Abstract
The introduction of semidwarf rice (Oryza sativa L.) led to record yield increases throughout Asia in the 1960s. The major semidwarfing allele, sd-1, is still extensively used in modern rice cultivars. The phenotype of sd-1 is consistent with dwarfism that results from a deficiency in gibberellin (GA) plant growth hormones. We propose that the semidwarf (sd-1) phenotype is the result of a deficiency of active GAs in the elongating stem arising from a defective 20-oxidase GA biosynthetic enzyme. Sequence data from the rice genome was combined with previous mapping studies to locate a putative GA 20-oxidase gene (Os20ox2) at the predicted map location of sd-1 on chromosome 1. Two independent sd-1 alleles contained alterations within Os20ox2:a deletion of 280 bp within the coding region of Os20ox2was predicted to encode a nonfunctional protein in an indica type semidwarf (Doongara), whereas a substitution in an amino acid residue (Leu-266) that is highly conserved among dioxygenases could explain loss of function of Os20ox2 in a japonica semidwarf (Calrose76). The quantification of GAs in elongating stems by GC-MS showed that the initial substrate of GA 20-oxidase activity (GA53) accumulated, whereas the content of the major product (GA20) and of bioactive GA1 was lower in semidwarf compared with tall lines. We propose that the Os20ox2 gene corresponds to the sd-1 locus.
The semidwarfing gene in rice (sd-1) is one of the most important genes deployed in modern rice breeding. Its recessive character results in a shortened culm with improved lodging resistance and a greater harvest index, allowing for the increased use of nitrogen fertilizers (1, 2). The sd-1 gene was first identified in the Chinese variety Dee-geo-woo-gen (DGWG), and was crossed in the early 1960s with Peta (tall) to develop the semidwarf cultivar IR8 (3), which produced record yields throughout Asia and formed the basis for the development of new high-yielding, semidwarf plant types (3). Since the 1960s, sd-1 has remained the predominant semidwarfing gene present in current rice cultivars.
Many aspects of plant growth and development involve the gibberellin (GA) hormone family. The widely used semidwarfing genes in wheat, Rht-B1b and Rht-D1b, encode a protein involved in GA-signal transduction (4). In addition, GA-responsive dwarf mutants have been identified in many species, often resulting from mutations in genes encoding GA biosynthetic enzymes (reviewed in ref. 5). In rice, mutants dx and d18 are defective in ent-kaurene oxidase and GA 3-oxidase activity, respectively, representing early and late steps in the GA biosynthetic pathway (6, 7). These dwarf plants have a low content of GA1 (a major growth active GA in rice) and can be rescued by application of active GAs such as GA3. Previous studies (8, 9) have suggested that the sd-1 semidwarf results from a deficiency in active GAs, although the methods used (enzyme-linked immunosorbent or bioassay) lack the precision of modern chemical methods of analysis. Furthermore, sd-1 plants retained responsiveness to GA3 during seedling, tillering, and heading growth stages (8, 10). These properties of sd-1 are consistent with a semidwarf phenotype that is the result of a partial block in GA biosynthesis. We have identified a putative GA 20-oxidase gene very close to the Sd-1 locus on chromosome 1. We show that semidwarf mutants contain alterations in this gene, leading to changes in GA content that are consistent with a block at this step in the GA biosynthetic pathway and, therefore, propose that the GA 20-oxidase gene corresponds to the Sd-1 locus.
Materials and Methods
Plant Material.
Kyeema is a tall indica type rice cultivar developed in the Australian breeding program at Yanco, Australia. Its pedigree includes Pelde and Kulu (Yanco varieties) and Della, a tall long grain (fragrant) variety from Louisiana (USA). The semidwarf, indica type Doongara was derived from a cross between a tall, long grain cultivar Dawn (Texas) and IR8, a semidwarf cultivar carrying sd-1 which was developed at the International Rice Research Institute (IRRI) in the 1960s (3). IR36 was developed from IR8 at IRRI and also carries sd-1. The dwarfing gene in short-stature japonica type Calrose76 (a mutant isolated in the tall Californian cultivar Calrose following irradiation) was shown to be allelic to sd-1 (11).
Analysis of GA Content.
Kyeema and Doongara plants were grown at 29°C day/22°C night temperatures during the summer season in a phytotron glasshouse. Stem material was harvested when the top internode of the stem had reached approximately 50% of final length, with average lengths of 139 mm in Kyeema, 101 mm in Doongara, 134 mm in Calrose, and 117 mm in Calrose76. The tissue was frozen in liquid N2 and stored at −80°C. Frozen stem segments were ground in liquid N2 with sand in a mortar and transferred to a beaker with 80% methanol. Extraction, purification, and analysis of GAs and abscisic acid (ABA) by GC-MS using selected ion monitoring was by methods that have been described (12).
PCR Amplification of Genomic DNA.
PCR primers for the putative GA 20-oxidase gene were designed on the basis of the genomic sequence of Nipponbare available on the Rice Genome Research Project (RGP) database (http://rgp.dna.affrc.go.jp). The predicted ORF was amplified from Kyeema with the following primers: forward primer, 5′-CAACTCACTCCCGCTCAACACAGC, and reverse primer, 5′-TTTGAAATGCAATGTCGTCCACC were used to amplify exon 1; forward primer, 5′-GCGCCAATGGGGTAATTAAAACG and reverse primer, 5′-GGCATTCCATTGTTTGTGATTGG were used for exon 2; forward primer, 5′-GTTTGTCCTTGTCGCGTTGCTCAG and reverse primer, 5′-TCTGTTCGTTCCGTTTCGTTCCG were used for exon 3. The 5′ region in Doongara containing a deletion in exon 1 and exon 2 (see Results) was amplified by using the forward primer for exon 1 and the reverse primer for exon 2. The PCR analysis (20 μl) contained 100 ng of template DNA, 1 × PCR buffer (Qiagen, Germany), 1.5 mM MgCl2, 0.2 mM dNTPs, 5 μM primer, and 1 unit of Taq polymerase (Hotstar, Qiagen). The reactions were heated to 94°C for 5 min followed by 35 cycles of amplification at 94°C for 30 sec, 55°C for 30 sec, and 72°C for 50 sec. PCR products were cloned into a pGEM-T Easy Vector (Promega), and at least two independent clones from each PCR product were sequenced by using an automated sequencing system (ABI 377, Applied Biosystems).
RT-PCR Analysis of OsGA20ox2 Transcripts.
To isolate the GA20ox2 cDNA, RNA was extracted according to Vries et al. (13) from elongating stems of Calrose and Calrose76 rice plants. The stem material comprised the upper internode and flag leaf sheath and was harvested at the stage where the panicle emerged from the flag leaf sheath. The cDNA was amplified by using a one-step RT-PCR kit (Qiagen) according to the manufacturer's protocols, with a 58°C annealing temperature and forward primer 5′-CAACTCACTCCCGCTCAACACAGC-3′ located 22-bp upstream from the translation start, and a reverse primer 5′-GTTCGTTCCGTTTCGGTTCCG-3′ eight-bp downstream from the TGA stop codon. The RT-PCR products were gel-purified and ligated into pGEM-T-Easy vector (Promega).
DNA Blot Analysis.
Genomic DNA was extracted by using the methods described by Dellaporta et al. (14). DNA probe from exon 2 of Os20ox2 was amplified from genomic DNA by using primer sequences described above. DNA hybridization was carried out with 10 μg of HindIII-digested genomic DNA by using methods described by Lagudah et al. (15). Filters were washed twice in 2× SSC, 0.1% SDS at 65°C for 20 min, once in 1× SSC, 0.1% SDS at 65°C for 20 min, and once in 0.5× SSC, 0.1% SDS at 65°C for 15 min.
Results
Genetic and Physical Analysis of the Sd-1 Locus.
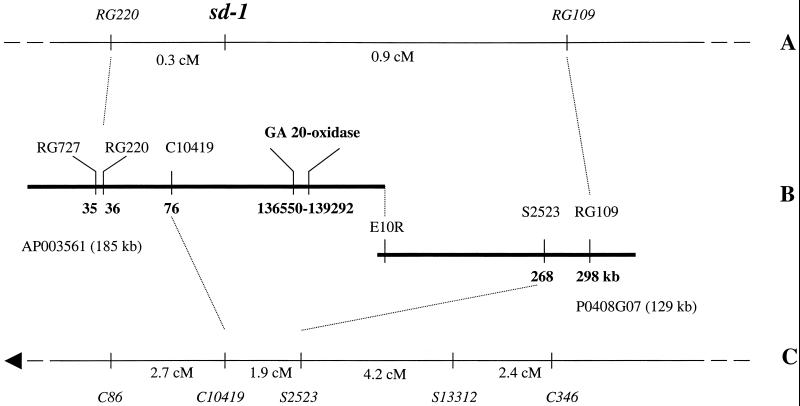
The rice sd-1 dwarfing gene was shown to be tightly linked to restriction fragment length polymorphism (RFLP) markers RG109 and RG220 on the long arm of chromosome 1 in at least three mapping studies. First, in 178 F2 segregants from a cross between Shiokari and a near-isogenic semidwarf line (ID47), the sd-1 gene was flanked by RG109 on the proximal (0.9 cM) and RG220 on the distal side (0.3 cM; ref. 16). Second, the analysis of 2,450 segregating lines derived from a cross between near-isogenic lines Taichung65 and Taichung65 (sd-1) separated sd-1 from RG109 by 0.2 cM (17). Third, in 50 F2 segregants from a cross between Kyeema and Doongara (sd-1), RG109 co-segregated with sd-1, whereas one recombinant was identified between RG220 and sd-1 (≈1 cM) (S. Garland, personal communication). In this study, we positioned RFLP markers RG109 and RG220 on a physical segment of chromosome 1 covered by a BAC contig of approximately 300 kb by using the physical map of Nipponbare available on the RGP database (http://rgp.dna.affrc.go.jp; Fig. 1). These markers were positioned near the ends of this contig spanning a physical distance of approximately 262 kb. Anchor markers C10419 and S2523, located within this region and separated by 192 kb, were mapped 1.9 cM apart on the high-resolution linkage map (RGP 2000). From this chromosomal segment and linkage map, we estimated that the ratio of physical to genetic distance was approximately 100 kb/cM in this region. When combined with data from the previous mapping studies, these results indicated that the sd-1 gene was located within a 262-kb interval flanked by markers RG109 and RG220.
Figure 1.
Genetic linkage maps and BAC contig of the sd-1 region on chromosome 1 of rice. (A) Genetic linkage map of sd-1 region (16). (B) Physical contig of DNA sequence flanking sd-1 consisting of BAC clones AP003561 and P0408G07 (RGP). (C) High-resolution genetic linkage map of the sd-1 region on chromosome 1 (RGP 2000). Arrow indicates the direction of the centromere.
Identification of a Putative GA 20-Oxidase in the sd-1 Region.
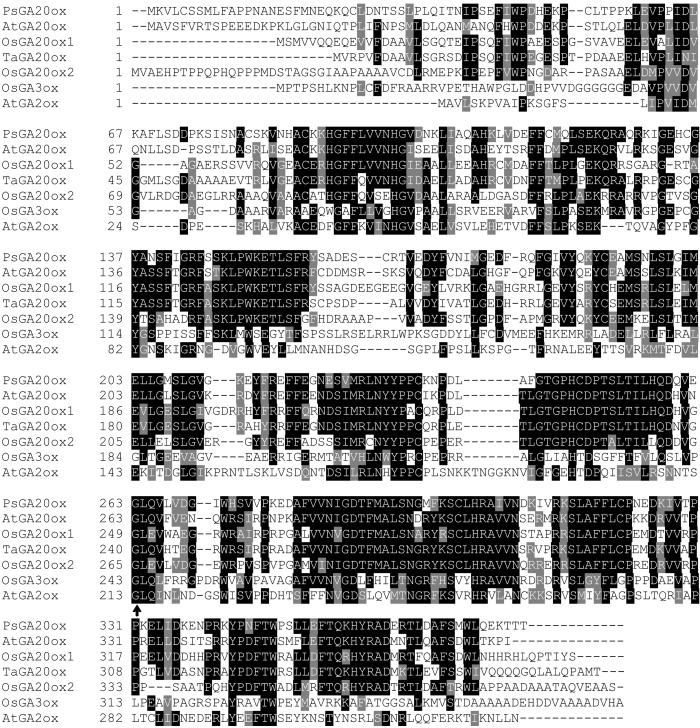
Because the sd-1 phenotype is consistent with the dwarfism that results from a deficiency in bioactive GA1, we investigated whether GA biosynthetic genes were present near the Sd-1 locus. The Rice Genome Automated Annotation system (RiceGAAS, RGP) was used to identify predicted coding regions within the interval flanked by RFLP markers RG109 and RG220, as well as within 500 kb of sequence on either side of these markers. Most of the 28 predicted coding regions located between RG109 and RG220 were related to hypothetical proteins of unknown function or retrotransposable elements. However, one predicted ORF (between positions 136550–139292 on BAC clone AP003561, Fig. 1) encoded a protein of 389 amino acids that was closely related to GA 20-oxidases previously isolated from Arabidopsis (GA5, 47% identity), pea (50% identity), rice (51% identity; refs. 18–20), and related GA 20-oxidase sequences from cereals such as wheat (Fig. 2). There was no other predicted coding region identified that contained significant amino acid sequence relatedness to proteins involved in GA biosynthesis in the regions either proximal or distal to the BAC contig. Genetic analysis has linked a mutant GA 20-oxidase gene to a semidwarf phenotype in Arabidopsis (18). Therefore, we investigated the possibility that the sd-1 allele is associated with a mutation in the putative GA 20-oxidase gene.
Figure 2.
Alignment of predicted dioxygenase proteins. Dark shading indicates amino acid residues that are conserved in the GA 20-oxidases. GenBank accession numbers are: pea, PsGA20ox, PSU58830; Arabidopsis, AtGA20ox, AAC39314; wheat, TaGA20ox, TAY14007; rice, OsGA20ox1, U50333; OsGA20ox2, AY114310; OsGA3ox, AB056519; AtGA2ox, AJ132435. Arrow indicates the highly conserved leucine residue.
Characterization of Mutant GA 20-Oxidase Genes in sd-1 Lines.
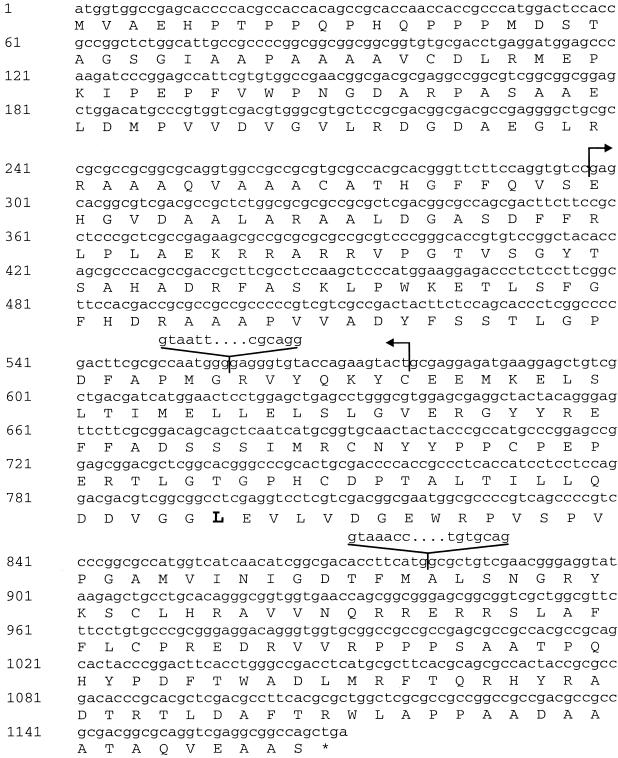
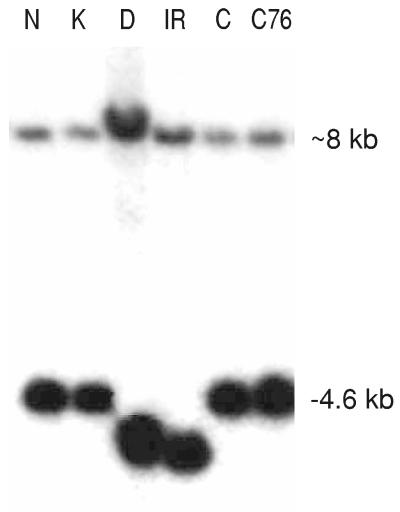
The putative GA 20-oxidase gene (Os20ox2) was amplified by PCR by using genomic DNA of cvs Kyeema (tall) and Doongara (sd-1) as template and primers developed from the Nipponbare genomic sequence (http://rgp.dna.affrc.go.jp). Sequence analysis of these PCR products demonstrated that the predicted ORF isolated from Kyeema was identical in sequence to Nipponbare. Intron positions (557–558 and 879–880 in Fig. 3) were confirmed by RT-PCR (see Materials and Methods) and were conserved in relation to GA 20-oxidase genes isolated from Arabidopsis and pea (18, 19). The corresponding sequence amplified from the semidwarf cultivar Doongara contained a 280-bp deletion within the coding region. This deletion spanned parts of exon 1 and exon 2 and was predicted to result in a truncated, nonfunctional polypeptide containing the first 99 amino acid residues of the GA 20-oxidase, followed by an additional 3 amino acids that were in the wrong reading frame before a termination codon (Fig. 3). Hybridization of a DNA probe derived from exon 2 to digested genomic DNA of Kyeema (tall), Doongara (sd-1), and IR36 (sd-1) resulted in an RFLP pattern which was consistent with a deletion in the putative GA 20-oxidase gene (Fig. 4). The RFLP analysis identified two gene members in the rice genome: a strong band (4.6 kb) of the predicted size corresponding to a single member at the sd-1 locus, and a weaker band (≈8 kb) probably identifying a related GA 20-oxidase gene (Os20ox1) located on chromosome 3 (GenBank U50333; ref. 20).
Figure 3.
Nucleotide and predicted amino acid sequence of GA 20-oxidase sequence isolated from chromosome 1 of rice. The positions located between 557–558 and between 879–880 are indicated. Arrows denote the position of the 280-bp deletion occurring within the coding sequence in Doongara (sd-1). The last nucleotide (cytosine) before the deletion start (position 297) in Kyeema is changed to an adenine residue in Doongara, resulting in a synonymous nucleotide substitution (Ser TCC → Ser TCA). The conserved leucine Leu-266 that was changed to phenylalanine in semidwarf mutant Calrose76 is highlighted.
Figure 4.
Hybridization of DNA probe of exon 2 derived from the putative GA 20-oxidase gene (Os20ox2) to (HindIII) digested genomic DNA of Nipponbare (N, tall), Kyeema (K, tall), Doongara (D, sd-1), IR36 (IR, sd-1), Calrose (C, tall), and Calrose76 (C76, sd-1 allele).
We investigated whether an independent semidwarf mutant, allelic to sd-1, also contained an altered Os20ox2sequence by amplifying sequences from Calrose (tall) and a semidwarf Calrose76 (11). The predicted amino acid sequence of Calrose was identical to the sequence of Nipponbare and Kyeema. The DNA sequence of Calrose76 was identical to Calrose except for a C to T transition at position 798 that resulted in a change of the predicted amino acid leucine (Leu-266) in Calrose to phenylalanine in Calrose76 (Fig. 3). This change represents a conservative substitution between hydrophobic amino acids, and although the side chain of phenylalanine is larger than that of leucine, such a substitution may not necessarily affect enzyme activity. However, a comparison of dioxygenase gene sequences from public databases revealed that Leu-266 was conserved in all GA 20-oxidase sequences. Furthermore, this residue was still conserved when 20-oxidase sequences were compared with more distantly related dioxygenases such as GA 3-oxidase and 2-oxidase sequences (refs. 7, 21; Fig. 2).
The deduced amino acid sequence of Os20ox2 was submitted to the SWISS-MODEL protein server (22), and a portion of 94 aa (starting at amino acid residue 244 in Fig. 2) was selected with a very close match to the crystal structure of a region of another dioxygenase, isopenicillin N synthase, from Aspergillus nidulans (results not shown). The α-carbon atoms of this region closely match those of the crystal structure, with an rmsd of 0.6–0.7 Å. This region includes the Leu-266 residue that is altered in the sd-1 Calrose76 mutant, and the corresponding residue in isopenicillin N synthase is also a leucine (Leu-231) that has been shown to interact by van der Waals contact with the substrate of the enzyme (23). The extremely high degree of conservation of this leucine residue, and its interaction with substrate in at least one case, suggest that any substitution will be likely to reduce or abolish enzyme activity.
Analysis of GA Contents in Elongating Stems.
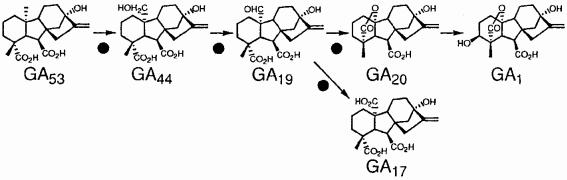
Most of the variation in final plant height between tall and semidwarf lines result from differences in stem length of the first (subtending panicle) and second (subtending flag leaf) stem internodes (24). We initially compared the GA contents of Kyeema (tall) and Doongara (sd-1) by using the uppermost stem internodes at equivalent stages of growth (about 50% of final length). The results (Table 1) showed a higher (about three-fold) content of GA53 in the semidwarf compared with the tall line, and lower (about two-fold) contents of GA20 and GA1, and possibly GA17, which is derived from GA19 (Table 1). There was no significant difference between semidwarf and tall lines in the contents of GA44 and GA19. This analysis also revealed that early 13-hydroxylated GAs predominated in the stem segments, and the peaks identified for GA9 and GA34 were just above the detection limits. The elevated content of GA53 in the semidwarf, and the reduced amount of GA20 (and GA1) were consistent with impaired GA 20-oxidase activity (Fig. 5) and dwarfing caused by a deficiency of GA1. Because sd-1 rice still contained considerable amounts of GA44, GA19, GA20, and GA1, yet is proposed to have a nonfunctional Os20ox2 gene, we conclude that there must be another functional GA 20-oxidase isozyme whose activity provides biosynthetic precursors to GA1 in the stem segment.
Table 1.
Contents of GAs in equivalent stem segments of tall vs. semidwarf rice plants
| Compound | Content (pg
segment−1)*
|
|||||
|---|---|---|---|---|---|---|
| Kyeema (tall) | Doongara (dwarf) | P† | Calrose (tall) | Calrose 76 (dwarf) | P† | |
| 13-OH GAs‡ | ||||||
| GA53 | 802 ± 54 | 2546 ± 133 | <0.001 | 1117 ± 72 | 2033 ± 146 | 0.005 |
| GA44 | 687 ± 34 | 661 ± 42 | 0.65 (ns) | 828 ± 109 | 445 ± 61 | 0.04 |
| GA19 | 4092 ± 269 | 4108 ± 401 | 0.97 (ns) | 4233 ± 141 | 2827 ± 171 | 0.003 |
| GA20 | 178 ± 13 | 73 ± 4 | <0.001 | 270 ± 27 | 131 ± 18 | 0.012 |
| GA1 | 146 ± 8 | 96 ± 6 | 0.005 | 169 ± 15 | 134 ± 13 | 0.15 (ns) |
| GA8 | <dl§ | <dl§ | nd | nd | ||
| GA17 | 395 ± 13 | 109 ± 20 | <0.001 | nd | nd | |
| Non-13-OH GAs¶ | ||||||
| GA9 | 7.9 | 6.1 | nd | nd | ||
| GA4 | interference | interference | nd | nd | ||
| GA34 | 7.1 | 5.7 | nd | nd | ||
Contents estimated using calibration curves for all GAs except for GA17 (estimated from the peak area of ion 492 relative to the corrected peak area of ion 436 of deuterated GA19) and GA53 (estimated on the basis of corrected 448/450 peak areas). Values are mean ± SE of three (Doongara, Calrose, Calrose 76) or four (Kyeema) independent samples, except for GA8, GA9, and GA34, which are single determinations.
Probability (t test) that the difference between the means is due to chance.
GAs are shown in metabolic order except for GA17, a tricarboxylic acid, which represents a branch off the biosynthetic pathway from GA19. GA1 is the only bioactive GA in this sequence.
Endogenous GA8 was below the detection limit.
GA9, GA4, and GA34 are equivalent to GA20, GA1, and GA8 in the 13-OH GAs. GA4 was not determined because interfering ions were present.
Figure 5.
The early 13-hydroxylation pathway of gibberellin biosynthesis in higher plants. Black circles indicate oxidation steps catalyzed by GA 20-oxidase (adapted from ref. 20).
We also compared GA contents in equivalent stem segments of Calrose and Calrose76. The results (Table 1) were similar to those above, in that there was an accumulation of GA53 and a deficiency of GA20 in the semidwarf variety, consistent with reduced activity of a GA 20 oxidase. The extent of these changes was slightly less than that observed for the Doongara/Kyeema pair, as was the degree of reduction in GA1 content of Calrose76 compared with Calrose (nonsignificant at P < 0.05). Calrose76 also had lower contents of GA44 and of GA19 than Calrose. These differences might be because of (i) other genetic differences between the Doongara/Kyeema pair, (ii) slight activity of the mutant GA20ox2 gene product in Calrose76, or (iii) experimental variation.
Discussion
The data from the public rice genome sequence was combined with previous mapping studies to locate a putative GA 20-oxidase gene (Os20ox2) at the expected position of sd-1 on chromosome 1. Os20ox2 was the only predicted ORF identified within a target interval of 262 kb that contained significant amino acid sequence relatedness to proteins involved in GA biosynthesis. Two independent semidwarf, allelic mutants contained alterations within Os20ox2: a deletion within Os20ox2 was predicted to encode a nonfunctional protein in Doongara (DGWG source of sd-1), whereas a change in an amino acid residue (Leu-266), which was highly conserved among dioxygenase sequences, could explain the loss of function of Os20ox2 in the semidwarf mutant Calrose76. The quantification of GAs in elongating stems revealed that the initial substrate of GA 20-oxidase (GA53) accumulated, whereas the content of the major product (GA20) was reduced in semidwarf compared with tall lines. These results were consistent with impaired GA 20-oxidase activity and dwarfing caused by a deficiency of GA1. Therefore, we propose that the semidwarf (sd-1) phenotype of rice is the result of reduced GA 20-oxidase activity, and that the defective GA 20-oxidase gene defines the sd-1 locus.
The determination of GA contents in elongating stems revealed very high levels of GA19, as observed previously in rice seedlings (20). In both semidwarfs, there was a lowered content of GA20, presumably a consequence of reduced 20-oxidase activity. In turn, the amounts of GA1, a major growth active GA, were only 65% (Doongara/Kyeema) or 80% (Calrose76/Calrose) of the corresponding tall values, although in the latter case the difference was not significant. This modest reduction in GA1 content is consistent with the relatively small reduction (approximately 25%) in the height of semidwarf vs. tall lines. The mutant GA 20-oxidase in Doongara should be a nonfunctional polypeptide, so our results imply that rice contains additional 20-oxidase activity that provided precursors contributing to approximately half of the GA1 content found in the stem internodes. A GA 20-oxidase mapping to chromosome 3 could account for the GA20 detected in stems (20). Similarly in Arabidopsis, the GA-responsive semidwarf mutant ga5 resulted from a defective GA 20-oxidase gene, and other gene members were predicted to supplement GA 20-oxidase activity resulting in a semidwarf phenotype (18).
The sd-1 gene in rice and Rht genes in wheat have played similar roles in height reduction associated with significant yield increases. The molecular bases for semidwarf stature seem to be different, with sd-1 resulting from a mutant GA 20-oxidase gene causing a deficiency of bioactive GA in elongating stems, whereas Rht genes are negative regulators of GA signaling that in the mutant are no longer modulated by GA (4). Unlike Rht semidwarf wheat, sd-1 semidwarf rice retains the ability to respond to applications of bioactive gibberellins. Most of the dwarf mutants identified in rice (d1 to d60), including those resulting from mutations in GA biosynthetic genes (dx and d18), are not used in crop improvement because they are associated with severe dwarfism, floret sterility, or abnormal plant and grain development (25). The agronomic success of sd-1 may be intimately linked to the developmental timing of its moderate dwarfing in relation to stem growth and panicle development. It will be of considerable interest to determine the detailed expression patterns of the two 20-oxidase isozymes in different parts of the rice plant, because this determination will underlie physiological interpretations of semidwarf productivity.
During the final stages of the review process of this manuscript, Monna et al. (26) and Sasaki et al. (27) described independent mutations in the GA20-oxidase gene located at the sd-1 locus, two of which confirmed the sequence alterations reported here.
Acknowledgments
We thank Dr. Laurie Lewin for his help in locating and supplying seed stocks, Dr. Steve Garland for sharing genetic mapping data, Drs. Tony Ashton and Paul Carr for help in protein structure analysis, and Lea-Ellen Hogie and Carol Harding for excellent technical assistance. W.S. was supported by Graingene, Australia.
Abbreviation
- GA
gibberellin
- RGP
Rice Genome Research Project
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY114310).
References
- 1.Jennings P R. Crop Sci. 1964;4:13–15. [Google Scholar]
- 2.Walcott J J, Laing D R. Aust J Exp Agric Anim Husb. 1976;16:578–587. [Google Scholar]
- 3.International Rice Research Institute. Annual Report for 1966. 1967. pp. 59–82. [Google Scholar]
- 4.Peng J, Richards D E, Hartley N M, Murphy G P, Devos K M, Flintham J E, Beales J, Fish L J, Worland A J, Pelica F, et al. Nature (London) 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- 5.Hedden P, Kamiya Y. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa S, Toyomasu T, Yamane H, Murofushi N, Ideda R. Plant Cell Physiol. 1996;37:363–368. [Google Scholar]
- 7.Itoh H, Uegushi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. Proc Natl Acad Sci USA. 2001;98:8909–8914. doi: 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Z, Li D. Intern Rice Res Notes. 1996;21:22–23. [Google Scholar]
- 9.Tang R S, Zhang Y H, Zhang J Y, Wu G N. Scientia Agric Sinica. 1991;24:51–56. [Google Scholar]
- 10.Mitsunga S, Tashiro T, Yamaguchi J. Theor Appl Genet. 1994;87:705–712. doi: 10.1007/BF00222896. [DOI] [PubMed] [Google Scholar]
- 11.Foster K W, Rutger J N. Genetics. 1978;88:559–574. doi: 10.1093/genetics/88.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green L S, Fergestad E M, Poole A, Chandler P M. Plant Physiol. 1997;114:203–212. doi: 10.1104/pp.114.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vries D, Hoge H, Bisseling T. In: Plant Molecular Biology Manual. Gelvin S B, Schilperoort R A, Verma D S, editors. Dordrecht, The Netherlands: Kluwer; 1988. pp. 1–13. [Google Scholar]
- 14.Dellaporta S L, Wood J, Hicks J B. Plant Mol Biol Rep. 1983;4:19–21. [Google Scholar]
- 15.Lagudah E S, Appels R, Brown A H D, McNeil D. Genome. 1991;34:375–386. [Google Scholar]
- 16.Maeda H, Ishii T, Mori H, Kuroda J, Horimoto M, Takamure I, Kinoshita T, Kamijima O. Breeding Sci. 1997;47:317–320. [Google Scholar]
- 17.Hahn J H, Yoon U H, Lee K S, Kim Y-W, Yun C H, Kim Y H, Kim H-I, Eun M Y. Proc. Plant Animal Genome Conf. VIII. 2000. p. 172. [Google Scholar]
- 18.Xu Y-L, Li L, Wu K, Peeters A J M, Gage D A, Zeevaart J A D. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin D N, Proebsting W M, Parks T D, Dougherty W G, Lange T, Lewis M J, Gaskin P, Hedden P. Planta. 1996;200:159–166. doi: 10.1007/BF00208304. [DOI] [PubMed] [Google Scholar]
- 20.Toyomasu T, Kawaide H, Sekimoto H, von Numers C, Phillips A L, Hedden P, Kamiya Y. Physiol Plant. 1997;99:111–118. [Google Scholar]
- 21.Thomas S G, Phillips A L, Hedden P. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 23.Roach P L, Clifton I J, Hensgens C M H, Shibata N, Schofield C J, Hajdu J, Baldwin J E. Nature (London) 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- 24.Shi C, Shen Z. Chin J Rice Sci. 1996;10:13–18. [Google Scholar]
- 25.Aquino R C, Jennings P R. Crop Sci. 1966;6:551–554. [Google Scholar]
- 26.Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y. DNA Res. 2002;9:1117. doi: 10.1093/dnares/9.1.11. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush G S, et al. Nature (London) 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]