Motor proteins use energy derived from the hydrolysis of ATP to move unidirectionally along microtubules and actin filaments. They play multifunctional roles in the cell, being intimately involved in transport processes, cell motility, and the organization and maintenance of cytoskeletal structures. During mitosis, the proper arrangement of chromosomes before cell division involves movement both toward and away from spindle poles, which is thought to be mediated by both plus- and minus-directed microtubule motors. In neurons, vesicles are transported along axonal microtubules both toward and away from the axon tip, carried by motors of opposite polarity. Given the functional importance of opposing motion, just what determines motor directionality has become a pressing question (1). It has been addressed most effectively by comparing two molecules from the kinesin family that move in opposite directions. Conventional kinesin and Ncd have similar dimeric structures, composed of a coiled-coil stalk attached to a pair of motor domains, which are closely homologous between the two species. Yet kinesin moves toward the plus end of a microtubule, whereas Ncd is a minus-directed motor. Establishing what makes these two molecules move in different directions might shed light on how motor proteins work.
If a number of motors are pulling one way, they enhance the likelihood that their teammates will join in.
The first intriguing result was obtained by using a chimera composed of the Ncd motor domain fused to the kinesin stalk region (2, 3). Unlike kinesin, Ncd is nonprocessive; an individual molecule is incapable of tracking a microtubule. So to test motor directionality, a gliding motility assay was used. Motors were adsorbed on a surface at sufficient density to enable dozens of molecules to interact with a single microtubule. Observing the motion of end-labeled microtubules under a microscope, it was found that the chimera propelled them across the surface in the opposite direction to the native Ncd protein. Clearly, the motor domain is not the sole determinant of directionality. An even more startling result was obtained recently by Endow and Higuchi (4), who made a mutant of Ncd with a single amino acid substitution in the neck region, which joins the motor domain to the stalk. In the gliding assay, the mutant drove microtubules in both directions. Typically, an individual microtubule traveled for several micrometers with its plus end leading, then abruptly reversed direction and traveled for a similar distance in the opposite sense. The speed was approximately the same in each direction and the reversals appeared to occur quite randomly. All very curious!
In this issue of PNAS, Badoual et al. (5) present a theoretical model that suggests that directionality in a gliding assay is a team property and cannot entirely be reduced to the characteristics of an individual motor molecule. They ascribe the ability of the mutant Ncd to push the microtubule both ways to an instability in the collective dynamics that arises when many motors work together.
Two different approaches may be taken to modeling the dynamics of molecular motors. One possibility is to base a model on the known structure and biochemistry of a specific motor protein. This approach has been productive for actomyosin, where the swinging lever arm model can account for many of the features of muscle contraction (6–8). Alternatively, if the aim is to determine the general types of behavior that motor systems can exhibit, a less specific formalism is more appropriate. Badoual et al. (5) take this second route.
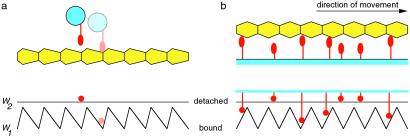
Motor proteins have two essential characteristics: they interact with cytoskeletal filaments; and this interaction is modulated as the hydrolysis reaction proceeds, catalyzed by the motor domain. Their operation can be captured, in essence, by a simplified two-state model called an “isothermal ratchet” (9, 10) (isothermal because molecular motors work at a fixed temperature, in contrast to the combustion engines with which we are familiar). As illustrated in Fig. 1a, a motor is considered to make stochastic transitions between abound state and a detached state, which correspond to two different nucleotide states. In the bound state, the motor is considered to experience an interaction potential with the filament which depends on position, rather than being thought of as fixed at a binding site. Because cytoskeletal filaments are polar polymers, this potential will, in general, be periodic but asymmetric.
Figure 1.
Simplified isothermal ratchet model of a motor protein considered by Badoual et al. A motor makes stochastic transitions between a bound and a detached state at fixed rates. The interaction potentials with the filament—different for each state—are indicated below. Attachment can occur at any location, but detachment is permitted only when the motor is in the vicinity of the potential minimum. (a) An individual processive motor can carry a cargo unidirectionally along a polar cytoskeletal filament. A motor that has just detached will be at the location of the potential minimum, as shown. Free diffusion will carry it past the potential peak to the right with greater probability than the more distant peak to the left. Consequently, when it rebinds, it is more likely to drift to the potential minimum to the right (as shown), than to the minimum to the left. The overall motion is therefore rightwards on average. (b) A team of motors can drive a nonpolar filament both forwards and backwards. Suppose that the filament is moving to the right. Then, owing to the detachment of motors that have recently passed through a potential well, there are more bound motors on the left-facing slope of the potential than on the right-facing slope (as shown). Sliding down the slope, these motors produce a force that maintains the rightward movement. An equivalent argument shows that the filament could just as well be driven to the left.
If this system were at chemical equilibrium, the ratios of the local binding and detachment rates would be fixed by the principle of detailed balance, and it can be proved that, in this case, there is no net movement (9). As expected from the laws of thermodynamics, motion cannot be had for free. But in the cell, the ATP hydrolysis reaction is maintained out of equilibrium. A reaction occurring at the nucleotide site might, for example, stimulate detachment of the motor when it is at a particular location on the filament. As explained in Fig. 1a, the motor then advances one way along the filament, albeit taking hesitant steps. It seems that certain single-headed kinesins, such as KIF1A, do operate this way (11). Most processive motors, like conventional kinesin, appear to be designed rather more effectively, and they move less stochastically (12). Their motion can be described by a slight alteration to the model, using two potentials with minima at different locations (9).
What happens when many motors work together? The fact that new phenomena can arise—that “more is different”—is most easily illustrated for the hypothetical case of a nonpolar filament. In this situation, an individual motor is incapable of moving directionally. But as shown in Fig. 1b, multiple motors can propel a filament in either direction. If a number of motors are pulling one way, they enhance the likelihood that their teammates will join in and pull in the same direction. This only happens, however, above a critical concentration of ATP, for which the rate of stimulated detachment is sufficiently high. The situation is analogous to phase transitions in condensed matter physics. In the ferromagnetic transition, for example, cooperative interactions exceeding a critical level cause many spins to point in the same direction. Indeed, the general mathematical properties at the critical point are closely related in both systems. But there are significant differences, too; the team of motors is a nonequilibrium system that is controlled by chemical kinetics, as opposed to an equilibrium system that is controlled by the temperature.
The prediction of bidirectionality was first made by Jülicher and Prost 7 years ago (13). They studied the case of an infinite number of motors. Badoual et al. (5) extend this analysis to the situation in which only a few motors are interacting with the filament—the experimentally relevant case. They suggest that the mutation in the Ncd neck modifies the effective interaction potential of the motor with the microtubule, so that it is almost symmetric (despite the microtubule's polarity). Then, according to their model, an individual motor would have no net bias when it interacts with a microtubule; but in a gliding assay, a team of Ncd mutants could propel microtubules in either direction. A filament would not travel one way for an indefinite period, however. Stochastic fluctuations in the numbers of motors pushing and pulling would eventually cause an abrupt switch to the alternative steady-state solution. One easily testable prediction of the model is that the frequency of switching should depend sensitively on the number of motors interacting with the microtubule; long microtubules should reverse direction much less often than shorter ones. A second prediction is that the bidirectionality should disappear if the ATP concentration is reduced below a critical value, and that the force-velocity relation measured at that critical point should display a characteristic nonlinear behavior.
Spontaneous bidirectional motion implies that there is a region of hysteresis in the force-velocity curve. This implication leads to an important insight: if the filament is connected to an elastic element, it should oscillate as the motors drive it first forwards then backwards (14). It would be interesting to try to detect such oscillations in a gliding assay by holding on to a microtubule with a flexible microneedle, for example. Oscillations occur in a number of natural motor protein systems. The vibration of insect flight muscle is too rapid to be controlled by nervous impulses on a cycle-by-cycle basis, and is thought to be generated by a dynamical instability of the actomyosin system, which might be based on the type of mechanism described by Badoual et al. (5). Similarly, the undulation of spermatazoid flagella might be caused by an oscillatory instability of teams of axonemal dynein motors (15).
Could this attractively simple model find applications beyond the domain of cytoskeletal motor proteins? One potential candidate is the bacterial flagellar motor. This compound structure, consisting of multiple copies of numerous gene products, can rotate both clockwise and counterclockwise at speeds of several hundred revolutions per second (16). In its natural environment, it switches stochastically between the two directions (17). The proportion of time spent in each state is sensitively controlled (18) by the concentration of a signaling protein—phosphorylated CheY—a feature that bacteria use to control the direction in which they swim. Torque is generated via interactions between a rotor—incorporating a ring of 34 FliM proteins—and about 12 stators, each consisting of two motor units, MotA and MotB. In one recent model (19), FliM is assumed to exist in two conformations, each of which produces torque in a different sense when it interacts with a stator. Allosteric interactions between FliM proteins lead to cooperative switching of the entire ring, accounting for the sudden, stochastic changes in the direction of rotation. The symmetric isothermal ratchet picture discussed by Badoual et al. (5) might provide an alternative description. The challenge would be to understand how the binding of CheYp, perhaps by subtly altering the effective potentials, might sensitively affect the relative probabilities of the two steady states.
The analysis of Badoual et al. (5) is a timely reminder that not all properties of a system can be reduced to the characteristics of the lowest level constituents, and that collective effects leading to phase transitions and instabilities are prevalent in biology. Indeed, recent research has indicated that certain biological systems might actually maintain themselves in the proximity of a phase transition, to take advantage of the unusually sensitive response that obtains there. Examples include the detection of ligands by membrane receptors (20), the detection of vibrations by mechanosensors (21, 22), and the regulation of enzymatic reactions (23). But it is important to remember that because the number of interacting components in a subcellular biological system is very small, fluctuations play a far more significant role than they do in most condensed-matter systems. We might expect that evolution has also put this element of chance to good effect, as appears to be the case for the Escherichia coli bacterium, which uses fluctuations of its motor as a basis for deciding where to go.
Footnotes
See companion article on page 6696.
References
- 1.Endow S A. Nat Cell Biol. 1999;1:163–167. doi: 10.1038/14113. [DOI] [PubMed] [Google Scholar]
- 2.Henningsen U, Schliwa M. Nature (London) 1997;389:93–96. doi: 10.1038/38022. [DOI] [PubMed] [Google Scholar]
- 3.Case R B, Pierce D W, Hom-Booher N, Hart C L, Vale R D. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- 4.Endow S A, Higuchi H. Nature (London) 2000;406:913–916. doi: 10.1038/35022617. [DOI] [PubMed] [Google Scholar]
- 5.Badoual M, Jülicher F, Prost J. Proc Natl Acad Sci USA. 2002;99:6696–6701. doi: 10.1073/pnas.102692399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huxley A F, Simmons R M. Nature (London) 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 7.Hill T L. Prog Biophys Mol Biol. 1974;28:267–340. doi: 10.1016/0079-6107(74)90020-0. [DOI] [PubMed] [Google Scholar]
- 8.Duke T A J. Proc Natl Acad Sci USA. 1999;96:2770–2775. doi: 10.1073/pnas.96.6.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jülicher F, Ajdari A, Prost J. Rev Mod Phys. 1997;69:1269–1281. [Google Scholar]
- 10.Astumian R D. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- 11.Okada Y, Hirokawa N. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 12.Visscher K, Schnitzer M J, Block S M. Nature (London) 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 13.Jülicher F, Prost J. Phys Rev Lett. 1995;75:2618–2621. doi: 10.1103/PhysRevLett.75.2618. [DOI] [PubMed] [Google Scholar]
- 14.Jülicher F, Prost J. Phys Rev Lett. 1997;78:4510–4513. [Google Scholar]
- 15.Camalet S, Jülicher F, Prost J. Phys Rev Lett. 1999;99:1590–1593. [Google Scholar]
- 16.Berg H C. Phil Trans R Soc London B. 2000;355:491–501. doi: 10.1098/rstb.2000.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner L, Caplan S R, Berg H C. Biophys J. 1996;71:2227–2233. doi: 10.1016/S0006-3495(96)79425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cluzel P, Surrette M, Leibler S. Science. 2000;287:1652–1657. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 19.Duke T A J, le Novère N, Bray D. J Mol Biol. 2001;308:541–553. doi: 10.1006/jmbi.2001.4610. [DOI] [PubMed] [Google Scholar]
- 20.Duke T A J, Bray D. Proc Natl Acad Sci USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camalet S, Duke T, Jülicher F, Prost J. Proc Natl Acad Sci USA. 2000;97:3183–3188. doi: 10.1073/pnas.97.7.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eguiluz V M, Ospeck M, Choe Y, Hudspeth A J, Magnasco M O. Phys Rev Lett. 2000;84:5232–5235. doi: 10.1103/PhysRevLett.84.5232. [DOI] [PubMed] [Google Scholar]
- 23.Berg O G, Paulsson J, Ehrenberg M. Biophys J. 2000;79:1228–1236. doi: 10.1016/S0006-3495(00)76377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]