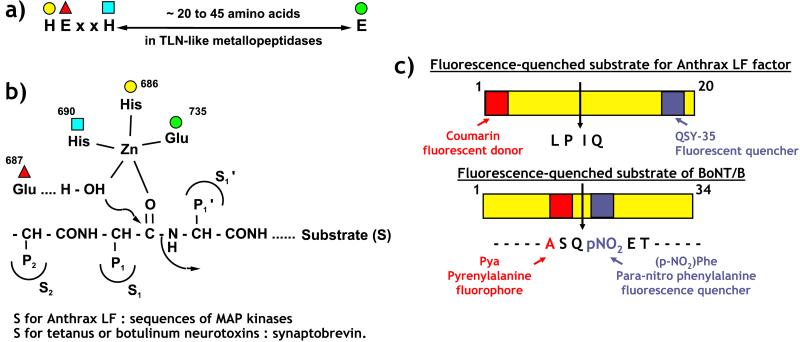
The anthrax toxins, edema (Edtx) and lethal (Letx) toxins, are major virulence factors of Bacillus anthracis and are responsible for the main symptoms of the disease. Soon after the toxins were discovered, i.v. injection of Letx into animals was shown to cause death (for review see Mock and Fouet, ref. 1). Letx is composed of protective antigen (PA), the cell binding component, and lethal factor (LF). It took more than 30 years of work to elucidate the enzymatic activity of LF. The finding that the C-terminal part of LF contains a well preserved zinc-metallopeptidase HExxH (2) consensus site was the first clue to LF activity (Fig. 1 a and b; ref. 3). This consensus sequence was described previously in the bacterial peptidase thermolysin (4) and various physiologically critical mammalian enzymes including angiotensin-converting enzyme (ACE; ref. 5) and neprylisin (6). This zinc binding motif has been found also in the neurotoxins released by the anaerobic bacteria Clostridium tetani and Clostridium botulinum (7), which are responsible for tetanus and botulism, respectively, and both are forms of muscle paralysis. However, anthrax toxins act on a wide variety of cell types, unlike the metalloprotease neurotoxins, which target the synaptosomes, and this was a major difficulty for the identification of the cellular target of LF. Finally, two different approaches led to the identification of the mitogen-activated protein kinase (MAPK) kinase (MKK) family of proteins as the specific substrate of LF proteolytic activity (8, 9); LF cleaves these proteins at the N terminus and thereby disrupts the transduction of extracellular signals into various biological responses including gene expression. Once the target of LF activity had been described, it was possible to search for specific inhibitors. Indeed, mutations in the consensus sequence 686HExxH690 (Fig. 1a), which abolish zinc-binding and peptidase activity of LF, cause complete loss of Letx toxicity both to sensitive cells in vitro (3) and after injection into rats in vivo (10). Recombinant strains of B. anthracis carrying these mutations do not express the lethal phenotype in experimental infections (11). Therefore, inhibition of LF metallopeptidase activity is a possible approach to designing new drugs. Such drugs could be used in association with antibiotics to treat anthrax. The need for specific therapeutics to treat anthrax was shown clearly during the recent bioterrorist attacks in the United States as underlined by Cummings et al. (12) in this issue of PNAS.
Figure 1.
(a) Consensus sequence present in zinc metallopeptidases such as thermolysin (TLN), lethal factor of anthrax toxins (LF), and tetanus and botulinum neurotoxins. (b) Organization of the amino acids from the consensus sequence around the zinc atom. The numbering corresponds to amino acids of LF. The glutamate (Glu-687) polarizes the water molecule, which induces the cleavage of the substrate. The nature of S2 and S1′ subsites (lipophilic in LF) are important for an efficient enzyme recognition and a rapid peptide cleavage. (c) Substrates [MKK fragment for LF and synaptobrevin fragment for botulinum neurotoxin type B (BoNT/B)] used for rapid screening of inhibitors. The fluorescence of the donor (coumarin or pyrenylalanine) is quenched almost totally by the acceptor [p-nitrophenylalanine (NOP) or QSY-35, a substituted nitro-benzoxadiazole] in the intact substrates. The peptide cleavage produces a large increase (6–50%) of the fluorescence.
The first step in the design of inhibitors is the development of a rapid, sensitive, and simple enzymatic assay for testing a large number of molecules. Fluorescent substrates have often been used for automated high-throughput screening tests. LF can cleave various proteins belonging to the MKK family, and the sequences of the cleavage sites are diverse. However, hydrophobic amino acids seem to be preferred in position P2 and P1′ and basic residues N-terminal to the cleavage site (8). Several peptides with these characteristics were tested. Selective cleavage by LF at a single site was verified by HPLC, and the sequences of the two metabolites formed were established by mass spectrometry and not by the more traditional method of comparing the retention times in HPLC of synthetic metabolites. The efficacies of cleavage of these substrates were determined, and one with a rate of cleavage ≈100-fold higher than the native MEK-1 substrate was selected (12).
Appropriate side chains were introduced into the N- and C-terminal positions of the substrate such that it carried a pair of fluorophores separated by 17 amino acids (Fig. 1c). A coumarin fluorophore was used as the donor and paired with QSY-35, a commercially available substituted nitro-benzoxadiazole that acts as quencher by nonradiative fluorescence resonance energy transfer (FRET). This phenomenon means that the fluorescence emitted by the coumarin is not relayed but switched off by the QSY-35 acceptor. The cleavage of this substrate by LF produces a fluorescence increase of between 9 and 16% at 100% cleavage. Although sufficient for high-throughput screening of LF inhibitors in 96-well plates, the assay using this substrate is much less efficient than that reported recently for botulinum neurotoxin type B (BoNT/B) (13). In this case, a pair of fluorophores [L-pyrenylalanine as donor and p-nitrophenylalanine (NOP) as quencher] are close together, separated by only two amino acids (Fig. 1c). As a consequence, quenching caused by collision between the two chromophores is almost complete, leading to an increase of 50-fold in the fluorescence after cleavage of the peptide. This substrate allows detection of less than 3.5 pg of botulinum neurotoxin, which is in the range of sensitivity of the currently used in vivo assay on mice (13). However, the in vivo assay is time-consuming and requires the sacrifice of a large number of animals (14).
The kinetic constants, Km and kcat, for the LF substrate and its stability have not been described, and thus the Ki of inhibitors identified by the assay cannot be calculated. Nevertheless, the rate of substrate cleavage and sensitivity of the assay are adequate for inhibitor screening, although the assay can be disturbed by the fluorescence of tested inhibitors. As a result, Cummings et al. are looking for other fluorophore/quencher pairs (12). As yet, no efficient inhibitor of LF has been described, which contrasts with tetanus and botulinum toxins for which molecules with a thiol group acting as a zinc-chelating reagent were reported recently to exhibit micromolar and nanomolar affinities, respectively (15, 16). However, the catalytic sites of LF and the clostridial neurotoxins are very similar structurally. The success with clostridial neurotoxins suggests that it will be possible to design potent inhibitors for LF.
A comparison of the sequences of the two neurotoxins and LF strongly suggested that the third zinc ligand is a glutamate. This hypothesis was confirmed recently by the determination of the structure of LF by crystallography (17). The three-dimensional organization of the LF active site can be superimposed on that of thermolysin, which is considered to be a typical example of a zinc metallopeptidase (2). It is interesting to observe that the S1′ and S2 subsites of LF are hydrophobic (17) supporting the selection of lipophilic side chains in the fluorescent substrate (Fig. 1 b and c) by Cummings et al. (12). Similar to botulinum neurotoxin type B (18), LF shows a long, deep groove in its three-dimensional structure (17). The grooves in these proteins can accommodate the extended conformations of long substrates [MKK for LF (17) and synaptobrevin for botulinum neurotoxin (18)]. The description of the LF structure is expected to facilitate the design of inhibitors.
Both LF and the neurotoxins use the same strategy to enter cells. The neurotoxins are two-chain proteins composed of a light chain linked to a heavy chain by a disulfide bridge. The heavy chain binds to a receptor at the surface of the nerve endings leading to internalization (19). The light chain then is released and inhibits neurotransmitter secretion by cleaving one component of the exocytosis machinery (reviewed in ref. 20). Cell penetration by Letx is mediated by PA, which first binds to the ubiquitous cell receptor ATR (21). PA then is cleaved by the cell protease furin and binds and delivers LF to the cytoplasm, where the proteolytic activity of LF interrupts the MAPK cascade (reviewed in ref. 1).
Therefore there is no obvious reason why potent inhibitors of LF could not be identified and optimized by using the high-throughput screening assay developed by Cummings et al. Nevertheless, in some cases it is more difficult than expected, as illustrated with tetanus toxin, for which it has not been possible to synthesize inhibitors with IC50 values lower than 10−6–10−7 M (15), although nanomolar blockers have been designed successfully for botulinum toxin type B (16). This discrepancy could be caused by differences in the mechanism of catalysis, which seems to require occupation of exocytes with tetanus toxin followed by an allosteric-like hydrolysis mechanism (22). The crystal structure of LF shows that a large conformationally ordered loop partly occludes the active site (17). It may be necessary to take this into account when designing LF inhibitors. Elucidation of LF substrate hydrolysis may be important for rationally designing potent and selective inhibitors. Such inhibitors would be of great value in several ways. A better understanding of anthrax pathogenesis undoubtedly requires clarification of the controversy about the role of macrophages in toxin-induced lethality. Letx has specific cytopathic effects on macrophages, and its disruption of MKK signaling is believed to induce lysis of these cells. However MKKs cannot be the only or most important substrates, because the MKKs in macrophage cell lines resistant to the lytic effect are degraded nevertheless. Specific inhibition of the proteolytic activity against MKKs may help identify other target proteins of LF. It is possible that components of the proteasome are LF substrates (23). It will be interesting to determine whether cells infected by B. anthracis undergo apoptosis as a result of the destruction of proteins inhibiting this lethal process.
In conclusion, the choice of LF as a drug target, through inhibition of the metallopeptidase activity, probably is a more promising approach than blocking the complex formed between PA and LF. Indeed it is difficult to inhibit protein–protein interactions by small molecules, because the inhibitor has to be adapted to the structural particularities involved in the binding of the two partners. Anthrax pathogenesis is complex and results from the massive multiplication of bacteria producing the toxin. Antibiotic treatment is effective only if administered early, before a lethal dose of toxins has been delivered to the infected host. Novel therapeutics are needed to be used in combination with antibiotics to control the disease. The design of drugs able to penetrate the cell cytosol and block the metallopeptidase activity of LF is a very promising approach to developing specific treatments. The work of Cummings et al. is a first step toward this goal.
Footnotes
See companion article on page 6603.
References
- 1.Mock M, Fouet A. Annu Rev Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 2.Jongeneel C V, Bouvier J, Bairoch A. FEBS Lett. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- 3.Klimpel K R, Arora N, Leppla S H. Mol Microbiol. 1994;13:1093–1100. doi: 10.1111/j.1365-2958.1994.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 4.Matthews BW. Acc Chem Res. 1988;21:333–340. [Google Scholar]
- 5.Wyvratt M J, Patchett A A. Med Res Rev. 1985;5:483–531. doi: 10.1002/med.2610050405. [DOI] [PubMed] [Google Scholar]
- 6.Roques B P, Noble F, Daugé V, Fournié-Zaluski M C, Beaumont A. Pharmacol Rev. 1993;45:81–146. [PubMed] [Google Scholar]
- 7.Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta B R, Montecucco C. Nature (London) 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 8.Vitale G, Bernardi L, Napolitani G, Mock M, Montecucco C. Biochem J. 2000;352:739–745. [PMC free article] [PubMed] [Google Scholar]
- 9.Duesbery N S, Webb C P, Leppla S H, Gordon V M, Klimpel K R, Copeland D T, Ahn N G, Oskarsson M K, Fukasawa K, Paull K D, Vande Woude G F. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 10.Brossier F, Mock M. Toxicon. 2001;39:1747–1755. doi: 10.1016/s0041-0101(01)00161-1. [DOI] [PubMed] [Google Scholar]
- 11.Brossier F, Weber-Levy M, Mock M, Sirard J-C. Infect Immun. 2000;68:1781–1786. doi: 10.1128/iai.68.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cummings R T, Salowe S P, Cunningham B R, Wiltsie J, Park Y W, Sonatore L M, Wisniewski D, Douglas C M, Hermes J D, Scolnick E M. Proc Natl Acad Sci USA. 2002;99:6603–6606. doi: 10.1073/pnas.062171599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anne C, Cornille F, Lenoir C, Roques B P. Anal Biochem. 2001;291:253–261. doi: 10.1006/abio.2001.5028. [DOI] [PubMed] [Google Scholar]
- 14.Kautter D A, Solomon H M. J Assoc Off Anal Chem. 1977;60:541–545. [PubMed] [Google Scholar]
- 15.Martin L, Cornille F, Turcaud S, Meudal H, Roques B P, Fournié-Zaluski M C. J Med Chem. 1999;42:515–525. doi: 10.1021/jm981066w. [DOI] [PubMed] [Google Scholar]
- 16. Roques, B. P., Anne, C., Turcaud, S. & Fournié-Zaluski, M. C. (2001) Fr. Patent 01.04895.
- 17.Pannifer A D, Wong T Y, Schwarzenbacher R, Renatus M, Petosa C, Bienkowska J, Lacyl D B, Collier R J, Park S, Leppla S H, Hanna P, Liddington R C. Nature (London) 2001;4:229–233. doi: 10.1038/n35101998. [DOI] [PubMed] [Google Scholar]
- 18.Hanson M A, Stevens R C. Nat Struct Biol. 2000;7:687–692. doi: 10.1038/77997. [DOI] [PubMed] [Google Scholar]
- 19.Montecucco C, Papini E, Schiavo G. FEBS Lett. 1994;346:92–98. doi: 10.1016/0014-5793(94)00449-8. [DOI] [PubMed] [Google Scholar]
- 20.Schiavo G, Matteoli M, Montecucco C. Physiol Rev. 2000;80:717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- 21.Bradley K A, Mogridge J, Mourez M, Collier R J, Young J A. Nature (London) 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 22.Cornille F, Martin L, Lenoir C, Cussac D, Roques B P, Fournié-Zaluski M C. J Biol Chem. 1997;272:3459–3464. doi: 10.1074/jbc.272.6.3459. [DOI] [PubMed] [Google Scholar]
- 23.Tang G, Leppla S H. Infect Immun. 1999;67:3055–3060. doi: 10.1128/iai.67.6.3055-3060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]