Abstract
The Arabidopsis thaliana SOS1 protein is a putative Na+/H+ antiporter that functions in Na+ extrusion and is essential for the NaCl tolerance of plants. sos1 mutant plants share phenotypic similarities with mutants lacking the protein kinase SOS2 and the Ca2+ sensor SOS3. To investigate whether the three SOS proteins function in the same response pathway, we have reconstituted the SOS system in yeast cells. Expression of SOS1 improved the Na+ tolerance of yeast mutants lacking endogenous Na+ transporters. Coexpression of SOS2 and SOS3 dramatically increased SOS1-dependent Na+ tolerance, whereas SOS2 or SOS3 individually had no effect. The SOS2/SOS3 kinase complex promoted the phosphorylation of SOS1. A constitutively active form of SOS2 phosphorylated SOS1 in vitro independently of SOS3, but could not fully substitute for the SOS2/SOS3 kinase complex for activation of SOS1 in vivo. Further, we show that SOS3 recruits SOS2 to the plasma membrane. Although sos1 mutant plants display defective K+ uptake at low external concentrations, neither the unmodified nor the SOS2/SOS3-activated SOS1 protein showed K+ transport capacity in vivo, suggesting that the role of SOS1 on K+ uptake is indirect. Our results provide an example of functional reconstitution of a plant response pathway in a heterologous system and demonstrate that the SOS1 ion transporter, the SOS2 protein kinase, and its associated Ca2+ sensor SOS3 constitute a functional module. We propose a model in which SOS3 activates and directs SOS2 to the plasma membrane for the stimulatory phosphorylation of the Na+ transporter SOS1.
Soil salinity is a prevalent abiotic stress for crop plants. Excess salts in the soil solution interfere with mineral nutrition and water uptake, and lead to the undue accumulation of toxic ions (1). Maladies associated to salt stress are membrane disorganization, impaired nutrient and water acquisition, metabolic toxicity, inhibition of photosynthesis, and production of reactive oxygen species. In most instances, ion toxicity results from immoderate Na+ uptake caused by its steep inward electrochemical gradient. Plant growth under salt stress depends, among other concomitant processes, on the re-establishment of proper cellular ion homeostasis. Low cytosolic Na+ content is preserved by the concerted interplay of regulated ion uptake, vacuolar compartmentation, and active extrusion to the extracellular milieu (2). Vacuolar partitioning of Na+ and other ions also contributes to the maintenance of cellular water relations in a hypertonic medium. Energy-dependent exclusion of Na+ from the cytosol is coupled to downhill reverse transport of H+ by Na+/H+ antiporters located in both the plasma membrane and tonoplast.
The Arabidopsis thaliana SOS1 protein is the first putative plasma membrane Na+/H+ antiporter to be described in plants (3, 4). Arabidopsis sos1 mutants were isolated in a genetic screen for plants hypersensitive to NaCl, together with sos2 and sos3 mutants (5). SOS2 is a Ser/Thr protein kinase in which two functional domains have been dissected (6). The N-terminal region contains the kinase catalytic domain, which has sequence similarity to the SNF1/AMP kinases. Interestingly, mammalian AMP-activated protein kinases are involved in protection against cellular stresses, such as heat shock, hypoxia, and oxidative stress (7). The C-terminal region of SOS2 has a regulatory function, and contains an autoinhibitory domain (the FISL domain) that interacts with SOS3 (6). SOS3 is a Ca2+-binding protein with strong similarity to the regulatory B subunit of the protein phosphatase calcineurin and to related proteins of the neuronal Ca2+ sensor family (5). The C-terminal regulatory domain of SOS2 interacts with and inhibits the N-terminal kinase domain. Removal of the regulatory domain, including the SOS3-binding domain, resulted in constitutive activation of the protein kinase. Hence, it has been postulated that SOS3 perceives the Ca2+ transients elicited by salt stress and activates SOS2 by relieving autoinhibition (6). There are, however, molecular and mechanistic details of SOS2/SOS3 function that remain obscure. For instance, the physiological substrate for the SOS2/SOS3 kinase complex has not been identified. The phenotypic similarities and the lack of additivity of sos1, sos2, and sos3 mutations suggest that the three SOS proteins function in the same process, affording Na+ tolerance to plants (5). Although SOS2 and SOS3 have been shown to modulate SOS1 gene expression (3), their effects on SOS1 mRNA levels may not be sufficient to explain the strong Na+ sensitivity of sos2 and sos3 mutants.
Here we show that the SOS pathway can be functionally reconstituted in yeast cells by the concerted expression of SOS1, SOS2 and SOS3. This experimental system has allowed us to demonstrate that the SOS2/SOS3 kinase complex promotes activation of the plasma membrane Na+ transporter SOS1 through phosphorylation. SOS3 is essential for both the kinase activity of SOS2 and its recruitment to the plasma membrane.
Materials and Methods
Yeast Strains and Media.
Saccharomyces cerevisiae strains WΔ3 (Δtrk1∷LEU2, Δtrk2∷HIS3), G19 (Δena1∷HIS3∷ena4), and ANT3 (Δena1∷HIS3∷ena4, Δnha1∷LEU2) have been described (8, 9). AXT3K (Δena1∷HIS3∷ena4, Δnha1∷LEU2, Δnhx1∷KanMX4) was constructed from ANT3 by replacement of the EcoRI/PstI fragment internal to NHX1 with an EcoRI/PstI fragment containing the selection marker KanMX. All strains are derivatives of W303-1B (MATα ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100). Na+ tolerance and growth in low K+ tests were performed in the alkali cation-free medium AP (10) supplemented with KCl and NaCl at the concentrations indicated. For ion content measurements, yeast cells were collected during exponential growth (OD550 ≈ 0.2) in liquid AP medium, and their Na+ and K+ contents were determined by atomic emission spectrophotometry after acidic extraction (10). Tolerance to high KCl concentrations was determined in YPD medium (1% yeast extract/2% peptone/2% glucose).
Plasmid Constructs.
Expression plasmids pSOS1-1 and pSOS1-2 were constructed by subcloning the SOS1 cDNA (4) under the control of the PGK1 promoter in the multicopy vector pYPGE15 (11) and the single copy plasmid YCp15, respectively. YCp15 was constructed by inserting the 2.0-kb SpeI–AlwNI fragment containing the expression cassette from pYPGE15 into the centromeric plasmid pCM189 (12), previously digested with SpeI and AlwNI. A C-terminal His6x-tagged version of SOS1 was obtained by PCR using the high-fidelity Pwo polymerase and the primers 5′-CTCGGTTACATTGAAAACCTC-3′ and 5′-ATGGTACCTCAATGATGATGATGATGATGACCTAGATCGTTCCTGA-3′. The SOS1:His6x cDNA was subcloned as an EcoRV–KpnI fragment into SmaI–KpnI sites of pYPGE15 and YCp15 to produce plasmids pSOS1–1His and pSOS1–2His. For expression of SOS2 and SOS3, a 2.1-kbp EcoRI fragment containing the SOS2 cDNA (13) was cloned in plasmid pAAR6 carrying the ADH1 promoter (14), whereas a 0.8-kbp XbaI–NotI fragment corresponding to the SOS3 cDNA (15) was placed under the control of the PMA1 promoter by ligation to the XhoI–NotI sites of vector pDR195 (16). To coordinate expression of SOS2 and SOS3 from a single plasmid, a HindIII-SphI fragment comprising the expression cassette PMA1:SOS3 was cloned in the centromeric plasmid pFL38 (17), then a EcoRI-PstI fragment containing the TRP1 gene was added, and finally the BglII fragment containing the URA3 gene was replaced for a BamHI fragment containing the ADH1:SOS2 expression cassette. This final construct was called pFL32T. Plasmids pFL3T and pFL2T were subsequently obtained from pFL32T by removing the SOS2 or the SOS3 expression cassette, respectively. Plasmid pFL2ΔT was constructed by subcloning the SOS2T/DΔ308 mutant allele (6) in pFL38 as described above for wild-type SOS2.
SOS Recruitment System (SRS).
An 1.3-kb fragment extending from +16 bp downstream of the ATG to the end of the SOS2 ORF was amplified with Pwo polymerase and primers: 5′-AGAGATCTTAGGAGGCAGAAGAGTGGGCAAG-3′ and 5′-GAGGATCCTCAAAACGTGATTGT-3′, which included BglII and BamHI restriction sites. The modified SOS2 was inserted into plasmid pADNS to produce a translational fusion downstream the hSos protein (18), resulting in plasmid pSRS2-1. A similar fusion was created with the mutant allele SOS2T/DΔ308. pSRS2–1 was digested with SacI and NotI to delete the 3′ part of SOS2 at position +282 and to replace it with the corresponding fragment from SOS2T/DΔ308, generating plasmid pSRS2-2. Plasmid pADNS-SosF producing membrane bound hSos was used as positive control (18). All plasmids used for SRS were transformed in the S. cerevisiae strain Cdc25-2 (MATα, cdc25-2, ade2, his3, leu2, lys2, trp1, ura3).
Membrane Fractionation and Immunological Methods.
For localization of the SOS1 protein, total membranes from AXT3K cells carrying plasmid pSOS1–2 were fractionated by centrifugation in a 10-step sucrose density gradient (18–54% wt/wt sucrose, in 4% increments) (9). SDS/PAGE and immunoblotting were performed by using standard protocols. Antibodies were used as follows: rabbit anti-SOS1 at 1:5,000 (H.S. and J.-K.Z., unpublished data), rabbit anti-PMA1 at 1:25,000 (19), and mouse anti-VPH1 at 1:500 (Molecular Probes). Horseradish peroxidase-coupled goat anti-mouse and horseradish peroxidase-coupled anti-rabbit (Sigma) were used at 1:10,000.
SOS1:His6x Purification.
Total membranes were obtained as described (20). Membrane proteins (2.5 mg) were solubilized in buffer A (50 mM NaH2PO4, pH 8.0/300 mM NaCl/20% glycerol/0.5% dodecyl-β-maltoside) with 10 mM imidazole, at 4°C for 30 min. Insoluble material was pelleted by centrifugation at 20,000 × g for 30 min, and the supernatant was incubated overnight with 250 μl of Ni–NTA resin (Qiagen). The resin was packed in a 1-ml column (BioRad) and washed four times with buffer A with 20 mM imidazole. Finally, bound protein was eluted with buffer A with 250 mM imidazole.
SOS1 Phosphorylation Assays.
As substrate for phosphorylation, 100 ng of SOS1:His6x purified by metal binding chromatography or 10 μg of total membrane protein purified from AX3TK cells transformed with plasmid pSOS1–1 or an empty vector were added as described for each experiment. Cell-free extracts from AXT3K cells carrying plasmids pFL2T, pFL3T, and pFL32T for expression of SOS2 and/or SOS3 were prepared as described (20). Construction and purification of the glutathione S-transferase (GST):SOS2T/DΔ308 translational fusion from bacteria has been described (6). Twenty micrograms of protein extracts or 100 ng of purified GST:SOS2T/DΔ308 were added to phosphorylation reactions. Phosphorylation of SOS1 was assayed in kinase buffer (20 mM Tris⋅HCl, pH 8.0/5 mM MgCl2/1 mM CaCl2/1 mM DTT). Reactions were started by adding 0.2 mM ATP with 1 μCi of [γ-32P]ATP (1 Ci = 37 GBq) to give 50 μl final reaction volume, incubated at 30°C for 30 min, and stopped with 15 μl of 4× SDS/PAGE sample buffer. Aliquots (10 μl) were resolved by SDS/PAGE and exposed to x-ray film.
Results
Coexpression of SOS2 and SOS3 Activates SOS1.
We have recently shown that the putative plasma membrane Na+/H+ antiporter SOS1 increases the salt tolerance of yeast cells devoid of endogenous Na+ transporters (4). Yeast AXT3K cells lack the Na+ efflux proteins ENA1–4 and NHA1, and the vacuolar Na+/H+ antiporter NHX1. Expression of SOS1 from a multicopy plasmid restored partial tolerance to NaCl (Fig. 1), which correlated with lower intracellular Na+ content (Fig. 2). These results, together with plasma membrane targeting of SOS1 (see below), indicate that SOS1 directs Na+ extrusion out of the cell.
Figure 1.
SOS2 and SOS3 increase the NaCl tolerance of yeast expressing the SOS1 ion transporter. AXT3K cells transformed with an empty vector (control) or expressing the indicated combination of Arabidopsis genes were grown overnight in liquid AP medium with 1 mM KCl. Five microliters of serial decimal dilutions were spotted onto plates of the same medium or supplemented with 70 mM NaCl. Plates were incubated at 28°C and photographed after 4 days. Plasmids used for expression of the SOS proteins were: pSOS1–1 for SOS1; pFL2T for SOS2; pFL3T for SOS3; pFL32T for SOS2 and SOS3; and pFL2ΔT for SOS2T/DΔ308.
Figure 2.
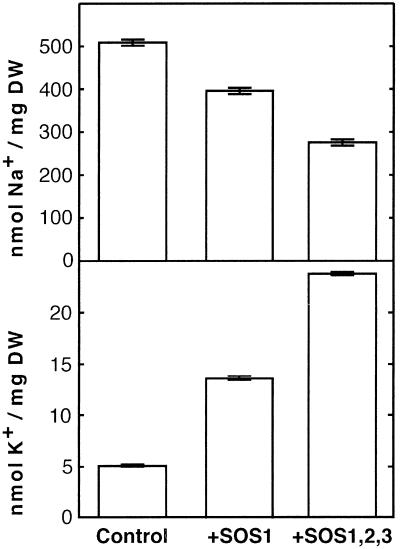
Expression of SOS genes reduces Na+ accumulation and improves K+ status. Cells of strain AXT3K transformed with an empty vector (control), or expressing SOS1 (+SOS1) or transformed with plasmids for the simultaneous expression of SOS1, SOS2, and SOS3 (+SOS1,2,3) were grown in liquid AP medium with 1 mM KCl and 50 mM NaCl. When cultures reached OD550 ≈ 0.2, cells were collected by filtration, and their Na+ and K+ contents were determined. Units are nmol of ion per mg dry weight of cell samples. Data shown are the average and SE of ion contents of three independent cultures of each strain.
Because the effect of SOS1 on the salt tolerance of the yeast mutant was rather small, we reasoned that SOS1 was not regulated properly in yeast, and that SOS1 might require other ancillary proteins for maximal activity. Because sos1 mutant plants share many phenotypic similarities with sos2 and sos3 mutants, SOS2 and SOS3 proteins were candidates to modulate the transport activity of SOS1. To test this hypothesis, we coexpressed SOS2 and SOS3 in AXT3K yeast cells, in the absence or presence of SOS1, and studied their effect on salt tolerance. As shown in Fig. 1, the concurrent expression of the three SOS proteins dramatically increased the Na+ tolerance of transformed cells, above levels imparted by SOS1 alone. Cells expressing the three SOS proteins were able to grow in media containing up to 200 mM NaCl (1 mM KCl), which is almost the tolerance of wild-type yeast cells in AP medium (data not shown). This enhanced tolerance was observed only when SOS1 was present, demonstrating that SOS2 and SOS3 were not unmasking an endogenous yeast activity. Moreover, any other combination of the SOS proteins failed to produce a substantial increase in tolerance. Omitting either SOS2 or SOS3 prevented the SOS1-dependent enhancement of Na+ resistance (Fig. 1). These results suggest that both the protein kinase SOS2 and the Ca2+ sensor SOS3 are necessary and together are sufficient for in vivo activation of the SOS1 transporter.
SOS2 is an inactive protein kinase whose activity depends on SOS3 binding (6, 21). The Ca2+ sensor SOS3 binds to a regulatory domain of SOS2 that also serves as an autoinhibitory domain (6). The allele SOS2T/DΔ308 combines a T168D mutation in the kinase activation loop of SOS2 with a C-terminal truncation at residue 308 that removes the autoinhibitory domain, yielding a hyperactive kinase that is independent of SOS3 (6) Expression of SOS2T/DΔ308 increased the Na+ tolerance of cells containing SOS1 in a SOS3-independent manner, although to a lesser extent than similarly expressed SOS2/SOS3 (Fig. 1).
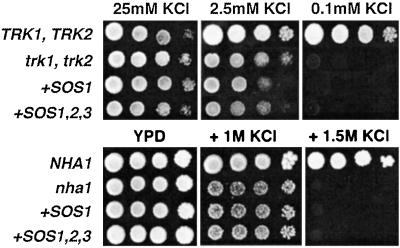
To elucidate the mechanism by which SOS1 and SOS2/SOS3 conferred greater Na+ tolerance to yeast cells, the Na+ content of cells expressing the three SOS genes was measured and compared with control AXT3K cells, and to cells expressing SOS1 alone (Fig. 2). Coexpression of the three SOS proteins further reduced Na+ accumulation, below levels of cells expressing SOS1 alone. Thus, enhanced Na+ efflux accounted for the increased tolerance afforded by the SOS2/SOS3 protein complex. As shown in Fig. 2, reduction in net Na+ content correlated with improved K+ status. This linkage could result from mutual exclusion of these ions or from an intrinsic K+ transport capacity of SOS1. Notably, sos1 plants are defective in K+ uptake at low external concentrations (22), thus raising the possibility that the primary substrate for SOS1 was K+ rather than Na+. However, SOS1 alone or coexpressed with SOS2/SOS3 failed to suppress the growth defect in low external K+ of mutant cells lacking TRK1 and TRK2 proteins (Fig. 3). TRK1 and TRK2 proteins comprise the high-affinity K+ uptake system of Saccharomyces cerevisiae and are required for growth at low external K+ concentrations (23). Similarly, SOS1 activated by SOS2/SOS3 did not substitute for the NHA1 protein regarding cell tolerance to high external KCl (Fig. 3). NHA1 is a plasma membrane alkali cation/proton antiporter that mediates rapid efflux of Na+ and K+ ions (24). K+ transport by SOS1, if any, seems negligible because no growth improvement was found at threshold K+ concentrations that permitted residual growth of trk1 trk2 (2.5 mM KCl), and nha1 (1 M KCl) mutant cells (Fig. 3). These results indicate that the primary activity of SOS1 in yeast cells is Na+ transport, both in its basal activity and after stimulation by SOS2/SOS3, and suggest that the K+ uptake deficiency of sos1 plants may arise from disturbed cation homeostasis.
Figure 3.
Activated SOS1 does not complement yeast mutants deficient in K+ transport. Cells of strains WΔ3 (trk1, trk2) and ANT3 (nha1) were transformed with SOS1 (+SOS1) or SOS1, SOS2, and SOS3 (+SOS1,2,3). Transformants of WΔ3 and ANT3 were grown overnight in AP medium supplemented with 25 mM KCl or YPD, respectively. Five microliters of serial decimal dilutions were spotted onto plates of the corresponding medium supplemented with the indicated KCl concentrations. Cells of strains W303 (TRK1, TRK2) and G19 (NHA1) were used as control as indicated. Plates were incubated at 28°C and photographed after 4 days.
SOS Proteins Interact at the Plasma Membrane.
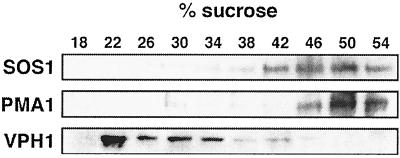
To confirm that SOS1 functions as a Na+ efflux transporter in yeast, its cellular localization was determined. Total membranes of AXT3K cells expressing SOS1 from the single copy plasmid pSOS1-2 were isolated and fractionated on a sucrose density gradient. SOS1 protein was immunodetected in Western blots with polyclonal antibodies. As shown in Fig. 4, SOS1 cofractionated with the plasma membrane ATPase PMA1, but not with tonoplast marker VPH1.
Figure 4.
Subcellular localization of SOS1. Total membrane extracts from AXT3K cells expressing SOS1 (plasmid pSOS1–2His) were fractionated on a 10-step sucrose gradient (18–54% wt/wt). Samples (25 μg protein) were resolved by SDS/PAGE and blotted. Western blots show the distribution of markers for the plasma membrane, PMA1, the vacuolar membrane, VPH1, and the SOS1 protein.
SOS3 is a myristoylated protein that is partially associated with cell membranes, but attempts to determine the precise subcellular localization of SOS3 were inconclusive (25). Because SOS1 localizes to the plasma membrane of plant (4) and yeast cells (Fig. 4), we sought to demonstrate interactions between the SOS proteins at the plasma membrane by using SRS (18). This system monitors binding between proteins residing at the plasma membrane and a bait fused to the human hSos protein, a functional homologue of the yeast Ras guanyl nucleotide exchange factor CDC25. When interaction occurs, recruitment of the human hSos protein to the plasma membrane restores Ras activation, allowing growth at 37°C of the thermosensitive yeast mutant cdc25-2. We used the wild-type SOS2 protein as bait fused to hSos and tested whether it was able to interact with SOS1 and SOS3 as preys at the plasma membrane. As depicted in Fig. 5, SOS2 by itself did not target hSos to the plasma membrane. However, the hSos:SOS2 fusion was efficiently recruited to the plasma membrane on coexpression of SOS3, allowing cdc25-2 cells to grow at 37°C. Interaction between SOS2 and SOS3 was independent of SOS1 and did not require salt stress. This result is evidence for plasma membrane localization of SOS3 in vivo. On the other hand, this experimental system failed to detect significant interaction between SOS1 and SOS2, which could be due to the inactive state of SOS2 in the absence of SOS3. To bypass the SOS3 requirement, we used the SOS2T/DΔ308 protein as bait fused to the human hSos because the SOS2T/DΔ308 mutant is hyperactive, SOS3-independent (6), and activates SOS1 (Fig. 1). However, the SOS2T/DΔ308 fusion protein also failed to show significant targeting to the plasma membrane in the presence of SOS1, suggesting that interaction of SOS1 and SOS2 in the absence of SOS3 is weak. These results indicate that SOS3 is essential for targeting of SOS2 to the plasma membrane, thereby facilitating subsequent interaction with SOS1.
Figure 5.
Interaction of SOS2 and SOS3 at the plasma membrane. Complementation of the cdc25-2 mutation through SOS2–SOS3 interaction. Cells of genotype cdc25–2 were transformed with the hSos:SOS2 translational fusion in plasmid pSRS2–1 (SOS2) or with a similar fusion using the SOS2T/DΔ308 variant in plasmid pSRS2–2 (SOS2T/DΔ308). These cells were subsequently transformed with plasmids containing the genes SOS1 and SOS3 as indicated for each lane. Plasmid pADNS–SosF (hSosF) was used as positive control. Transformants were grown on YPD plates at 23°C or 37°C. Only cells containing both the SOS3 gene and the hSos:SOS2 chimera grew at 37°C, as did control hSosF-transformed cells.
SOS1 Phosphorylation by the SOS2/SOS3 Kinase Complex.
Phosphorylation by the SOS2/SOS3 complex was the likely mechanism involved in the activation of the SOS1 transporter. A search for putative phosphorylation sites in SOS1 identified several consensus sequences at the C-terminal hydrophilic domain that could be recognized by the SOS2 kinase (data not shown) (21). Phosphorylation assays were carried out by using cell membranes from SOS1-expressing yeast cells and protein extracts of cells expressing SOS2, SOS3, SOS2/SOS3, or transformed with an empty vector. A distinctive phosphorylated band with the expected molecular mass of SOS1 (≈127 kDa) was apparent in the sample where both SOS2 and SOS3 were present in the phosphorylation reaction (Fig. 6A). This band was not detected when protein extracts lacked SOS2, SOS3, or both. To further confirm that the phosphorylated band among plasma membrane proteins corresponded to SOS1, the phosphorylation assay was repeated by using purified SOS1:His6x as substrate. Again, a band migrating at the same position as the purified SOS1:His6x protein appeared phosphorylated only when incubated with protein extracts containing SOS2 and SOS3 (Fig. 6B). Together these results indicate that the SOS2/SOS3 kinase complex may phosphorylate SOS1 and that SOS3 is essential for the kinase activity of SOS2. To demonstrate that SOS1 is the genuine substrate for the SOS2 protein kinase, a mutant SOS2T/DΔ308 kinase fused to GST was purified from bacteria and tested for phosphorylation of the SOS1:His6x protein purified from yeast membranes by affinity chromatography. As shown in Fig. 6C, the SOS2T/DΔ308 kinase catalyzed strong phosphorylation of SOS1.
Figure 6.
The SOS2/SOS3 kinase complex phosphorylates SOS1. Crude membranes extracts (10 μg protein) containing SOS1 (A) or purified SOS1:His6x protein (100 ng; B) were incubated with 20 μg of protein extracts from AXT3K cells carrying an empty plasmid (lane 1), or plasmid pFL2T for expression of SOS2 (lane 2), plasmid pFL3T for expression of SOS3 (lane 3), or plasmid pFL32T for expression of both SOS2 and SOS3 (lane 4). (C) Purified SOS1:His6x protein (100 ng) was incubated with 100 ng of purified GST:SOS2T/DΔ308 fusion protein (lane 5). Control samples without the GST:SOS2T/DΔ308 protein (lane 6) or the SOS1:His6x protein (lane 7) are also shown. All reactions were carried on kinase buffer for 30 min. Aliquots of kinase reactions were resolved by SDS/PAGE and exposed to x-ray films. Additional lanes with 100 ng of purified SOS1:His6x were included for protein staining and assessment of correspondence with phosphorylated bands (not shown). Arrows indicate the 127-kDa bands pertaining to phosphorylated SOS1.
Discussion
The convenient and powerful yeast system has been extensively used for the isolation and functional characterization of individual proteins from various organisms. Expression in yeast has been particularly useful for studying plant transport proteins (26). We show here that a multicomponent response pathway regulating ion homeostasis in plant cells can be functionally reconstituted in yeast. By this method, both the necessary and sufficient elements of the response pathway were assessed in vivo. The Arabidopsis SOS pathway, as demonstrated here, is comprised of a Ca2+ sensor, SOS3, that transduces a Ca2+ signal to recruit and activate an effector kinase, SOS2, which in turn phosphorylates and stimulates the output protein, the ion transporter SOS1.
Reconstitution of the SOS response pathway also permitted a better dissection of the molecular interactions between its constituents. For instance, SOS3 was previously shown to be an essential regulatory subunit of the protein kinase SOS2 that relieved the kinase domain of SOS2 from autoinhibition (6). We show here that SOS3 mediates the activity of SOS2 in two complementary ways, kinase activation and protein targeting. Moreover, the protein kinase activity of SOS2 had been demonstrated by using synthetic peptides, but no physiological substrate for SOS2 had been identified (21). Here we show that the ion transporter SOS1 is a genuine target for the SOS2 kinase. In the yeast system, SOS1 shows very low activity and negligible phosphorylation in the absence of SOS2 or SOS3 (Figs. 1 and 6). Phosphorylation of SOS1 by the SOS2/SOS3 kinase complex brings about substantial activation of SOS1, as inferred from increased tolerance to NaCl and enhanced Na+ exclusion. Consistent with previous results (6), SOS3 is indispensable for the phosphorylation of SOS1 by the SOS2 kinase in vitro (Fig. 6). These results demonstrate that the SOS1 ion transporter, the SOS2 protein kinase and its associated Ca2+ sensor SOS3 compose a functional module that controls cellular Na+ homeostasis. These findings also provide a molecular basis for the phenotypic similarities between sos1, sos2, and sos3 mutant plants (5), because they imply that SOS1 would be locked in an inactive state in both sos2 and sos3 mutants.
Results with the SRS show that recruitment of SOS2 to the plasma membrane is another essential function of SOS3 (Fig. 5). The hyperactive and SOS3-independent kinase SOS2T/DΔ308 strongly phosphorylated SOS1 in vitro (Fig. 6C) but could not fully substitute for the SOS2/SOS3 complex regarding SOS1 activation in vivo, even when SOS2T/DΔ308 was greatly overexpressed (Fig. 1). Together, these results indicate that the SOS2/SOS3 complex must colocalize with SOS1 in the plasma membrane to achieve efficient activation of the ion transporter. Although N-myristoylation was shown to be essential for SOS3 functionality in planta, no significant difference in membrane association was observed between wild-type SOS3 and nonmyristoylated SOS3 mutant, nor was the purported target membrane identified, at least in part because SOS3 had to be overexpressed to permit protein detection (25). The role of myristoylation in subcellular targeting is complex and the mere presence of myristate in a protein does not constitute a signal for membrane binding. SOS3 has significant sequence similarity to recoverin-like Ca2+ sensors (15). Ca2+ binding to recoverin triggers a conformational change that exposes an N-terminal myristoyl group, thereby allowing the anchoring of the recoverin–rhodopsin kinase complex to the plasma membrane (27). As evidenced by the SRS, SOS3 recruits the SOS2 kinase to the plasma membrane in vivo. Membrane localization of SOS2/SOS3 is independent of SOS1 and does not require salt-induced Ca2+ signaling. The SOS2/SOS3 complex may also localize to the cell membrane of nonstressed A. thaliana plants because both SOS2 and SOS3 are expressed under physiological growth conditions, although at low levels (13, 25). We cannot rule out, however, that in plant cells SOS3 interaction with the plasma membrane and/or recruitment of SOS2 may be triggered or enhanced by the Ca2+ transients elicited by salt stress.
Besides their Na+ sensitivity, the Arabidopsis mutants sos1, sos2, and sos3 bear a defect on K+ nutrition at low external concentrations (28). Although this puzzling phenotype is exacerbated in sos1 plants relative to sos2 and sos3, we have shown that SOS1 behaves in yeast as a Na+ transporter with no discernible K+ transport capacity even when activated by SOS2/SOS3 (Fig. 3). In fact, SOS1 is very specific for Na+ ions because it does not transport Li+, a common feature of many Na+ transporters (data not shown). This strict Na+ selectivity of heterologously expressed SOS1 may not be necessarily applicable to plants. Recently, however, it has been reported that K+ currents and membrane potentials were indistinguishable in sos1 mutants and wild-type plants without Na+, whereas a pretreatment with NaCl impaired the K+ permeability of sos1 root cell membranes but not in wild-type plants.§ Thus, mounting evidence suggests that the K+ uptake system is not directly affected by the sos1 mutation but it becomes impaired by abnormal cytosolic levels of Na+ in sos1 mutants. Moreover, SOS1 has been implicated in the long-distance transport of Na+ (4). Malfunction of the net Na+/K+ exchange achieved at the xylem–symplast boundary through the concerted action of Na+/H+ and K+/H+ antiporters could also distort K+ acquisition and translocation by the plant (29).
From previous results (28) and those reported here, the following model emerges. The SOS1 transporter resides in the plasma membrane, in a resting state with low basal activity in nonsalinized plants. Some or all of the SOS3 protein is also anchored to the plasma membrane in association with the SOS2 kinase before salinity-induced Ca2+ signaling. Shifting to a saline environment elicits a Ca2+ transient that, on Ca2+ binding by SOS3, transduces into the activation of the protein kinase SOS2, which in turn phosphorylates SOS1. The SOS1 transporter thus modified becomes stimulated and extrudes Na+ with greater efficiency to re-establish ion homeostasis. In addition to these biochemical changes, activation of SOS2/SOS3 results in the up-regulation of SOS1 gene expression (3), but the associated transcription factor remains to be identified.
The function of the SOS response pathway is central to Na+ homeostasis and the salinity tolerance of A. thaliana and, presumably, other higher plants. The successful reconstitution of the SOS pathway described in this paper will open new avenues for dissecting functional relationships between its constituents and for detailed structure-function studies with the SOS1 transporter.
Acknowledgments
We thank Dr. Ramon Serrano for PMA1 antibody. This work was supported by Spanish Ministerio de Ciencia y Tecnologia Grant BIO2000-0938 (to F.J.Q. and J.M.P.) and by National Institutes of Health Grant R01GM59138 (to J.-K.Z.).
Abbreviations
- SRS
SOS Recruitment System
- GST
glutathione S-transferase
Footnotes
Zhi, Q. & Spalding, E. P., Annual Meeting of the American Society of Plant Biologists, Providence, RI, July 21–25, 2001, abstr. 158.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hasegawa P M, Bressan R A, Zhu J-K, Bohnert H J. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 2.Blumwald E, Aharon G S, Apse M P. Biochim Biophys Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 3.Shi H, Ishitani M, Kim C, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi S, Quintero F J, Pardo J M, Zhu J-K. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J-K. Plant Physiol. 2000;124:941–948. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Halfter U, Ishitani M, Zhu J-K. Plant Cell. 2001;13:1383–1400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchelhill K I, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters L A, Kemp B E. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 8.Madrid R, Gomez M J, Ramos J, Rodriguez-Navarro A. J Biol Chem. 1998;273:14838–14844. doi: 10.1074/jbc.273.24.14838. [DOI] [PubMed] [Google Scholar]
- 9.Quintero F J, Blatt M R, Pardo J M. FEBS Lett. 2000;471:224–228. doi: 10.1016/s0014-5793(00)01412-5. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Navarro A, Ramos J. J Bacteriol. 1984;159:940–945. doi: 10.1128/jb.159.3.940-945.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunelli J P, Pall M L. Yeast. 1993;9:1299–1308. doi: 10.1002/yea.320091203. [DOI] [PubMed] [Google Scholar]
- 12.Garí E, Piedrafita L, Aldea M, Herrero E. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Ishitani M, Halfter U, Kim C O, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ammerer G. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Zhu J-K. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 16.Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer W B V. FEBS Lett. 1995;370:264–268. doi: 10.1016/0014-5793(95)00853-2. [DOI] [PubMed] [Google Scholar]
- 17.Bonneaud N, Ozier-Kalogeropoulos O, Li G Y, Labouesse M, Minvielle-Sebastia L, Lacroute F. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 18.Aronheim A, Zandi E, Henneman H, Elledge S J, Karin M. Mol Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monk B C, Montesinos C, Ferguson C, Leonard K, Serrano R. J Biol Chem. 1991;266:18097–18103. [PubMed] [Google Scholar]
- 20.Serrano R. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- 21.Halfter U, Ishitani M, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S J, Ding L, Zhu J-K. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez-Navarro A. Biochim Biophys Acta. 2000;1469:1–30. doi: 10.1016/s0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 24.Bañuelos M A, Sychrová H, Bleykasten-Grosshans C, Souciet J L, Potier S. Microbiology. 1998;144:2749–2758. doi: 10.1099/00221287-144-10-2749. [DOI] [PubMed] [Google Scholar]
- 25.Ishitani M, Liu J, Halfter U, Kim C, Shi W, Zhu J-K. Plant Cell. 2000;12:1667–1678. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dreyer I, Horeau C, Lemaillet G, Zimmermann S, Bush D R, Rodríguez-Navarro A, Schachtman D P, Spalding E P, Sentenac H, Gaber R F. J Exp Bot. 1999;50:1073–1087. [Google Scholar]
- 27.Ames J B, Ishima R, Tanaka T, Gordon J I, Stryer L, Ikura M. Nature (London) 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J-K. Curr Opin Plant Biol. 2001;4:401–406. doi: 10.1016/s1369-5266(00)00192-8. [DOI] [PubMed] [Google Scholar]
- 29.Lacan D, Durand M. Plant Physiol. 1996;110:705–711. doi: 10.1104/pp.110.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]