Abstract
The glycopeptide antibiotics vancomycin and teicoplanin are vital components of modern anti-infective chemotherapy exhibiting outstanding activity against Gram-positive pathogens including members of the genera Streptococcus, Staphylococcus, and Enterococcus. These antibiotics also provide fascinating examples of the chemical and associated biosynthetic complexity exploitable in the synthesis of natural products by actinomycetes group of bacteria. We report the sequencing and annotation of the biosynthetic gene cluster for the glycopeptide antibiotic A47934 from Streptomyces toyocaensis NRRL15009, the first complete sequence for a teicoplanin class glycopeptide. The cluster includes 34 ORFs encompassing 68 kb and includes all of the genes predicted to be required to synthesize A47934 and regulate its biosynthesis. The gene cluster also contains ORFs encoding enzymes responsible for glycopeptide resistance. This role was confirmed by insertional inactivation of the d-Ala-d-lactate ligase, vanAst, which resulted in the predicted A47934-sensitive phenotype and impaired antibiotic biosynthesis. These results provide increased understanding of the biosynthesis of these complex natural products.
Glycopeptide antibiotics (GPAs) have been mainstays of antimicrobial chemotherapy since their discovery in the mid 1950s. These antibiotics act exclusively on Gram-positive bacteria by forming a tight and specific noncovalent complex with the d-Ala-d-Ala terminus of the peptidoglycan, inhibiting cell wall growth and crosslinking (1, 2). Clinical resistance to GPAs was first described in the enterococci in 1988 (3), and resistance has now manifested itself in the more virulent streptococci and staphylococci (reviewed in ref. 4). GPA resistance is now a significant worldwide phenomenon that has severely impacted the health care sector both in increased mortality and morbidity, and economically (5). The predominant mechanism of resistance is the synthesis of cell wall peptidoglycan terminating in d-Ala-d-lactate, which dramatically decreases the affinity of these antibiotics for their target (6).
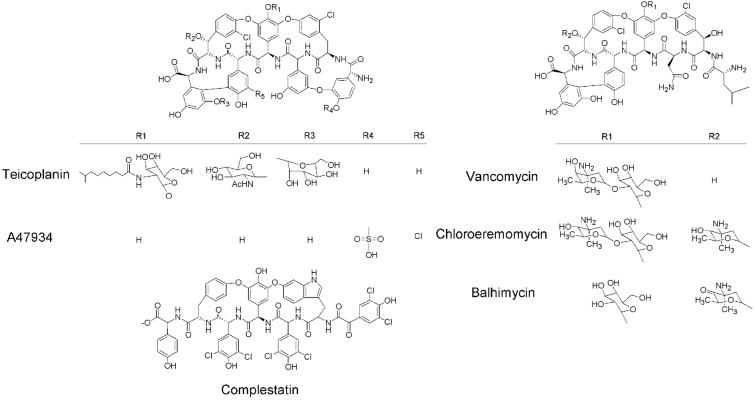
GPAs are exclusively obtained through fermentation, yet despite their importance, their biosynthesis is not well understood. They are comprised of a heptapeptide core consisting of both common and unusual amino acids (4). Crosslinking of the amino acids through aryl ether and carbon–carbon bonds provides rigidity to the peptide. There are two major structural classes of GPAs based on the identity of the core peptide, and the two clinically used GPAs, vancomycin and teicoplanin, exemplify both classes (Fig. 1). An additional class of structurally homologous secondary metabolites with anticomplement activity is exemplified by complestatin (Fig. 1). Structural diversity in these natural products is achieved through changes in the peptide backbone, and selective amino acid halogenation, glycosylation, lipidation, methylation, and sulfonylation. In principle, understanding of the genetic and mechanistic basis of these modifications could lead to the expansion of structural diversity through genetic engineering of GPA-producing bacteria.
Figure 1.
Structure of glycopeptide antibiotics. Glycopeptide antibiotics incorporate one of two core heptapeptide structures and additional structural diversity is derived from modification by a variety of halogenase and transferase activities. Also shown is complestatin, a glycopeptide antibiotic homologue. The biosynthetic gene clusters of complestatin, chloroeremomycin, and balhimycin have been sequenced.
The sequences of biosynthetic gene clusters have been reported for the vancomycin analogues chloroeremomycin (7) and balhymicin (8), and the GPA-like complestatin (9). The antibiotic A47934, produced by Streptomyces toyocaensis NRRL15009, incorporates the teicoplanin peptide backbone, is sulfonated on the N-terminal p-hydroxyphenylglycine residue, and is not glycosylated (Fig. 1). We report herein the sequencing and arrangement of the biosynthetic gene cluster for the GPA A47934. The cluster includes all of the requisite genes for A47934 biosynthesis, self-resistance, gene regulation, and export, and is thus the most comprehensive GPA gene cluster yet reported.
Materials and Methods
Bacterial Strains and Culture Conditions.
S. toyocaensis NRRL15009 spores were maintained in 20% glycerol at −70°C and seeded into a defined vegetative medium as described (10). Cells were grown at 30°C and 225 rpm and subsequently inoculated into spring-baffled 50- or 250-ml flasks containing 16 or 50 ml, respectively, of Tryptone Soy Broth (TSB; Oxoid, Basingstoke, U.K.) or Streptomyces antibiotic medium (SAM; ref. 11). Escherichia coli SURE2 (Stratagene), NovaBlue (Novagen), and TOP10 (Invitrogen) were grown in LB broth at 37°C with either ampicillin (final concentration 100 μg/ml) or apramycin selection (50 μg/ml), as appropriate.
Genomic and Plasmid DNA Isolation.
Genomic DNA was isolated by modification of existing methodology (12). S. toyocaensis cultures of 15 ml of TSB/0.5% glycine were incubated at 30°C and 225 rpm for 24–36 h. Mycelia were washed in SET buffer (20 mM Tris, pH 7.5/75 mM NaCl/25 mM EDTA) and lysed in buffer supplemented with lysozyme (2 mg/ml final concentration) at 25°C for 5–10 min. SDS was then added to 1.2% followed by 600 μg/ml proteinase K and incubated at 55°C for 2 h. NaCl was added to 1.25 M and the solution mixed thoroughly and cooled to 37°C. The preparation was chloroform-extracted and DNA was precipitated with isopropanol, washed with 70% (vol/vol) ethanol, and dissolved in 25 mM Tris (pH 7.5)/2 mM EDTA, to a concentration of 500 μg/ml. Plasmid DNA was isolated from overnight LB liquid cultures containing the appropriate antibiotic selection by using the QIAprep Spin Miniprep kit (Qiagen).
Construction and Screening of the S. toyocaensis Cosmid Library.
‘S. toyocaensis genomic DNA was partially digested using Sau3AI and 35–40-kb fragments were isolated following sucrose gradient centrifugation. DNA fragments of the appropriate size were cloned into the BamHI-digested cosmid pWE15 vector and packaged into λ phage by using the Gigapack III XL Packaging Extract Kit (Stratagene). The cepA gene was [α-32P]dATP-labeled and used to identify cosmids containing glycopeptide biosynthetic genes by colony hybridization. Positive clones were confirmed by Southern hybridization. Two cosmids, pCepC1 and pCepC4, were identified in this manner. The cosmid library was rescreened using a [α-32P]dATP-labeled DNA fragment from staS and positive clones were subsequently screened by PCR using vanAst-specific primers (5′-GAGATATACATATGGCCAGACTGAAGATCGG-3′ and 5′-TGACATAAGCTTCAGAGCGAGGAGACGGTGA-3′) to identify cosmid pCepC5.
Partial Sau3AI digestion of pCepC1 and pCepC4 generated 1-kb DNA fragments that were cloned into BamHI-digested pUC19 for DNA sequencing. pCepC5 was sheared under N2 pressure to give DNA fragments of 1.8 to 2.5 kb, which were then cloned into the pCR4Blunt-TOPO vector by using the Zero Blunt TOPO Kit for Shotgun Sequencing (Invitrogen).
Sequencing and Annotating the A47934 Biosynthetic Cluster.
Plasmid DNA obtained from the subclone libraries of the three cosmids pCepC1, pCepC4, and pCepC5 were sequenced at the MOBIX Central Facility (Hamilton, ON) and the Dana-Farber/Harvard Cancer Center High-Throughput DNA Sequencing Facility. DNA sequences were manually edited and compiled using seqman software (DNAstar, Madison, WI). DNA stretches of the A47934 biosynthetic cluster that were missing or sequenced only once were sequenced either directly off of the appropriate cosmid by using specific primers or by sequencing subclones of the DNA fragments created by PCR amplification. The entire A47934 biosynthetic gene cluster was sequenced with an average 7-fold and minimum 2-fold redundancy. Annotation of ORFs and gene functions was performed manually by using FRAMEPLOT 2.3.2 (National Institute of Health, Japan) and blast (National Institute for Biotechnology Information). The completed A47934 biosynthesis cluster has been deposited in GenBank (accession no. STO22459).
Insertional Inactivation of vanAst.
The vanAst inactivation plasmid pBlutsr-ddl3.0-Am was created from pBluescript KS+. Thiostrepton resistance was added by subcloning tsr from pIJ702, and apramycin resistance (Am) was added by subcloning PCR-amplified aac(3)-IV from pOJ260. A S. toyocaensis genomic DNA fragment containing the vanAst gene was subcloned from pFD666-ddl3.0 (13). S. toyocaensis protoplasts were prepared as described (14). Typical transformations involved 1 × 108 protoplasts, using alkaline-denatured plasmid DNA (up to 3 μg of plasmid; ref. 15) obtained after passage of plasmids through the E. coli GM48 (dam−, dcm−). Regeneration of transformed protoplasts was allowed to take place overnight after which 3 mg of apramycin sulfate was overlaid in 500 μl of TSB and individual colonies were picked after 3–4 days. Generation of the vanAst∷Am mutants were confirmed by Southern analysis and PCR using the specific primers: 5′-GAG ATA TAC ATA TGG CCA GAC TGA AGA TCG G-3′ and 5′-TGA CAT AAG CTT CAG AGC GAG GAG ACG GAG A-3′.
Complementation of the vanAst∷Am strain was performed using the integrating plasmid pJN7 that contained the vanH,A,X operon from the vancomycin producer Amycolatopsis orientalis (13) and the φC31 integrase from pSET152 (16). Transformants exhibiting thiostrepton resistance were subsequently tested for resistance to A47934 and vancomycin by using a disk diffusion method on solid R2YE media (17).
A47934 production was monitored during liquid culture growth by Bacillus subtilis susceptibility assay as described (11).
Results and Discussion
Cloning and Sequencing of the A47934 Biosynthetic Cluster.
The A47934 biosynthetic gene cluster was cloned from a cosmid library of S. toyocaensis NRRL15009 genomic DNA probed with the cepC gene from the chloroeremomycin producer A. orientalis (7). This gene encodes the nonribosomal peptide synthetase (NRPS) that recognizes the C-terminal 3,5-dihydroxyphenylglycine that is common to both the vancomycin and teicoplanin class of glycopeptide antibiotics. Two overlapping cosmids, pCepC1 and pCepC4, were identified in this screen and an additional cosmid, pCep5 was isolated using probes designed from a region on pCepC4 containing the genes ORF 17 and the self-resistance gene vanAst.
Analysis of the A47934 Biosynthetic Gene Cluster.
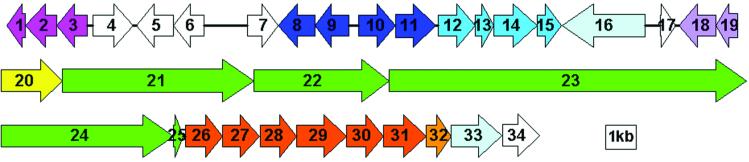
The A47934 biosynthesis cluster includes approximately 68 kb of DNA encoding 34 genes predicted to be responsible for assembly and export of the antibiotic and for self-resistance and gene regulation (Fig. 2, Table 1). The DNA approximately 20 kb downstream of ORF 34 and 2 kb upstream of ORF 1 has been sequenced and no genes predicted to be associated with A47934 biosynthesis, or resistance, are located in these regions. The genes associated with the A47934 biosynthetic cluster include orthologues of several genes identified in the biosynthesis of the glycopeptides chloroeremomycin, balhimycin, and the glycopeptide-like complestatin, and also include a number of unique elements important to the teicoplanin group of antibiotics. Comparison of all four glycopeptide gene clusters sequenced to date provides significant insight into the biochemistry of these important natural products.
Figure 2.
Organization of A47934 cluster. The 34 ORFs indicated above result from the sequencing of three S. toyocaensis cosmids containing the A47934 biosynthetic cluster. Functions associated with each of the genes are outlined in Table 1.
Table 1.
Summary of proteins encoded by the A47934 biosynthetic gene cluster
| ORF | Start/stop, bp | A47934 Protein | Cep homologue* | Com homologue† | Bal homologue‡ | Proposed function/homology |
|---|---|---|---|---|---|---|
| 1 | 914–288 | VanXst | — | — | — | d-Ala-d-Ala dipeptidase |
| 2 | 1951–911 | VanAst§ | — | — | — | d-Ala-d-lactate ligase |
| 3 | 2931–1939 | VanHst | — | — | — | Lactate dehydrogenase |
| 4 | 3059–4399 | MurX | — | — | — | d-Ala-d-Ala adding enzyme |
| 5 | 5578–4412 | StaO | — | — | — | FemABX homologue |
| 6 | 6509–5667 | StaP | — | — | — | Putative membrane protein |
| 7 | 8101–9102 | StaQ | — | ComG | — | Transcriptional regulator |
| 8 | 10324–9224 | Hmo | Hmo | Hmo | ORF 6 | p-Hydroxymandelate oxidase |
| 9 | 11417–10308 | HmaS | HmaS | HmaS | ORF 5 | p-Hydroxymandelate synthetase |
| 10 | 11592–12746 | Pdh | Pdh | Pdh | — | Prephenate dehydrogenase |
| 11 | 12908–14266 | HpgT | HpgT | HpgT | PgaT | p-Hydroxy- and 3,5-dihydroxyphenylglycine aminotransferase |
| 12 | 14412–15584 | DpgA | DpgA | — | DpgA | 3,5-Dihydroxyphenylacetyl-CoA synthase |
| 13 | 15596–16273 | DpgB | DpgB | — | DpgB | Enhances DpgA activity |
| 14 | 16270–17586 | DpgC | DpgC | — | DpgC | 3,5-Dihydroxyphenylacetyl-CoA oxygenase |
| 15 | 17583–18386 | DpgD | DpgD | — | DpgD | Enhances DpgA activity |
| 16 | 21475–18767 | StaR | — | — | — | Putative flavoprotein |
| 17 | 21820–22284 | StaS | — | — | — | Putative DNA binding protein |
| 18 | 23466–22363 | VanSst | — | — | — | Transmembrane histidine kinase |
| 19 | 24148–23453 | VanRst | — | — | — | Two-domain response regulator |
| 20 | 24407–26350 | StaU | CepM | ComL | — | ABC transporter |
| 21 | 26347–32559 | StaA | CepA | ComA | BpsA | Peptide synthetase (modules 1–2) |
| 22 | 32562–37037 | StaB | CepA | ComB | BpsA | Peptide synthetase (module 3) |
| 23 | 37067–49267 | StaC | CepB | ComC | BpsB | Peptide synthetase (modules 4–6) |
| 24 | 49287–54854 | StaD | CepC | ComD | BpsC | Peptide synthetase (module 7) |
| 25 | 55192–56367 | StaE | CepD | ComE | ORF1 | Hypothetical protein |
| 26 | 55192–56367 | StaF | CepE | ComI | OxyA | P450-related oxidase |
| 27 | 56388–57542 | StaG | — | — | — | P450-related oxidase |
| 28 | 57532–58728 | StaH | CepF | ComJ | OxyB | P450-related oxidase |
| 29 | 58733–60271 | StaI | CepH | — | BhaA | Nonheme halogenase |
| 30 | 60315–61493 | StaJ | CepG | — | OxyC | P450-related oxidase |
| 31 | 61538–62845 | StaK | — | ComH | — | Nonheme halogenase |
| 32 | 62842–63654 | StaL | — | — | — | Sulfotransferase |
| 33 | 63734–65320 | StaM | — | — | — | Putative nonheme iron dioxygenase |
| 34 | 65783–67039 | StaN | CZA382.28 | ComF | — | Integral membrane ion transporter |
Biosynthesis of Nonproteinogenic Amino Acids.
In addition to the common amino acid Tyr, the teicoplanin group of glycopeptide antibiotics incorporate the nonproteinogenic amino acids 4-hydroxyphenylglycine (HPG), 3,5-dihydroxyphenylglycine (DHPG), and β-hydroxytyrosine (β-OHTyr) (Fig. 1). Chlorination of Tyr, HPG, and β-OHTyr at amino acid positions 2, 5, and 6 are predicted to occur following peptide assembly (18). The mechanism of HPG synthesis from the precursors prephenate and Tyr has been shown to require four gene products in the chloroeremomycin producer A. orientalis, Pdh, Hmo, HmaS, and HpgT (19, 20), and genes encoding orthologous proteins have been identified in the biosynthetic gene clusters of the glycopeptide-like antibiotic complestatin (9). Pdh is proposed to convert prephenate to p-hydroxyphenylpyruvate in the only biochemically uncharacterized step. p-Hydroxyphenylpyruvate is decarboxylated and hydroxylated by HmaS to yield p-hydroxymandelate, which is oxidized to p-hydroxybenzoylformate by Hmo followed by transamination by HpgT, using Tyr as the amino donor, to yield HPG. Orthologous gene products in S. toyocaensis are predicted from the clustered ORFs 8–11 and are expected to provide similar functions in HPG biosynthesis (Table 1). The S. toyocaensis orthologous genes are grouped in a similar fashion to those in the complestatin biosynthetic cluster in contrast with the equivalent chloroeremomycin genes, which are distributed throughout the cluster.
The biochemical details of DHPG biosynthesis have been investigated in the chloroeremomcyin producer A. orientalis, and involve four characterized enzymes and one additional transamination step (21, 22). DpgA is a polyketide synthase that generates 3,5-dihydroxyphenylacetyl-CoA from four molecules of malonyl-CoA, with the assistance of DpgB and DpgD, proteins that exhibit dehydratase activity. The oxidase DpgC converts 3,5-dihydroxyphenylacetyl-CoA to 3,5-dihydroxyphenylglyoxylate, which generates DHPG on transamination by HpgT (21). Orthologous genes encoding these enzymes in S. toyocaensis, ORFs 12–15, are found arranged as they are in other glycopeptide clusters (Table 1).
The third unusual amino acid found in A47934 is β-OHTyr. Analysis of the ORFs in the chloroeremomycin and balhimycin biosynthesis clusters suggests that synthesis of β-OHTyr from Tyr likely follows a novel mechanism recently characterized in the biosynthesis of novobiocin, requiring the action of three gene products: an NRPS homologue, a P450 hydroxylase, and a thioesterase (23). These enzymes covalently tether the precursor Tyr (NRPS), oxidize the amino acid at the β-position (P450 hydroxylase), and release the amino acid product by thioester hydrolysis (hydrolase). Although these genes are present in the biosynthetic gene clusters of the vancomycin class glycopeptides, chloroeremomycin and balhimycin, which each incorporate two molecules of β-OHTyr at amino acid positions 2 and 6, no such set of genes exists in the A47934 biosynthesis cluster, which incorporates only one β-OHTyr in position 6 (Fig. 1). Analysis of the A47934 gene cluster reveals two genes that each have the potential to hydroxylate the Tyr residue at position 6. First is a predicted flavoprotein (ORF 16, StaR), which could hydroxylate Tyr once incorporated into the heptapeptide backbone. The second candidate is a putative non-heme iron dioxygenase (ORF 33, StaM) with homology to CmlA in the chloramphenicol biosynthetic cluster (24). Resolution of the mechanism of Tyr β-hydroxylation in A47934 will require further study.
Nonribosomal Peptide Synthesis.
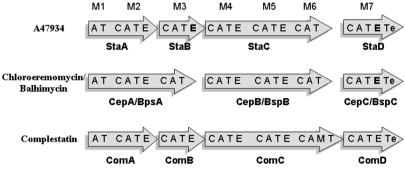
The A47934 biosynthesis cluster contains four NRPS genes, staA, staB, staC, and staD, that are predicted to encode enzymes that catalyze the assembly of the heptapeptide backbone. Organization of the NRPS-specific adenylation (A), thiolation (T), condensation (C), and epimerization (E) domains within the four proteins is shown in Fig. 3 and was determined through identification of consensus sequences (25–27). Modules in the four NPRS proteins are arranged in a 2–1-3–1 fashion with respect to amino acid activation, analogous to the arrangement found in the complestatin biosynthetic cluster and divergent from the 3–3-1 gene arrangement in chloroeremomycin and balhimycin biosynthesis.
Figure 3.
Organization of the NRPS subunits in glycopeptide-producing bacteria. Organization of the adenylation (A), condensation (C), epimerization (E), thiolation (T), and thioesterase (Te) domains for each of the sequenced glycopeptide antibiotic and the glycopeptide-like complestatin NRPS systems is illustrated. E domains shown in bold type represent domains that are presumed to be nonfunctional.
The organization of the E domains predicts a stereochemistry of HOOC-l-HPG–d-Tyr–d-DHPG–d-HPG–d-HPG–l-β-OHTyr–d-DHPG-NH2 (l-d-d-d-d-l-d), which is inconsistent with the known stereochemistry of d-d-l-d-d-l-l derived from the three-dimensional structures of glycopeptides (28, 29). Thus, the predicted stereochemistries of the amino acids at positions 1, 3, and 7 are discordant with the NRPS sequence analysis. This is not uncommon, and unexpected placement of E domains in other NRPS systems has been documented (26). The absence of an E domain in module 1 is paralleled in the chloroeremomycin NRPS, in which the corresponding residue in the final peptide is d-Leu. In vitro studies using a recombinant A-domain from CepA showed a 6-fold preference for l-Leu over d-Leu activation in vitro, suggesting that downstream epimerization of this amino acid by an unidentified E domain is required to generate the active antibiotic (30). This E domain may lie out of place in the biosynthetic cluster, or exist somewhere else in the chromosome. Alternatively, the natural system may activate d-Leu even though the in vitro-defined kinetic parameters suggest that the adenylation domain favors l-Leu. The same questions can now be raised for the A47934 NRPS system where the stereochemical fate of the N-terminal HPG activated by StaA is undetermined.
The other two disagreements between bioinformatic-predicted stereochemistry and known stereochemistry can be resolved by classifying two predicted E domains as nonfunctional. Inspection of the His motif conserved in E domains and linked with racemase activity (31, 32) reveals that the conserved second His has been mutated to Pro. Thus, we predict that this motif in StaB is inactive, consistent with the observed l stereochemistry of the DHPG at this position.
Of interest is the presence of an unusual E domain between the final T domain and the thioesterase domain in StaD, which incorporates the C-terminal l-DHPG in A47934. This E domain includes a degenerate sequence with homology to conserved regions of both E and C domains, but is not predicted to be active given the l stereochemistry of the amino acid incorporated in this position. Homologous E domains are found similarly positioned in CepC and BspC in the chloroeremomycin and balhimycin clusters, respectively, which also incorporate an amino acid of l stereochemistry in the C-terminal position. A similar domain is found in ComD of the complestatin cluster, which does incorporate an amino acid of d stereochemistry at this position. It remains to be determined whether this is the result of selective activation of the d-isomer, or a cryptic racemase activity in this hybrid E/C domain.
Oxidation, Crosslinking, Halogenation, Sulfonylation, and Export of A47934.
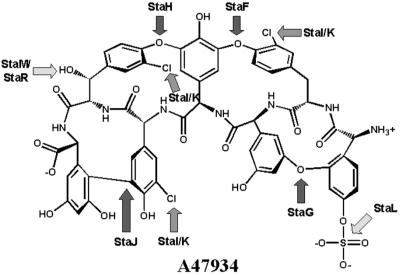
The hallmark of glycopeptide antibiotics is the presence of aromatic ether and carbon–carbon crosslinks that rigidify the molecule. In all presently available glycopeptide biosynthesis gene clusters there is one predicted P450 oxygenase for each amino acid crosslink. A47934 contains four such crosslinks: three ether linkages (between amino acids 1 and 3, 2 and 4, and 4 and 6) and one carbon–carbon linkage (between amino acids 5 and 7) (Fig. 1). The predicted assignment of oxygenase enzyme specificity for A47934 biosynthesis can be based on sequence homologies to the crosslinking enzymes in the balhimycin producer A. mediterranei, and associated gene inactivation studies by Bischoff et al. (33, 34). Null mutants of oxyA in the balhimycin producer resulted in peptides with only the 4–6 ether linkage between HPG and β-OHTyr, whereas oxyB mutants had no linkages and oxyC mutants had both the 4–6 and the 4–2 ether linkages (8, 33, 34). Thus, OxyA is predicted to catalyze formation of the 2–4 linkage, OxyB the 4–6 linkage, and OxyC the 5–7 carbon–carbon linkage between HPG and DHPG. The A47934 biosynthetic cluster reveals genes with high homology to each of the characterized oxygenases from the balhimycin biosynthesis cluster, facilitating tentative assignment of oxygenase function (Fig. 4): StaF crosslinking amino acids 2–4, StaH crosslink 4–6, and StaJ crosslink 5–7. StaG, with no orthologous A. mediterranei oxygenases, is predicted to catalyze the formation of the teicoplanin class-specific crosslink between HPG at position 1 and DHPG at position 3.
Figure 4.
Predicted sites of action of the A47934-modifying enzymes. The sites of A47934 modification as predicted by the annotation of the sequence data are indicated. StaF, StaG, StaH, and StaJ crosslink the indicated amino acids, while StaI and StaK catalyze halogenations. The predicted sulfonylation action of StaL is unique to A47934. StaM or StaR may be responsible for hydroxylation of Tyr at position 6.
A47934 has three chlorinated amino acids at positions 2 (3-Cl-Tyr), 4 (3-Cl-HPG), and 5 (3-Cl-β-OHTyr) and the biosynthetic gene cluster encodes two putative halogenases, StaI and StaK, which are predicted to be required for amino acid halogenation. StaI is homologous to ORF 10 and BhaA from the chloroeremomycin and balhimycin biosynthetic gene clusters, respectively (88% amino acid similarity). This latter enzyme has been shown to be responsible for chlorination of the Tyr/β-OHTyr residue at positions 2 and 6 (18) and thus StaI may perform a similar function. However, it is difficult to make a clear prediction of the specificity of each halogenase or whether both StaI and StaK are required for A47934 biosynthesis based on amino acid similarities with the previously sequenced halogenases.
Another modification unique to A47934 biosynthesis is sulfonylation of the N-terminal HPG. StaL (ORF 32) shows high homology to 3′-phosphoadenosine 5′-phosphosulfate (PAPS)-dependent eukaryotic sulfotransferases and is predicted to catalyze sulfonylation of A47934.
staT encodes a putative ABC transport protein that is found with all sequenced glycopeptide biosynthesis clusters. Similarly, staN encodes an equally conserved putative integral membrane ion transporter. In view of the predicted membrane association and similarity to other transport systems, these proteins, either in concert or independently, may play a role in antibiotic export from the cell interior to the environment.
Antibiotic Resistance and Gene Regulation.
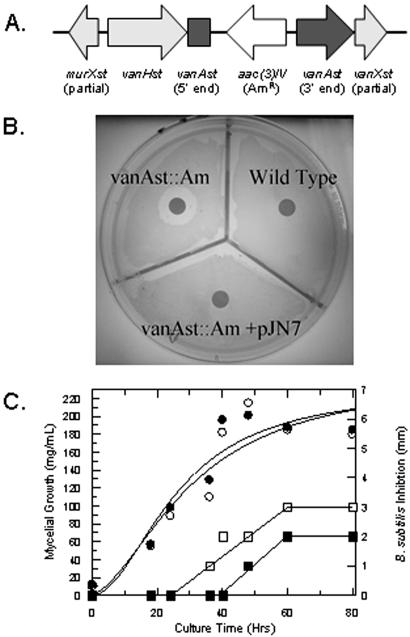
We have previously demonstrated the presence of the vanH, vanA, and vanX resistance genes in S. toyocaensis and other glycopeptide producing bacteria, precisely in the same orientation and with the same enzymatic activities as the orthologous genes that confer high-level glycopeptide resistance in Enterococci (VRE) (35). The genes for vanHst, an α-ketoacid reductase required for d-lactate biosynthesis (36), vanAst, a d-Ala-d-lactate ligase (13, 37) and vanXst, a d-Ala-d-Ala-specific dipeptidase (38), located at the 5′-end of the A47934 biosynthetic cluster, represent the first time antibiotic resistance genes have been unambiguously coupled to the biosynthesis genes in a glycopeptide-producing organism. These genes are both necessary and sufficient for host resistance to A47934 by predicted conversion d-Ala-d-Ala in the peptidoglycan layer to d-Ala-d-lactate. The importance of this mechanism was established by insertional inactivation of vanAst with an apramycin-resistance gene (Fig. 5A), which resulted in loss of resistance to A47934 (Fig. 5B). Furthermore, inactivation of vanAst resulted in a delay of over 16 h in A47934 production to a period when cells have ceased growing and are predicted to be insensitive to the action of the antibiotic on peptidoglycan synthesis (Fig. 5C). Incorporation of a single copy of the orthologous vanH,A,X gene cluster from the vancomycin producer A. orientalis C329.2 in trans at the unique φC31 site of the S. toyocaensis genome, rescued the A47934 sensitivity phenotype, thereby validating its role in resistance (Fig. 5B).
Figure 5.
Insertional inactivation of vanAst. (A) Orientation of the apramycin-resistance cassette in the resistance cluster. (B) A47934 inhibits growth of the vanAST∷Am disruption mutant but not wild-type S. toyocaensis. The sensitivity can be reversed by supplying the vanH,A,X cluster in trans on pJN7. (C) Liquid culture growth of the vanAST∷Am S. toyocaensis mutant (●) shows no difference with wild-type S. toyocaensis (○). However, antibiotic production is delayed approximately 16 h in the disruption strain (■) vs. wild type (□).
Associated with the vanHst,Ast,Xst cluster are three ORFs: murXst, encoding a predicted d-Ala-d-Ala adding enzyme (35), staO, encoding a protein with high homology to the femABX family responsible for production of the pentaglycine inter-muramyl chain peptide (39), and staP, a putative membrane protein with no assigned function.
Glycopeptide-resistance gene regulation in VRE requires a two-component regulatory system consisting of a soluble response regulator, VanR, and a membrane-associated His kinase, VanS (40). The A47934 biosynthetic gene cluster does include vanR and vanS homologues (ORFs 18 and 19, vanRst and vanSst). In VRE, these genes are in close proximity to the vanH,A,X cluster; however, in S. toyocaensis, they are separated by approximately 20 kb. The vanRst and vanSst genes from S. toyocaensis show high homology to a two-component regulatory system that is adjacent to a vanH,A,X gene cluster and orthologues of staO and staP genes found in the glycopeptide nonproducer Streptomyces coelicolor (12).
In addition to the vanRst and vanSst two-component system, two additional ORFs predicted to encode elements of gene regulation are founding associated with the A47934 biosynthetic gene cluster. StaQ is a predicted transcriptional regulator found upstream of the biosynthesis genes for which there is a homologous protein upstream of the complestatin cluster. Finally, staS encodes a putative DNA-binding protein of unknown function and is unique to the A47934 cluster.
Conclusions
The sequence analysis of the A47934 biosynthesis cluster gives significant insight into many aspects of glycopeptide antibiotic biosynthesis, regulation, and resistance. This will facilitate further studies on the regulation of glycopeptide biosynthesis and its interplay with resistance. Furthermore, this work sets the stage for metabolic engineering of the aglycone core in an effort to rationally design new structural diversity into this important family of antibiotics.
Acknowledgments
This work was supported by Canadian Institutes of Health Research Grant MT-14981 (to G.D.W.) and National Institutes of Health Grant GM 49338 (to C.T.W.). G.D.W. is the recipient of a Canada Research Chair in Biochemistry.
Abbreviations
- HPG
p-hydroxyphenylglycine
- DHPG 3,5-dihydroxyphenylglycine
NRPS, nonribosomal peptide synthetase
- GPA
glycopeptide antibiotic
- β-OHTyr
β-hydroxytyrosine
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. STO22459).
References
- 1.Barna J C J, Williams D H. Annu Rev Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 2.Nicas T I, Cooper R D G. In: Biotechnology of Antibiotics. Strohl W R, editor. New York: Dekker; 1997. pp. 363–392. [Google Scholar]
- 3.Leclercq R, Derlot E, Duval J, Courvalin P. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 4.Pootoolal J, Neu J, Wright G D. Annu Rev Pharmacol Toxicol. 2002;42:381–408. doi: 10.1146/annurev.pharmtox.42.091601.142813. [DOI] [PubMed] [Google Scholar]
- 5.Murray B E. N Engl J Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 6.Walsh C T, Fisher S L, Park I-S, Prohalad M, Wu Z. Chem Biol. 1996;3:21–28. doi: 10.1016/s1074-5521(96)90079-4. [DOI] [PubMed] [Google Scholar]
- 7.van Wageningen A M, Kirkpatrick P N, Williams D H, Harris B R, Kershaw J K, Lennard N J, Jones M, Jones S J, Solenberg P J. Chem Biol. 1998;5:155–162. doi: 10.1016/s1074-5521(98)90060-6. [DOI] [PubMed] [Google Scholar]
- 8.Pelzer S, Sussmuth R, Heckmann D, Recktenwald J, Huber P, Jung G, Wohlleben W. Antimicrob Agents Chemother. 1999;43:1565–1573. doi: 10.1128/aac.43.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu H T, Hubbard B K, Shah A N, Eide J, Fredenburg R A, Walsh C T, Khosla C. Proc Natl Acad Sci USA. 2001;98:8548–8553. doi: 10.1073/pnas.151246498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall C G, Wright G D. Biochem Biophys Res Commun. 1996;219:580–583. doi: 10.1006/bbrc.1996.0276. [DOI] [PubMed] [Google Scholar]
- 11.Neu J, Wright G D. FEMS Microbiol Lett. 2001;199:16–20. doi: 10.1111/j.1574-6968.2001.tb10644.x. [DOI] [PubMed] [Google Scholar]
- 12.Pospiech A, Neumann B. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 13.Marshall G C, Broadhead G, Leskiw B, Wright G D. Proc Natl Acad Sci USA. 1997;94:6480–6483. doi: 10.1073/pnas.94.12.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsushima P, Batlz R H. Microbiology. 1996;142:261–267. doi: 10.1099/13500872-142-2-261. [DOI] [PubMed] [Google Scholar]
- 15.Oh S H, Chater K F. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bierman M, Logan R, O’Brien K, Seno E T, Rao N, Schoner B E. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 17.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M. Genetic Manipulations of Streptomyces, A Laboratory Manual. Norwich, U.K.: The John Innes Foundation; 1985. [Google Scholar]
- 18.Puk O, Huber P, Bischoff D, Recktenwald J, Jung G, Sussmuth R D, van Pee K H, Wohlleben W, Pelzer S. Chem Biol. 2002;9:225–235. doi: 10.1016/s1074-5521(02)00101-1. [DOI] [PubMed] [Google Scholar]
- 19.Choroba O W, Williams D H, Spencer J B. J Am Chem Soc. 2000;122:5389–5390. [Google Scholar]
- 20.Hubbard B K, Thomas M G, Walsh C T. Chem Biol. 2000;7:931–942. doi: 10.1016/s1074-5521(00)00043-0. [DOI] [PubMed] [Google Scholar]
- 21.Pfeifer V, Nicholson G J, Ries J, Recktenwald J, Schefer A B, Shawky R M, Schroder J, Wohlleben W, Pelzer S. J Biol Chem. 2001;276:38370–38377. doi: 10.1074/jbc.M106580200. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Tseng C C, Hubbard B K, Walsh C T. Proc Natl Acad Sci USA. 2001;98:14901–14906. doi: 10.1073/pnas.221582098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Walsh C T. Chem Biol. 2001;8:301–312. doi: 10.1016/s1074-5521(01)00009-6. [DOI] [PubMed] [Google Scholar]
- 24.He J, Magarvey N, Piraee M, Vining L C. Microbiology. 2001;147:2817–2829. doi: 10.1099/00221287-147-10-2817. [DOI] [PubMed] [Google Scholar]
- 25.Stachelhaus T, Mootz H D, Marahiel M A. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 26.Konz D, Marahiel M A. Chem Biol. 1999;6:R39–R48. doi: 10.1016/S1074-5521(99)80002-7. [DOI] [PubMed] [Google Scholar]
- 27.Challis G L, Ravel J, Townsend C A. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 28.Schafer M, Sheldrick G M, Schneider T R, Vertesy L. Acta Crystallogr D. 1998;54:175–183. doi: 10.1107/s0907444997008895. [DOI] [PubMed] [Google Scholar]
- 29.Schafer M, Schneider T R, Sheldrick G M. Structure (London) 1996;4:1509–1515. doi: 10.1016/s0969-2126(96)00156-6. [DOI] [PubMed] [Google Scholar]
- 30.Trauger J W, Walsh C T. Proc Natl Acad Sci USA. 2000;97:3112–3117. doi: 10.1073/pnas.040560597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stachelhaus T, Mootz H D, Bergendahl V, Marahiel M A. J Biol Chem. 1998;273:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 32.Stachelhaus T, Walsh C T. Biochemistry. 2000;39:5775–5787. doi: 10.1021/bi9929002. [DOI] [PubMed] [Google Scholar]
- 33.Bischoff D, Pelzer S, Holtzel A, Nicholson G J, Stockert S, Wohlleben W, Jung G, Sussmuth R D. Angew Chem Int Ed Engl. 2001;40:1693–1696. doi: 10.1002/1521-3773(20010504)40:9<1693::aid-anie16930>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 34.Bischoff D, Pelzer S, Holtzel A, Nicholson G J, Stockert S, Wohlleben W, Jung G, Sussmuth R D. Angew Chem Int Ed Engl. 2001;40:4688–4691. doi: 10.1002/1521-3773(20011217)40:24<4688::aid-anie4688>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 35.Marshall C G, Lessard I A, Park I, Wright G D. Antimicrob Agents Chemother. 1998;42:2215–2220. doi: 10.1128/aac.42.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall C G, Zolli M, Wright G D. Biochemistry. 1999;38:8485–8491. doi: 10.1021/bi982843x. [DOI] [PubMed] [Google Scholar]
- 37.Marshall C G, Wright G D. J Bacteriol. 1998;180:5792–5795. doi: 10.1128/jb.180.21.5792-5795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lessard I A, Pratt S D, McCafferty D G, Bussiere D E, Hutchins C, Wanner B L, Katz L, Walsh C T. Chem Biol. 1998;5:489–504. doi: 10.1016/s1074-5521(98)90005-9. [DOI] [PubMed] [Google Scholar]
- 39.Hegde S S, Shrader T E. J Biol Chem. 2001;276:6998–7003. doi: 10.1074/jbc.M008591200. [DOI] [PubMed] [Google Scholar]
- 40.Arthur M, Molinas C, Courvalin P. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshall C G, Wright G D. FEMS Microbiol Lett. 1997;157:295–299. doi: 10.1111/j.1574-6968.1997.tb12788.x. [DOI] [PubMed] [Google Scholar]
- 42.Bentley S D, Chater K F, Cerdeno-Tarraga A M, Challis G L, Thomson N R, James K D, Harris D E, Quail M A, Kieser H, Harper D, et al. Nature (London) 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]