 |
| SW480 | Unstable | n.d. | 2.40 ± 0.95 |
| Adenoma 8103 | Stable | 1.66 ± 0.48 | 1.72 ± 0.52 |
| Adenoma 8106 | Stable | 1.64 ± 0.53 | 1.64 ± 0.49 |
| Adenoma 8111 | Stable | 1.64 ± 0.49 | 1.67 ± 0.47 |
| Adenoma 8112 | Stable | 1.62 ± 0.49 | 1.70 ± 0.51 |
| Adenoma 8114 | Stable | 1.64 ± 0.49 | 1.66 ± 0.48 |
| Adenoma 8116 | Stable | 1.62 ± 0.53 | 1.62 ± 0.49 |
All data presented are for chromosome 18. Chromosome 7 data were similar. n.d. denotes no normal tissue controls for adenocarcinoma cell lines.
Individual adenomas are identified by their histology number.
Our analysis of adenocarcinoma cell lines allowed us to establish that (i) CI can be detected by interphase FISH and (ii) chromosomes 7 and 18 specifically are unstable in cells with karyotypic instability. With this knowledge in hand, we used interphase FISH to analyze the karyotype of human adenomas. The average number of signals per nucleus was 1.67 for chromosome 7 (n = 5 tumors) and 1.68 for chromosome 18 (n = 6 tumors), with standard deviations ranging from 0.47 to 0.52 signals per nucleus. Chromosome 18 data for a representative set of individual tumors are displayed in Table 2. The standard deviations obtained from interphase FISH on adenomas are near the 50% confidence intervals for the karyotypically stable cell lines. More importantly, the standard deviations in the number of chromosomes per cell for the primary adenomas are far below the 99.9% confidence intervals of the standard deviations for the CI adenocarcinoma cell lines and are therefore highly significantly different (P < 0.001). Based on these data, early adenomas from the human colon appear to have stable karyotypes.
We examined the same tumors for evidence of MI. Specific microsatellites were chosen on the basis of their earlier characterization as unstable loci in human colorectal tumors (25). Because no normal tissue was available as a negative control for each tumor, we used DNA from MI cell lines as a positive control. MI was defined as the presence of more than two copies for any given locus. The microsatellites were unstable in the cell lines tested, but no instability could be detected in any of the early benign adenomas (data not shown). This result is consonant with the recent report of Wang et al. (26), who found that most sporadic colorectal cancers also fail to display global genomic instability at the DNA sequence level.
Conclusions
Genomic instability is proposed to be a driving force in the initiation of human colonic tumorigenesis. To investigate this hypothesis and determine how closely human colon cancer can be modeled in mice, we sought to analyze the earliest stages of tumorigenesis in the human and mouse intestine. We have found that adenomas from B6 Min/+ mice have a stable karyotype and stable microsatellites. Further, we have found that genomic instability, at least CI or MI, is not required in the early stages of tumorigenesis in the human colon. Our observations are consistent with the hypothesis that selection for loss of APC function, and not an increase in mutation rate, drives the process of early tumorigenesis in the intestine (27). The absence of genomic instability in these benign adenomas is also consistent with the hypotheses that loss of APC/Apc function is sufficient as the sole genetic event that initiates neoplastic growth (28) and that any genomic instability in sporadic colorectal tumors arises after loss of APC function (29).
Shih et al. (8) recently reported that very early human colorectal adenomas show widespread allelic imbalance (8). Although our studies do not directly assay LOH in early human adenomas, we have demonstrated that any such instances of LOH do not involve CI. One explanation for the observations of Shih et al. in this context is that allelic imbalance does not reflect genomic instability, but instead reflects stable genomic changes (e.g., subchromosomal deletions) that are clonally expanded in early adenomas. Stable genomic changes at the chromosomal and subchromosomal level have been identified in sporadic adenomas (30) and adenocarcinomas (22, 31). There is no evidence, however, that these changes result from genomic instability.
An alternate explanation for the allelic imbalances detected by Shih and his colleagues is somatic recombination; a single recombination event near the centromere will lead to LOH along the entire length of the chromosome arm. Indeed, selection for a high rate of recombination has been proposed as being among the earliest events in the etiology of human bladder cancer (32). Recombination can be distinguished from nondisjunction by monitoring LOH on both arms of a metacentric chromosome. The study of Shih et al. was limited to single arms of each of five chromosomes (1p, 5q, 8p, 15q, and 18q) (8).
Our study of Rb9/Rb9 Min/+ Tyrc-2J/+ mice shows definitively that LOH in B6 Min/+ mice can occur by somatic recombination. This result contrasts with two previous reports that characterized the mechanism of LOH as nondisjunction and nondisjunction with reduplication (9, 12). Interestingly, homozygosity for the Min mutation has been reported to induce karyotypic instability in embryonic stem (ES) cells (33, 34). Nevertheless, the apparent diploid state of the Min adenoma indicates that karyotypic instability is not a general feature of the Min/Min state and may instead represent an intrinsic difference between ES cells and adult intestinal stem cells. The lack of accumulated karyotypic abnormalities in the cells of Min/Min tumors may involve checkpoint functions not active in ES cells (34).
In conclusion, our results indicate that although genomic instability can enhance the formation of adenomas in the human and mouse intestine (e.g., in individuals with defects in DNA mismatch repair), it is not a required feature of these lesions. Beyond CI and MI lies homologous somatic recombination. Indeed, the genomewide allelic imbalances seen in early adenomas may be the result of stable karyotypic changes or homologous somatic recombination events. Homologous somatic recombination is a process that destroys heterozygosity in diploids, while at the same time maintaining a balanced karyotype. It remains to be seen whether this process lies under environmental or genetic control. If so, variable factors that control homologous recombination in the soma of humans will be variable risk factors for certain neoplasms.
Acknowledgments
We thank Drs. Marcia Haigis, Alfred Knudson, Ilse Riegel, and Norman Drinkwater for critical reading of the manuscript; Chuck Harris for help in optimizing the interphase FISH assay; Dr. Andy Thliveris for help obtaining histological samples; Antonis Kirmizis for the gift of HT29, SW480, and HCT116 cells; the laboratory of Norman Drinkwater for maintaining the RPCI-23 B6 bacterial artificial chromosome library; and Jane Weeks and Harlene Edwards for help in preparing histological samples. This work was supported by the National Cancer Institute (Grant CA07075 to the McArdle Laboratory and Grants CA58085 and CA63677 to W.F.D.). K.M.H. was supported by National Institutes of Health Predoctoral Training Grant 5T32GM07133. This is publication No. 3601 from the Laboratory of Genetics.
Abbreviations
- APC
adenomatous polyposis coli
- B6
C57BL/6
- CI
chromosomal instability
- FISH
fluorescent in situ hybridization
- LOH
loss of heterozygosity
- Min
multiple intestinal neoplasia
- MI
microsatellite instability
References
- 1.Stoler D L, Chen N, Basik M, Kahlenberg M S, Rodriguez-Bigas M A, Petrelli N J, Anderson G R. Proc Natl Acad Sci USA. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 3.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 4.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 6.Strand M, Prolla T A, Liskay R M, Petes T D. Nature (London) 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 7.Cahill D P, Lengauer C, Yu J, Riggins G J, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nature (London) 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 8.Shih I M, Zhou W, Goodman S N, Lengauer C, Kinzler K W, Vogelstein B. Cancer Res. 2001;61:818–822. [PubMed] [Google Scholar]
- 9.Luongo C, Moser A R, Gledhill S, Dove W F. Cancer Res. 1994;54:5947–5952. [PubMed] [Google Scholar]
- 10.Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Proc Natl Acad Sci USA. 1995;92:4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smits R, Kartheuser A, Jagmohan-Changur S, Leblanc V, Breukel C, de Vries A, van Kranen H, van Krieken J H, Williamson S, Edelmann W, et al. Carcinogenesis. 1997;18:321–327. doi: 10.1093/carcin/18.2.321. [DOI] [PubMed] [Google Scholar]
- 12.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin M F, Taketo M M. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 13.Su L K, Kinzler K W, Vogelstein B, Preisinger A C, Moser A R, Luongo C, Gould K A, Dove W F. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 14.Lane P W, Eicher E M. J Hered. 1985;76:476–477. doi: 10.1093/oxfordjournals.jhered.a110152. [DOI] [PubMed] [Google Scholar]
- 15.Davisson M T, Akeson E C. Cytogenet Cell Genet. 1987;45:70–74. doi: 10.1159/000132432. [DOI] [PubMed] [Google Scholar]
- 16.Jacoby R F, Schlack S, Cole C E, Skarbek M, Harris C, Meisner L F. Gastroenterology. 1997;112:1398–1403. doi: 10.1016/s0016-5085(97)70156-2. [DOI] [PubMed] [Google Scholar]
- 17.Midgley C A, White S, Howitt R, Save V, Dunlop M G, Hall P A, Lane D P, Wyllie A H, Bubb V J. J Pathol. 1997;181:426–433. doi: 10.1002/(SICI)1096-9896(199704)181:4<426::AID-PATH768>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 18.Merritt A J, Gould K A, Dove W F. Proc Natl Acad Sci USA. 1997;94:13927–13931. doi: 10.1073/pnas.94.25.13927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker S M, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliott E A, Yu J, Ashley T, Arnheim N, et al. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 20.Herter P, Kuhnen C, Muller K-M, Wittinghofer A, Muller O. J Cancer Res Clin Oncol. 1999;125:297–304. doi: 10.1007/s004320050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts R B, Min L, Washington M K, Olsen S J, Settle S H, Coffey R J, Threadgill D W. Proc Natl Acad Sci USA. 2002;99:1521–1526. doi: 10.1073/pnas.032678499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ried T, Heselmeyer-Haddad K, Blegen H, Schröck E, Gert A. Genes Chromosomes Cancer. 1999;25:195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Griffin C A, Lazar S, Hamilton S R, Giardiello F M, Long P, Krush A J, Booker S V. Cancer Genet Cytogenet. 1993;67:14–20. doi: 10.1016/0165-4608(93)90038-n. [DOI] [PubMed] [Google Scholar]
- 24.Efron C. Ann Stat. 1979;7:1–26. [Google Scholar]
- 25.Bapat B V, Madlensky L, Temple L K, Hiruki T, Redston M, Baron D L, Xia L, Marcus V A, Soravia C, Mitri A, et al. Hum Genet. 1999;104:167–176. doi: 10.1007/s004390050931. [DOI] [PubMed] [Google Scholar]
- 26.Wang T L, Rago C, Silliman N, Ptak J, Markowitz S, Willson J K, Parmigiani G, Kinzler K W, Vogelstein B, Velculescu V E. Proc Natl Acad Sci USA. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomlinson I P, Novelli M R, Bodmer W F. Proc Natl Acad Sci USA. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamlum H, Papadopoulou A, Ilyas M, Rowan A, Gillet C, Hanby A, Talbot I, Bodmer W, Tomlinson I. Proc Natl Acad Sci USA. 2000;97:2225–2228. doi: 10.1073/pnas.040564697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Homfray T F, Cottrell S E, Ilyas M, Rowan A, Talbot I C, Bodmer W F, Tomlinson I P. Hum Mutat. 1998;11:114–120. doi: 10.1002/(SICI)1098-1004(1998)11:2<114::AID-HUMU3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 30.Muleris M, Zafrani B, Validire P, Girodet J, Salmon R J, Dutrillaux B. Cancer Genet Cytogenet. 1994;74:104–108. doi: 10.1016/0165-4608(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 31.Thiagalingam S, Laken S, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B, Lengauer C. Proc Natl Acad Sci USA. 2001;98:2698–2702. doi: 10.1073/pnas.051625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Tilborg A A, de Vries A, de Bont M, Groenfeld L E, van der Kwast T H, Zwarthoff E C. Hum Mol Genet. 2000;9:2973–2980. doi: 10.1093/hmg/9.20.2973. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan K B, Burds A A, Swedlow J R, Bekir S S, Sorger P K, Nathke I S. Nat Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 34.Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es J H, Breukel C, Wiegant J, Giles R H, et al. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]