Abstract
The Arabidopsis thaliana genome contains at least 50 predicted AtCMPG genes. The encoded protein family is defined by a common domain possessing four strictly conserved amino acid residues [Cys, Met, Pro, and Gly (CMPG)] that designate the family. Two members, AtCMPG1 and AtCMPG2, with high sequence similarity to the previously described, immediate-early pathogen-responsive PcCMPG1 gene from Petroselinum crispum were selected for analysis of their expression modes and defense-related promoter elements. Among the most striking similarities with PcCMPG1 were immediate-early transcriptional activation on infection or treatment with a pathogen-derived elicitor and the functional importance of a W-box-containing AtCMPG1 promoter element. Remarkably, this strongly pathogen/elicitor-responsive element, F, did not respond to wounding, in contrast to the AtCMPG1 promoter itself. Comparative analysis, both within the A. thaliana genome and across species, provided further insight into the large structural diversity of W-box-containing elements. Possible roles of AtCMPG proteins in regulatory processes are discussed with reference to a large variety of family members, partly with assigned functions, from plants as well as animals.
The family of CMPG proteins was recently identified on the basis of extensive sequence similarity within one characteristic domain among otherwise seemingly unrelated regions (1). A domain consensus sequence was inferred from the original representative, PcCMPG1, in the context of more than 20 homologs from Arabidopsis thaliana and other plant species. This consensus included several strictly conserved amino acid residues, beginning with the four residues (Cys, Met, Pro, and Gly) designating the family. Particular interest in the PcCMPG1 gene and the encoded protein arose from its suspected function as a transcriptional regulator in pathogen defense (1). With the genomic sequence of A. thaliana almost fully established (2), a comprehensive analysis of the entire CMPG family in this species is now possible, as is a comparison of selected AtCMPG genes with PcCMPG1.
Among the characteristic features of PcCMPG1 is the unusually rapid, immediate-early induction of the encoding mRNA by a pathogen-derived elicitor or by cycloheximide (1). Particularly noteworthy is an exceptional type of pathogen- or elicitor-responsive W-box-containing element within the PcCMPG1 gene promoter, which does not respond to wounding and may therefore bear potential for new approaches to disease resistance breeding in crop plants (1). Here we analyze a structurally and functionally related AtCMPG1 gene promoter element, compare the expression patterns of the pathogen-responsive AtCMPG1 and AtCMPG2 genes with those of PcCMPG1 in parsley, and discuss possible functions of the CMPG protein family.
Materials and Methods
Plant Material.
A. thaliana ecotype Columbia 0 plants were grown in illuminated phytochambers (≈500 μE⋅m−2⋅s−1) either under short-day conditions (8 h) for leaf development or under long-day conditions (14–16 h) for flower induction. Parsley (Petroselinum crispum; ref. 3) and A. thaliana (ecotype Columbia 0, line At7) cells (4) were propagated as previously reported.
Elicitor Treatment and Infections.
A crude Pmg elicitor preparation (5), synthetic Pmg-derived Pep25 elicitor (6) or a synthetic flagellin-derived elicitor (7) were added to cultured cells as indicated, using final concentrations of 50, 0.3, or 2.3 μg/ml, respectively. Pseudomonas syringae pv. tomato strain DC3000 was cultivated and vacuum-infiltrated into leaves by using a titer of 108 colony-forming units (cfu)/ml (8). Homogenized mycelium of Alternaria brassicicola strain MUCL20297 (9) was applied in droplets and conidiosporangia of Peronospora parasitica pv. Cala2 (1) were spray-inoculated onto leaf surfaces.
Transgenic Plants.
Plants were transformed using the floral-dip method (10), Agrobacterium tumefaciens GV3101 (11), and the binary Ti vector pGPTV (12) harboring promoter fragments upstream of the GUS reporter gene. Seeds from transformed plants were surface-sterilized with 70% and 95% ethanol, stored at 4°C for 2 days, and germinated on 0.8% agar (1). Plants from the T2 and T3 generation were analyzed for β-glucuronidase (GUS) expression (13).
Molecular Methods.
The AtCMPG1 gene was isolated by hybridization of a filter containing DNA from 9,216 bacterial artificial chromosome (BAC) clones (14) with a random 32P-labeled cDNA probe. Recombinant DNA techniques were conducted according to standard protocols (15, 16). Rapid amplification of cDNA ends (RACE) was performed using the 5′/3′-RACE Kit (Boehringer Mannheim) according to the manufacturer's instructions. Total extracted RNA or digested genomic DNA (10 μg) was separated on 1.3% formaldehyde-agarose or 0.8% agarose gels, respectively, and transferred onto Nylon membrane (17). cDNAs were 32P-labeled by random priming and used for hybridization under stringent conditions (18). Promoter fragments were translationally fused either directly to the GUS reporter gene or with the 35S minimal promoter in between (1). Parsley or A. thaliana protoplasts (≈2 × 106) were transfected (19) using 10 μg of ScaI-linearized plasmid and assayed for GUS activity (13) 6 h post-transfection. The gcg software package (WISCONSIN PACKAGE VERSION 10.0, Genetics Computer Group, Madison, WI) was used for DNA sequence analysis. The findpatterns program of the same package was used for promoter element searches within 499 bp upstream of all ORFs contained in the MIPS database (http://mips.gsf.de/) as annotated by November 2001.
Protein Sequence Analysis.
All protein sequences were obtained by blast analysis (20) of the TAIR (http://www.arabidopsis.org/) and GenBank databases (http://www.ncbi.nlm.nih.gov/) using an E value cut-off of 1 × 10−5. A neighbor-joining tree of uncorrected pair-wise distances between protein sequences (alignment available on request) was constructed using clustalw(21). Gapped sites were excluded.
Results
Structural Analysis of Selected AtCMPG Genes.
When this study was initiated, the A. thaliana genome sequence was only partially complete. Of all predictable AtCMPG genes accessible in the databases, the two closest relatives of PcCMPG1, designated AtCMPG1 and -2, were chosen for further analysis. Whereas the entire sequence was available for AtCMPG2, AtCMPG1 existed only in the form of a 476-bp expressed sequence tag (EST) and was cloned and sequenced from a BAC library. The deduced AtCMPG1 (48.2 kDa) and AtCMPG2 (46.8 kDa) proteins are 45% and 30% sequence identical, respectively, to PcCMPG1. Both are most probably encoded by single-copy genes, as indicated by genomic DNA-blot and database analyses. The transcriptional start sites of the two intronless genes, as determined by RACE, are located 117 bp (AtCMPG1) and 94 bp (AtCMPG2) upstream of the predicted translational start sites.
mRNA Expression Modes.
RNA gel-blot analysis indicated very similar relative mRNA abundance for AtCMPG1 and -2. Both were very low or undetectable in all major plant organs or cell types, except roots, suspension-cultured cells, and, to a much lesser extent, senescing leaves (data not shown). Expression in roots was probably due to infectious soil conditions and was not detectable in plants grown in sterilized, liquid MS (Murashige–Skoog) culture medium under long-day (16 h), low-light conditions. Other possible causes may have been the inevitable wounding of roots during soil removal before analysis or dark/light effects.
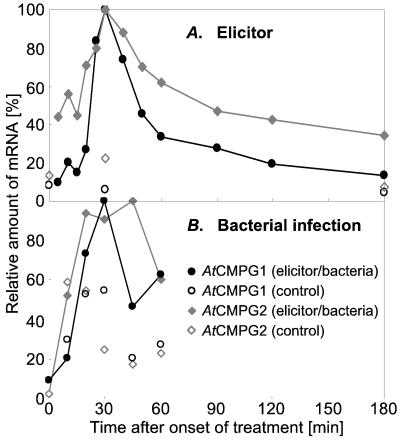
On treatment of cultured A. thaliana cells with a pathogen-derived (Pmg) elicitor preparation, both mRNAs accumulated rapidly, strongly, and transiently with similar time courses (Fig. 1A). AtCMPG1 mRNA started from a lower background than AtCMPG2 mRNA and increased and decreased even more steeply around a common peak at about 30 min after addition of elicitor. Similar, strong induction of both mRNAs by 10 μM cycloheximide indicated an immediate-early response (22, 23). Similar changes occurred in A. thaliana leaves on infection with either an avirulent or a virulent race of the bacterial pathogen, P. syringae pv. tomato DC3000 (with or without the avirulence gene avrRPM1), as shown in Fig. 1B for the response to avirulent bacteria.
Figure 1.
Timing of AtCMPG1 and AtCMPG2 mRNA induction in Pmg elicitor-treated, suspension-cultured cells (A) and in P. syringae-infected leaves of A. thaliana (B) [incompatible interaction; compatible interaction essentially similar (not shown)].
Transgenic Plants.
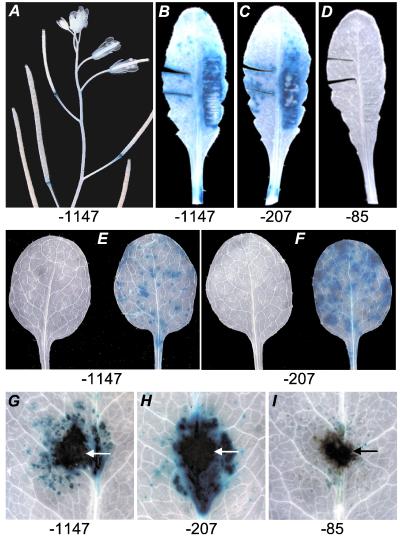
Major portions of the two gene promoters, comprising 1147, 207, or 85 bp of AtCMPG1, or 1382 or 522 bp of AtCMPG2, were translationally fused to the GUS reporter gene and the constructs used for generating transgenic A. thaliana plants. Representative results from the analysis of at least five independent AtCMPG1 lines each are shown in Fig. 2. Constitutive expression was very low throughout the plants, except roots (not shown) and receptacles (Fig. 2A), as well as the tips of siliques in the case of transgenic AtCMPG2 plants. Wounding (Fig. 2 B–D) or infection with either one of two exogenously applied pathogens (Fig. 2 E–I) strongly induced GUS activity in AtCMPG1 plants bearing the −1147 or the −207 fragment, whereas the −85 fragment gave a very weak response. Although injection of the bacterial pathogen, P. syringae pv. tomato, caused similar effects, this inoculation method was inevitably associated with wounding, and mock injection of a sterile MgCl2 solution also induced GUS activity (see also Fig. 1B), rendering the bacterial contribution equivocal under these assay conditions.
Figure 2.
Expression modes of AtCMPG1 promoter/GUS constructs in transgenic A. thaliana plants. Promoter fragments of different lengths up to the indicated positions (below panels) were tested for endogenous GUS expression (A; 6-week-old plant) and for responsiveness either to wounding [B–D; 5-week-old plant 4 h after wounding by cutting (Left) or squeezing with forceps (Right)] or to fungal infections by P. parasitica pv. Cala 2 (E and F, 10-day-old primary leaves) or A. brassicicola (G–I; 5-week-old leaves). Infected leaves were harvested 1 or 4 days postinoculation, respectively. Arrows mark the positions of fungal mycelium. Mock-inoculated leaves are shown on the Left in E and F. The fragments tested were the same as constructs A–C in Fig. 3 (see below).
Promoter Deletions and Mutations.
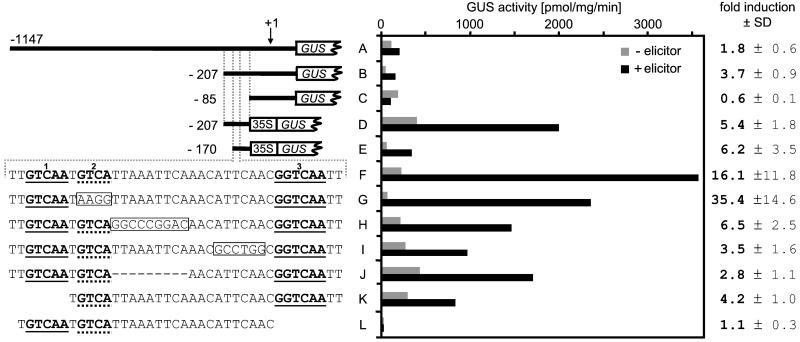
The apparent, critical part of the AtCMPG1 gene promoter between positions −85 and −207 contains a short region with considerable similarity to the major pathogen/elicitor-response element of the PcCMPG1 gene promoter (1). In AtCMPG1, this region comprises three W-box core sequences (TGAC; ref. 24). These were tested for functional significance by using the parsley protoplast assay (Fig. 3). Although direct translational fusions with the GUS reporter gene gave only low induction values (Fig. 3, constructs A–C), the results indicated an increase in inducibility on shortening of the fragment from 1147 (A) to 207 bp (B), and a complete loss on removal of the W-box-containing region (C). These effects were more pronounced when the putative element-containing fragment (positions −85 to −207) was combined with the 35S minimal promoter from the cauliflower mosaic virus. Relative to this construct (D), deletions from the 5′ (E) and additionally from the 3′ end (F; positions −132 to −170) yielded increasingly strong elicitor responsiveness, with highest values displayed by the putative element proper (F). Mutation of the second of the three W-box core motifs within F (G) further increased, rather than decreased, the effect. By contrast, various other mutations (H–L), including internal block mutations (H and I), an internal deletion (J), or removal of either one of the 5′- and 3′-located W-box motifs (K and L), had markedly negative effects.
Figure 3.
Functional analysis of the AtCMPG1 gene promoter. Elicitor (Pep25) responsiveness of promoter/GUS constructs was measured using the parsley protoplast transfection assay. Absolute GUS activity (bars) and fold-induction values are means from at least six independent transfections. Constructs A–C are direct translational fusions with the reporter gene, and constructs D–L contain the 35S minimal promoter in between.
Qualitatively, but not quantitatively, similar results were obtained using protoplasts from cultured A. thaliana cells and a system-adapted, flagellin-based peptide elicitor (7). However, higher background activities and smaller elicitor effects than achieved with the parsley system greatly impaired the sensitivity of the assay. Nevertheless, the results confirmed the association of elicitor responsiveness with element F.
Based on these results, F was tested in transgenic plants by the same procedure as used above in Fig. 2. Plants harboring F in dimerized form in front of the 35S minimal promoter and the GUS reporter gene responded strongly to fungal infection, but not to wounding, similar to the results shown in Fig. 2 G and H. The monomeric form gave no visible response under these conditions, possibly because of the low sensitivity of the method. Using monomers or dimers of E17 for comparison, the results were very similar to those obtained with F.
Analogous attempts to identify an elicitor-responsive element on the AtCMPG2 gene promoter were unsuccessful. Although this promoter contains a total of 11 W-box core motifs within 1,000 bp from the transcription start site, neither large fragments (up to positions −1342 and −522) nor small fragments containing either all or 3′- or 5′-deleted versions of the structurally most suspicious element, located at positions −43 to −87 (TCTCGTCAAAAAAGTGAAATTTGACGTCACCAAAGTTTGACTCTT), showed any elicitor responsiveness in the standard parsley or A. thaliana protoplast assays. None of the 11 W-box core motifs is embedded in a sequence similar to F.
Frequency of F in A. thaliana Gene Promoters.
Moreover, no F-like sequence, other than F itself in AtCMPG1, was found in a database comprising all predicted A. thaliana gene promoter regions up to 499 bp from the translational start sites. Twelve mismatches had to be permitted before two additional 39-bp regions could be identified, both of which, however, lacked W boxes. A search for the basic W-box arrangement in F, as in GTCAATGTCA(N19)GGTCAA, yielded only F itself. Within this arrangement, considerable relaxation of the W-box spacing, as in GTCA(N1–4)GTCA(N17–21)GTCA, was required to arrive at a total of 32 promoter sequences containing three W-box cores with similar spacing, but otherwise little consensus left. However, neither AtCMPG2 nor PcCMPG1 were among this latter group.
AtCMPG3–6.
These results prompted us to select four additional AtCMPG genes, with varying degrees of sequence similarity to AtCMPG1 and -2, for comparison with respect to both the occurrence of W-box repeats in the promoter and expression modes. The predicted phylogenetic relationships among all six deduced AtCMPG proteins are shown below in the context of the next paragraph (Fig. 4). In contrast to AtCMPG2 (see above), none of these four additional AtCMPG genes contained W-box repeats (within positions up to −1000) with a spacing approaching that observed for F. The mRNA expression levels in cultured A. thaliana cells were either much lower than those of AtCMPG1 or -2 (Fig. 1A) and remained largely unaffected by elicitor (AtCMPG3 and -4), or were even too low to prove or disprove elicitor-induced changes (AtCMPG5 and -6). In A. thaliana plants, at least some expression could be demonstrated by reverse transcription (RT)-PCR for all six AtCMPG mRNAs.
Figure 4.
Predicted phylogenetic relationships among all proteins from A. thaliana and other species that follow the criteria for sequence similarity defined in the text. See text also for explanation of symbols (filled and open circles) and definition of the three structural types ①–③. Numbers above branches indicate frequencies within 100 bootstrap replicates (shown only for values >50%). Dark shading, functionally identified proteins; light shading, proteins reported to be putative kinases.
The AtCMPG Family.
On the basis of the more complete A. thaliana data, we extended our similarity search (March 2002) to all predicted proteins from this species. Fig. 4 depicts the apparent phylogenetic relationship of those 46 identified members of the AtCMPG family, as well as 18 presently known homologs from this and other plant and animal species, that share ≥40% sequence identity with AtCMPG1 across a continuous stretch of ≥50 aa. These criteria were chosen such that all six AtCMPGs analyzed, as well as PcCMPG1, were included in the survey.
With few exceptions, the 64 proteins (58 plant, 6 animal) can be assigned to one of three structural types, largely overlapping with clade membership, where the CMPG domain is located in the N-terminal (type 1), central (type 2), or C-terminal part of the protein (type 3). In five of the six AtCMPG isoforms investigated in this study, as well as in PcCMPG1, the CMPG domain is N-terminally positioned.
Discussion
These results demonstrate the existence of a sizable, hitherto unrecognized family of CMPG proteins in A. thaliana. As expected from the initial comparative approach, at least two genes from this family, AtCMPG1 and -2, share the special property of immediate-early pathogen/elicitor responsiveness with the reference gene from parsley, PcCMPG1 (1). However, the apparent, contrasting behavior of the four additionally analyzed genes, AtCMPG3–6, including the closest structural relative of AtCMPG1, AtCMPG5, suggests that primary structure similarity is not necessarily correlated with gene regulatory response. In this regard, the CMPG family appears to resemble other large families of early or immediate-early inducible proteins with diverse regulatory functions—e.g., the MYB (25) and the Trp/Arg/Lys/Tyr-containing (WRKY) (26) protein families, both of which are involved in gene regulation of multiple metabolic domains.
Fig. 4 was confined to 64 structurally conserved proteins (see above), including 46 members of the AtCMPG family and 18 related proteins from A. thaliana and several other species. Gradual lowering of the stringency criteria yielded four additional AtCMPG proteins (25–40% sequence identity with AtCMPG1 within stretches of ≥50 aa) and numerous homologs from other plant or animal species, including various groups of RING- and zinc-finger proteins (27, 28), U box-containing proteins (29), and other functionally assigned proteins with about 25–35% sequence identity to AtCMPG1 (again within stretches of ≥50 aa). The AtCMPG family proper thus comprises at least 50 members.
Nearly all proteins of Fig. 4 not only contain highly conserved CMPG domains (Fig. 5), but also exhibit identical spacing of the strictly conserved amino acids: Cys-Xxx6-Met-Xxx2-Pro-Xxx5-Gly (indicated by filled circles in Fig. 4). In a few cases, open circles indicate either the insertion of one (AAG50227, NP_062693, AAD33400, and NP_176000) or six (AAK43499) additional amino acids between Cys and Met, or a shift of the Pro residue within the otherwise unaltered spacing (XP_092267: Cys-Xxx6-Met-Xxx6-Pro-Xxx1-Gly). Two different types of notable exception are the A. thaliana proteins AAF26976 and AAF02851. AAF02851 shares high sequence similarity with all true CMPG proteins within the domain, but lacks one of the conserved residues (Cys) and hence is not a member of the AtCMPG family, whereas AAF26976, despite its particularly high overall sequence similarity to AtCMPG1 and PcCMPG1, lacks almost precisely the entire CMPG domain, which is thus indirectly confirmed as a structural entity. RT-PCR demonstrated low but unequivocal expression also for AAF26976.
Figure 5.
CMPG domain of AtCMPG1 (Upper) and consensus sequence derived from all 50 AtCMPG proteins (≥50% conserved; shaded and marked with dots, fully conserved).
The presence of the domain and the occurrence as well as spacing of all four strictly conserved, nominal Cys, Met, Pro, and Gly residues robustly define the CMPG family. This family circumscription includes a few proteins that have elsewhere been classified as CPI domain-containing (Cys/Pro/Ile) proteins (30), albeit by less stringent criteria, or as AtPUB (plant U-box) proteins based on moderate sequence similarity to the multiubiquitin chain assembly factor (E4 ligase) UFD2 from yeast (31, 32). However, the CPI and AtPUB families are less inclusive than membership assigned by the CMPG criteria.
Several previously identified protein families, largely comprising DNA-binding transcription factors, contain two or more distinct, functionally assigned domains that often cross-connect entire families (33). By contrast, the CMPG family is characterized by a single, uniquely occurring domain, for which a functionally unifying theme, however, has yet to be ascertained. Most obvious is the frequent occurrence (shaded in Fig. 4) of established or putative receptor protein kinase- and ubiquitinylation-related proteins, as well as several RING- or zinc-finger proteins with slightly lower sequence similarity. All of them act through protein–protein interactions, suggesting that the CMPG domain as the common structural element may be responsible for this shared property. However, although this property may well be a mechanistically unifying theme, only subsets of family members seem to share specific metabolic functions, such as involvement in pathogen defense. As the latter probably applies to AtCMPG1, AtCMPG2, and PcCMPG1, their immediate-early induction by elicitor may indicate an involvement in very early steps of pathogen defense-related gene regulation either as transcriptional regulators with or without kinase activity or as protein degradation factors with or without ubiquitin ligase activity. Both phosphorylation by MAP kinases (34, 35) and ubiquitin-mediated degradation (36, 37) of regulatory proteins have been reported to be key elements in pathogen defense, although the actual molecular targets are still elusive.
Of all CMPG genes occurring in A. thaliana, AtCMPG1 is the closest relative of PcCMPG1 in terms of both structure and implicit function. Most importantly in the context of our long-term interest, this includes the occurrence of structurally as well as functionally related pathogen- and elicitor-responsive promoter elements, F and E17 (Fig. 6), which responded efficiently in the parsley protoplast assay, and are unique on the respective promoters. Both elements contain two functionally important W-box motifs, albeit in different relative positions, and one additional 9-bp motif that is likely to be of similar functional importance (Fig. 3, construct I; ref. 1). With full allowance for positional permutations of the individual components, E17 can be regarded as a reduced version of F, with F possessing a fourth, centrally located, functionally unassigned 10-bp sequence that is lacking in E17. In contrast to its obvious dispensability in E17, deletion of 9 bp of this region from F caused a strong reduction of elicitor responsiveness (Fig. 3, construct J), similar to the deletion of either one of the two functional W boxes. Notably, no less than 12 bp from the central part of F, including most of this 10-bp region, are fully identical with the 5′ half of yet another strongly elicitor-responsive but W-box-free element from the parsley PcPR2 gene (D box; Fig. 6 and ref. 38), indicating that this sequence also has a role in pathogen defense.
Figure 6.
Structural comparison of F with E17 and D box. Functional W-box core motifs are given in bold, underlined letters; dotted line, nonfunctional core motif. Shaded areas in E17 and D box indicate sequence identity with F.
W-Box core motifs, including tandem or multiple repeats within individual elements (24, 39, 40), occur with high frequency in pathogen- or elicitor-responsive gene promoters (26, 41). Most match the consensus sequence (C/T)TGAC(C/T) in either orientation. Among these promoters is that of another immediate-early elicitor-responsive gene from parsley, PcWRKY1, encoding the W-box-binding protein WRKY1 (39). This possibly autoregulated promoter likewise contains three closely spaced W-box motifs (WABC; ref. 39), two of which are essential for elicitor responsiveness. Their relative positions again differ from those in F and E17, and there is little sequence similarity outside the W boxes.
Thus, all three elicitor-response elements analyzed so far from immediate-early activated genes feature two closely spaced, functional W-box motifs, possibly indicating a specific asset of this particular type of response within otherwise highly variable arrangements. If F, E17, and WABC indeed represented such a specific type of W-box-containing response element, they would be ideal tools for the identification of those transcriptional regulators that are expected to act as direct links between infection-induced, intracellular signaling cascades and the consequential, extensive metabolic reprogramming of cells during pathogen defense (42, 43). W-box-binding WRKY proteins (39) would be among the most likely candidates, likewise with a high probability of falling into functionally distinct subtypes (26).
Within A. thaliana, the closest relative of AtCMPG1 in terms of nucleotide sequence is AtCMPG5 (67% identity of the coding regions; 55% identity of the deduced proteins). And yet, the AtCMPG1 and -5 promoters (up to 1,200 bp from the respective translation start sites) show almost no similarity at all. One notable exception is a 13-bp sequence (ATTCAATGGTCAA) 234 bp upstream of the predicted ATG start codon on the AtCMPG5 promoter, which is almost fully identical with the 3′ part of F and the 5′ part of E17 (one or two mismatches, respectively), including one functional W-box motif in each case. However, the isolated sequence, when analyzed as a 21-bp, 3′-extended version, was unresponsive to elicitor in both protoplast assays (data not shown), in agreement with numerous other cases where sequences of similar length bearing only one potentially functional W-box motif responded either very poorly or not at all to elicitor (e.g., fragments K and L in Fig. 3; refs. 1 and 39).
Taken together, these data verify F as a structurally unique member of a specific class of W-box-containing promoter elements. Neither the closest structural (AtCMPG5) nor putative functional (AtCMPG2) paralogs of AtCMPG1 contain strikingly similar elements. Although extensive searches throughout the A. thaliana genome indicated the frequent occurrence of all three closely spaced motif combinations constituting F {three W-box core motifs; two W-box cores with one D box motif in between; two W-box cores with the CATTCAAC sequence in between [e.g., GTCA(N1–20)CATTCAAC(N1–20)GTCA]}, the overall spacing and sequence similarity to F, or E17, was very low in all cases (data not shown).
This structural uniqueness, together with the remarkable lack of wound responsiveness, a strongly synergistic action in various artificial element combinations (ref. 38 and preliminary results), and the exceptional combination of very rapid, transient, localized, and specific, yet broadly acting, pathogen responsiveness, may render F, as well as E17 (1), particularly suited for designing new types of gene technology-based disease resistance mechanisms in crop-plant breeding.
Acknowledgments
We thank Dr. Kurt Stüber for valuable help with the promoter element search, Dr. Willem Broekaert for a sample of A. brassicicola, and Drs. Bill Martin and Imre E. Somssich for critical reading of the manuscript.
Abbreviations
- CMPG
Cys/Met/Pro/Gly-containing
- GUS
β-glucuronidase
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the Munich Information Center for Protein Sequences database, http://mips.gsf.de/ [accession nos. At1g66160 (AtCMPG1) and At5g64660 (AtCMPG2)].
References
- 1.Kirsch C, Logemann E, Lippok B, Schmelzer E, Hahlbrock K. Plant J. 2001;26:217–227. doi: 10.1046/j.1365-313x.2001.01015.x. [DOI] [PubMed] [Google Scholar]
- 2.The Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 3.Kombrink E, Hahlbrock K. Plant Physiol. 1986;81:216–221. doi: 10.1104/pp.81.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trezzini G F, Horrichs A, Somssich I E. Plant Mol Biol. 1993;21:385–389. doi: 10.1007/BF00019954. [DOI] [PubMed] [Google Scholar]
- 5.Ayers A R, Ebel J, Valent B, Albersheim P. Plant Physiol. 1976;57:760–765. doi: 10.1104/pp.57.5.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nürnberger T, Nennstiel D, Jabs T, Sacks W R, Hahlbrock K, Scheel D. Cell. 1994;78:449–460. doi: 10.1016/0092-8674(94)90423-5. [DOI] [PubMed] [Google Scholar]
- 7.Felix G, Duran J D, Volko S, Boller T. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 8.Debener T, Lehnacker H, Arnold M, Dangl J L. Plant J. 1991;1:289–302. doi: 10.1046/j.1365-313X.1991.t01-7-00999.x. [DOI] [PubMed] [Google Scholar]
- 9.Broekaert W F, Terras F R G, Cammue B P A, Vanderleyden J. FEMS Microbiol Lett. 1990;69:55–60. [Google Scholar]
- 10.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 11.Van Larebeke N, Engler G, Holsters M, Van den Elsacker S, Zaenen I, Schilperoort R A, Schell J. Nature (London) 1974;252:169–170. doi: 10.1038/252169a0. [DOI] [PubMed] [Google Scholar]
- 12.Becker D, Kemper E, Schell J, Masterson R. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- 13.Jefferson R A, Kavanaugh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mozo T, Dewar K, Dunn P, Ecker J R, Fischer S, Kloska S, Lehrach H, Marra M, Martienssen R, Meier-Ewert S, Altmann T. Nat Genet. 1999;22:271–275. doi: 10.1038/10334. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 17.Koetsier P A, Schorr J, Doerfler W. BioTechniques. 1993;15:260–262. [PubMed] [Google Scholar]
- 18.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 19.Van de Löcht U, Meier I, Hahlbrock K, Somssich I E. EMBO J. 1990;9:2945–2950. doi: 10.1002/j.1460-2075.1990.tb07486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herschman H R. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 23.Abel A, Theologis A. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rushton P J, Tovar Torres J, Parniske M, Wernert P, Hahlbrock K, Somssich I E. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- 25.Stracke R, Werber M, Weisshaar B. Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 26.Eulgem T, Rushton P J, Robatzek S, Somssich I E. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 27.Joazeiro C A P, Weissman A M. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 28.Takatsuji H. Cell Mol Life Sci. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama K-I. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 30.Amador V, Monte E, Garcia-Martinez J-L, Prat S. Cell. 2001;106:343–354. doi: 10.1016/s0092-8674(01)00445-7. [DOI] [PubMed] [Google Scholar]
- 31.Azevedo C, Santos-Rosa M J, Shirasu K. Trends Plant Sci. 2001;6:354–358. doi: 10.1016/s1360-1385(01)01960-4. [DOI] [PubMed] [Google Scholar]
- 32.Aravind L, Koonin E V. Curr Biol. 2000;10:R132–R134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 33.Riechmann J L, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe O J, Samaha R R, et al. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 34.Romeis T. Curr Opin Plant Biol. 2001;4:407–414. doi: 10.1016/s1369-5266(00)00193-x. [DOI] [PubMed] [Google Scholar]
- 35.Asai T, Tena G, Plotnikova J, Willmann M R, Chiu W-L, Gomez-Gomez L, Boller T, Ausubel F M, Sheen J. Nature (London) 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 36.Austin M J, Muskett P, Kahn K, Feys B J, Jones J D G, Parker J E. Science. 2002;295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- 37.Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 38.Rushton P J, Reinstadler A, Lipka V, Lippok B, Somssich I E. Plant Cell. 2002;14:749–762. doi: 10.1105/tpc.010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eulgem T, Rushton P J, Schmelzer E, Hahlbrock K, Somssich I E. EMBO J. 1999;18:4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang P Z, Chen Z X. Plant Sci. 2001;161:655–664. [Google Scholar]
- 41.Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton K A, Dangl J L, Dietrich R A. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- 42.Somssich I E, Hahlbrock K. Trends Plant Sci. 1998;3:86–90. [Google Scholar]
- 43.Batz O, Logemann E, Reinold S, Hahlbrock K. Biol Chem. 1998;379:1127–1135. doi: 10.1515/bchm.1998.379.8-9.1127. [DOI] [PubMed] [Google Scholar]