Abstract
Telomerase adds telomeric repeats to chromosome 3′ ends, forestalling the cellular senescence, apoptosis, and genomic instability that result from telomere loss caused by incomplete DNA replication. The telomerase ribonucleoprotein is dedicated to synthesis of tandem, simple-sequence repeats by virtue of its specialization for copying only a specific template region within the integral RNA. Here, using circularly permuted variants of Tetrahymena thermophila telomerase RNA, we identify the features that allow recognition of the template region within the RNA. We engineered a template-less telomerase ribonucleoprotein that can position and reverse transcribe an exchangeable RNA oligonucleotide template accurately. Only a short “template-recognition” element sequence tag is required to direct efficient use of adjacent 5′ residues as a template for telomeric repeat synthesis. Our findings reveal molecular requirements for template selection by telomerase and physically resolve templating from other RNA functions in catalysis.
Telomerase, a specialized reverse transcriptase (RT), copies a small template region within its RNA subunit to extend chromosome 3′ ends (1). A minimal, catalytically active recombinant telomerase ribonucleoprotein (RNP) can be produced in rabbit reticulocyte lysate by expressing the telomerase RNA and the telomerase RT protein (TERT) from mammals (2–4) or the ciliate Tetrahymena thermophila (5). Telomerase RNAs vary widely in length and primary sequence, but elements of conserved secondary structure have been predicted for ciliate and vertebrate RNAs by phylogenetic analysis (6, 7). The primary sequences of TERTs also are highly divergent, sharing only RT active site motifs and a few telomerase-specific motifs common to TERTs from all or phylogenetically related subsets of species (8–11).
Unlike viral RTs, which copy a variety of single-stranded nucleic acid templates, telomerase can copy only the template region of its stably associated RNA. The molecular mechanism underlying one aspect of internal template use, template-boundary definition, has been examined previously by using telomerases from Kluyveromyces lactis and Tetrahymena. Different mechanisms establish the template 5′ boundary in these organisms: RNA secondary structure is required in K. lactis (12) and protein–RNA interaction is required in Tetrahymena (10, 13). Left remaining is the broader question of how a telomerase RNP recognizes and accurately positions the template in the first place. The correct template region must be selected from within the larger telomerase RNA and delivered to the RT active site, an aspect of template selection that we refer to as “template recognition.”
What features of the RNP could accomplish template recognition? One possibility is that the template is recognized in a sequence-specific manner. This simple mechanism seems unlikely given the relatively high accessibility of template bases to chemical modification (14) and the tolerance of endogenous Tetrahymena telomerase RNP catalytic activity for template sequence changes (15, 16). Instead, the template region could be specified positionally, determined by linkage through the RNA backbone to nearby bases that make sequence-specific RNP contacts. Such an interaction could place the template in the active site yet leave the template itself accessible for substrate hybridization and mobile for transit through the active site.
To examine how the telomerase RNP selects a portion of its RNA as the template for telomere synthesis, we introduced discontinuities in the backbone of the Tetrahymena telomerase RNA at positions 5′ and/or 3′ of the template. If TERT recognizes the template by binding an adjacent RNA element, breaks in the backbone between the bound element and the template should inhibit template use. We show that telomerase recognizes its template via an adjacent 3′ motif and can do so even when the template is not a part of the stable RNP.
Materials and Methods
RNA Expression Constructs and Synthesis.
We cloned two copies of the T. thermophila telomerase RNA gene in tandem, separated by a linker. Transcription templates for circularly permuted (cp) and body RNAs were generated by PCR from this construct by using a forward primer with the minimal T7 promoter. We found no difference in cp RNA function by using linkers of different lengths and used linker 5′-TTTTGGATCC-3′ for all experiments here. The cp25 RNA has an extended stem II and a deletion of the terminal loop, with the sequence 5′-GCGAGAACUG…CAGAUCUCGC-3′ (nucleotides 31–37 and 19–25 of the wild-type RNA are underlined).
Wild-type, cp, and body RNAs were transcribed by using T7 RNA polymerase. Each transcript began with a GTP for efficient transcription even when not encoded by the wild-type telomerase RNA (inserted GTPs are green in Fig. 1A). Insertion of nonendogenous G residues into the unpermuted RNA at equivalent positions had little if any effect on telomerase activity. After transcription, RNAs were gel-purified, eluted, precipitated, and resuspended in H2O. Trans-template RNAs were synthesized chemically (Dharmacon, Lafayette, CO), gel-purified, and used at 40 μM unless noted otherwise.
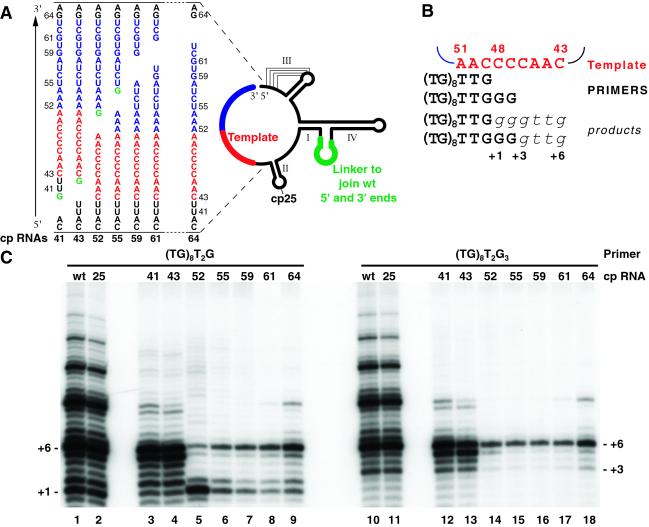
Figure 1.
Use of cp telomerase RNAs. (A) RNA permutations used in this study. (Right) Schematic of T. thermophila telomerase RNA secondary structure as derived from phylogenetic analysis (6, 38) with modifications for circular permutation at position 64. The stems and pseudoknot are indicated by roman numerals. Wild-type 5′ and 3′ ends are joined by a 10-nt linker (green). The line drawing also indicates the position of the break in cp25. (Left) Detailed view of the 5′ and 3′ ends of the cp RNAs disrupted near the template, shown with the template (red) and template-recognition element (TRE, blue; see below) indicated. Each cp RNA begins with its 5′ end immediately above the gap and continues for the full length of a wild-type RNA to its new 3′ end below the gap. To promote transcription by T7 RNA polymerase, a 5′ GTP (green) was introduced in cp RNAs 41, 43, 53, and 55. (B) Telomerase RNA template region, primers, and products. The template (red) and primers (capital letters) used in this study are shown. Nucleotides added to either primer in the first round of repeat synthesis (italics) are numbered relative to the products of primer (TG)8T2G. (C) Telomerase activity with cp RNAs. Equal amounts of immunopurified telomerase RNPs containing the indicated cp or wild-type (wt) RNA were assayed for extension of the indicated DNA primers. The first-repeat addition product lengths are indicated.
RNP Assembly and Activity Assays.
Four picomoles of telomerase RNA and 5 μg of yeast tRNA were assembled with 10 μl of rabbit reticulocyte lysate translation reaction (Promega) containing His/hemagglutinin-tagged T. thermophila TERT (17) for 15 min at 30°C. Under the binding conditions used, TERT associated with similar levels of wild-type, cp, or body RNAs. The resulting RNPs were immunopurified by using hemagglutinin antibody bound to protein G beads (Amersham Pharmacia; ref. 10). Equivalent amounts of assembled RNP (≈10 fmol) were assayed in each reaction unless noted otherwise. Functional trans-template RNA association with the telomerase RNP was independent of reticulocyte lysate.
Activity assay reactions contained 0.5× T2MG (1× T2MG contains 20 mM Tris⋅HCl, pH 8, 1 mM MgCl2, 10% glycerol, and 2 mM DTT), primer (1 μM), [α-32P]dGTP (3.25 μM), and dTTP (250 μM unless noted otherwise). Assays were carried out for 20 min at 30°C in TTB (50 mM Tris acetate, pH 8/10 mM spermidine/5 mM β-mercaptoethanol/2 mM MgCl2). Reactions were stopped with a solution containing a radiolabeled oligonucleotide loading control, extracted, and precipitated. Products were electrophoresed on denaturing 12% (19:1) polyacrylamide gels and visualized by autoradiography or quantitated by a phosphorimager (Fuji). We observed a higher Km for primer when assaying RNPs that contained a cp instead of wild-type RNA, probably because of a lack of cooperativity between primer binding at the anchor site and template. The 1 μM primer concentration used in assays shown here is saturating for all enzymes.
Results
cp RNAs Retain Different Levels of Template Function.
We designed a series of telomerase RNAs joined at the wild-type 5′ and 3′ ends and broken in the RNA backbone at various internal positions (Fig. 1A). Purified wild-type and cp RNAs were assembled with TERT expressed in reticulocyte lysate, and then the telomerase RNPs were immunopurified and assayed. Each RNP was assayed with two primers differing at their 3′ ends to direct the formation of either a 3- or 5-bp hybrid with the template [primers (TG)8T2G and (TG)8T2G3; see Fig. 1B]. We have indicated the addition of nucleotides within the first repeat as +1 to +6 relative to primer (TG)8T2G. The first nucleotide added to primer (TG)8T2G occurs at +1, whereas the first nucleotide added to primer (TG)8T2G3 occurs at +3. Products labeled +1 to +6 result from first-repeat synthesis, whereas those longer than +6 result from processive repeat addition.
As a control for insertion of the linker sequence joining the wild-type ends, we designed a cp telomerase RNA with new 5′ and 3′ ends base-paired at the distal end of stem II (designated cp25 RNA according to the wild-type RNA position at the new 5′ end; Fig. 1A). The distal end of stem II is dispensable for wild-type activity of recombinant Tetrahymena telomerase (17). In activity assays with either primer, telomerase RNPs containing either wild-type or cp25 RNA generated similar levels of telomerase activity and nearly identical product profiles (Fig. 1C, lanes 1–2 and 10–11).
RNAs discontinuous in the single-stranded regions adjacent to the 5′ or 3′ boundaries of the template also supported substantial levels of telomerase activity (Fig. 1C). Particularly striking is the strong first-repeat synthesis and even moderate repeat addition processivity attained with cp41 and cp43 RNAs, disrupted 2 or 0 nts from the 5′ template boundary, respectively (lanes 3–4 and 12–13). These RNAs lack local covalent connectivity between the template and the motif that interacts with the TERT RNA-binding domain, which is composed of stem II and the flanking single-stranded nucleotides (13, 18). The activity observed with telomerase RNPs containing cp41 and cp43 RNAs suggests that local backbone linkage of the template with 5′ adjacent RNA residues that compose the high-affinity TERT- binding site is not essential for template recognition or use despite the importance of this linkage in template-boundary definition (10, 13, 19).
Telomerase RNAs with backbone discontinuity 3′ of the template also supported telomerase activity (Fig. 1C, lanes 5–9 and 14–18) although at a lower level than cp25, cp41, or cp43 RNAs. An RNA with a discontinuity immediately 3′ of the template (cp52) demonstrated less processive nucleotide addition within a repeat as well as synthesis of aberrant products longer than +6 (lanes 5 and 14) particularly when assayed with the primer forming a shorter template hybrid [(TG)8T2G; lane 5]. Moving the backbone break slightly 3′ (cp55 and cp59; lanes 6–7 and 15–16) restored a first-repeat addition product profile more similar to that of the wild-type enzyme. Movement of the interruption to 9 or 12 nt 3′ of the template (cp61 and cp64) restored a detectable level of repeat-addition processivity as well (lanes 8–9 and 17–18). Movement of the interruption further 3′ did not improve either total activity or repeat-addition processivity (data not shown). These results suggest the unanticipated conclusion that efficient copying of the template requires local backbone connectivity between the template and a 3′ adjacent motif.
Telomerase Can Use Oligonucleotides as Trans-Template RNAs.
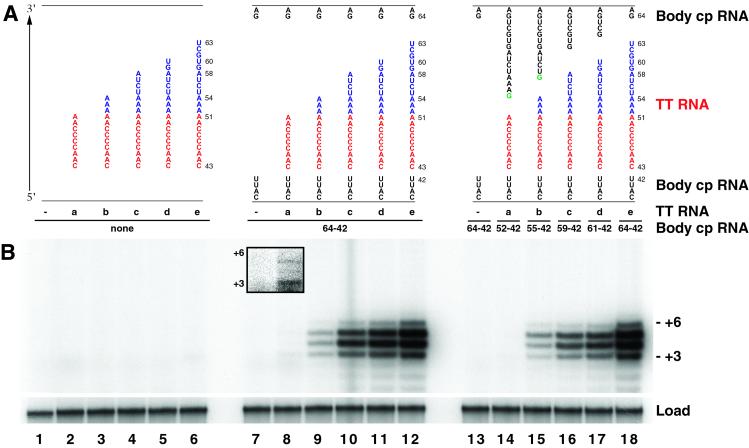
If the residues 3′ of the template are not only necessary but also sufficient to direct template recognition, they should promote copying of the adjacent sequence even if the template is unlinked completely from the remainder of the telomerase RNA. To test this possibility, we disrupted the RNA backbone at both ends of the template simultaneously. We designed a series of trans-template RNA oligonucleotides (TTa–TTe; Fig. 2A) corresponding at minimum to wild-type RNA positions 43–51 (TTa, the template region alone) and at maximum to wild-type RNA positions 43–63 (TTe, the template with the 3′ sequence extension maximally stimulatory for template use in the cp RNA). We assessed the ability of each of these trans-template RNAs to support product synthesis by TERT in the presence or absence of the remainder of a template-less telomerase RNA. TERT was expressed in reticulocyte lysate and assembled with a telomerase RNA “body” permuted and truncated such that it lacked the template and adjacent residues (e.g., body RNA cp64–42 has its 5′ end at wild-type position 64 and its 3′ end at wild-type position 42, lacking residues 43–63). Immunopurified RNPs containing TERT and body RNA were assayed in the presence of purified trans-template RNA, primer, and nucleotides [results with primer (TG)8T2G3 are shown in Fig. 2B; a similar pattern of results was obtained with primer (TG)8T2G, data not shown].
Figure 2.
Use of trans-template RNAs by telomerase RNPs. (A) Sequences of template-containing trans-template RNAs and of body RNA 5′ and 3′ ends, depicted as described for Fig. 1A. Trans-template RNAs (red and blue) begin with the 5′ end at wild-type RNA position 43 and extend to five different 3′ positions. (B) Telomerase activity supported by the body and trans-template RNA pairs shown in A. RNPs containing TERT and body RNA were immunopurified and assayed for extension of (TG)8T2G3 in the presence of 40 μM trans-template RNA. Each of the trans-template RNAs, labeled a–e from shortest to longest, was assayed with TERT RNPs containing no body RNA (lanes 1–6), body RNA cp64–42 (lanes 7–12), or the body RNA matched to the individual trans-template RNA such that all of the nucleotides of the wild-type RNA are present considering both RNAs (lanes 13–18). (Right) First-repeat addition product lengths are indicated. (Inset) Lanes 7 and 8 from a different exposure of the same gel. A loading control (load) was added to each sample before precipitation to verify sample recovery.
No product synthesis was detected in reactions that contained TERT and trans-template RNAs but no body RNA (Fig. 2B, lanes 2–6). Likewise, when TERT was assembled with any body RNA and assayed without a trans-template RNA, no activity was evident (lanes 7 and 13 and data not shown). In contrast, when an RNP consisting of TERT and cp64–42 body RNA was assayed in the presence of any of the trans-template RNAs, product synthesis indeed was observed (lanes 8–12). Activity with TTa was weak but consistently detectable for RNP containing cp64–42 body RNA (Inset shows a different exposure of lanes 7 and 8). The level of activity improved substantially if trans-template RNAs included additional 3′ residues up to wild-type position 63 (TTb–TTe, lanes 9–12). A trans-template RNA including the pseudoknot (wild-type positions 43–102 in total) did not appear to stimulate either substantially more activity than TTe or any repeat-addition processivity (data not shown). In most cases, identical results were obtained when each trans-template RNA was assayed with a telomerase RNP that contained cp64–42 body RNA (lanes 7–12) or body RNAs extended at their 5′ ends such that all wild-type telomerase RNA residues were present when considering the two RNAs combined (lanes 14–18). The single exception was TTa-templated activity, which was more readily detectable with RNP containing cp64–42 RNA rather than cp52–42 RNA (lanes 8 and 14). This difference may reflect a steric hindrance by the 5′ G nucleotide on cp52–42 (inserted to promote transcription, see Materials and Methods).
We conclude that a wild-type template region separated from the telomerase RNA body can be copied efficiently. The presence of an endogenous RNA element 3′ of the copied template, hereafter referred to as the TRE (shown in blue), dramatically enhances template use if provided in cis with the template (Fig. 2B, lanes 8–12) but not if provided in trans as part of the body RNA (compare lanes 14–17 with lane 18). Activity on a trans- template RNA strictly requires elements within the template-less telomerase body RNA, as well as TERT. We note that copying a template external to the remainder of the telomerase RNA generates a product profile different from that of the wild-type enzyme (see Discussion).
Trans-Template RNAs Dissociate from a Stable TERT–Body RNA Complex.
Wild-type, cp, and body RNAs have nanomolar dissociation constants for TERT (ref. 18; data not shown). Titration of TTe in activity assays indicated that product synthesis saturated only with very high concentrations of trans-template RNA, with a Km of 10 μM. In all the experiments described above, trans-template RNAs were used at a 40 μM final concentration, which supported near maximal activity for each trans-template RNA. This concentration represents an ≈1,000-fold excess of trans-template RNA over body RNP in an activity assay reaction. This observation suggests that a stable RNP composed of TERT and a body RNA positions and copies a trans-template RNA that associates only weakly with the RNP.
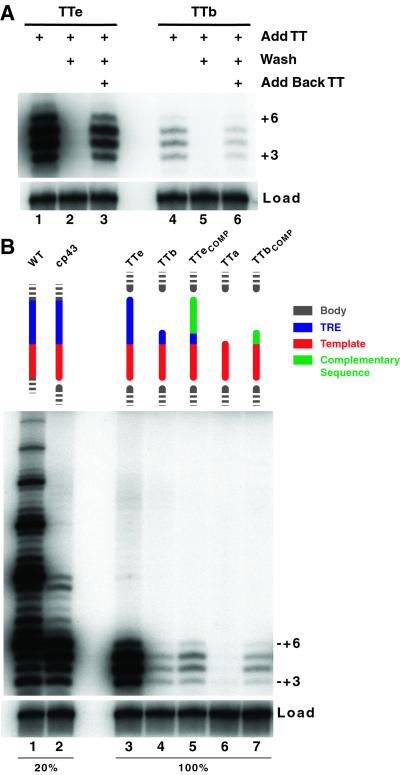
We directly tested whether trans-template RNAs dissociate and reassociate with a body RNA-containing telomerase RNP under activity assay conditions. To do this, we measured the activity of telomerase complexes formed in the presence of 40 μM trans-template RNA with or without removing unbound trans-template before the activity assay. Telomerase RNP with cp64–42 body RNA was immobilized on beads via the hemagglutinin-epitope tag on TERT and combined with TTe or TTb and primer (TG)8T2G3. This pool of RNP was split, with part assayed without washing (Fig. 3A, lanes 1 and 4) and the rest washed in assay buffer to remove unbound trans-template RNA. When the washed telomerase complexes were assayed by the addition of primer and nucleotides, no activity was detected (lanes 2 and 5). However, when the washed complexes were supplied with fresh trans-template RNA in addition to primer and nucleotides, telomerase activity was restored (lanes 3 and 6). Similar results were obtained independent of the presence or absence of primer in the initial trans-template RNA-binding reaction (data not shown). The decrease in telomerase activity observed with washing resulted in part from incomplete recovery of the beads carrying the TERT/cp64–42 RNP. These results reveal that the TERT–body RNA complex is significantly more stable than the association of this RNP with a trans-template oligonucleotide. The lower affinity for the template in trans relative to the template that is part of a cp RNA may explain why trans templates did not support repeat-addition processivity while the cp RNAs did.
Figure 3.
Trans-template binding. (A) Test of trans-template dissociation and reassociation. Immunopurified RNP containing TERT and cp64–42 RNA was incubated with primer (TG)8T2G3 and 40 μM TTe or TTb in assay buffer lacking dNTP substrates. This reaction was split and either assayed by the addition of dNTPs (lanes 1 and 4), washed in assay buffer, and then assayed by the addition of dNTPs and primer (lanes 2 and 5) or washed and assayed by the addition of dNTPs, primer, and 40 μM trans-template RNA (lanes 3 and 6). A loading control (load) was added to each sample before precipitation to verify sample recovery. (B) Sequence specificity of TRE function. Immunopurified RNPs containing TERT and wild-type (WT) telomerase RNA (lane 1), cp43 RNA (lane 2), or cp64–42 body RNA (lanes 3–8) were assayed for extension of (TG)8T2G3 with 40 μM of the indicated trans-template RNA. (Right) Assays in lanes 1 and 2 were performed with 20% of the amount of RNP used in assays in lanes 3–7. First-repeat addition product lengths are indicated. A loading control (load) was added to each sample before precipitation to verify sample recovery. Residues in green are of sequence complementary to wild type. TTeCOMP, CAACCCCAAAAAAGAUCACGA; TTbCOMP, CAACCCCAAUUU.
Telomerase containing cp64–42 body RNA with TTe directed ≈20% of the level of product synthesis catalyzed by telomerase RNP containing wild-type or cp RNA, as judged by quantitation of first-repeat synthesis (Fig. 3B, lanes 1–3; note that 20% as much enzyme was assayed in lanes 1 and 2 as in lanes 3–7). This quantitation may underrepresent trans-template RNA-dependent active site function. For example, the reduced formation of +6 product with trans-template RNAs and cp RNAs discontinuous 5′ of the template leads to a product distribution of lower specific activity per nucleotide added. Changes in repeat-addition processivity may bias apparent activity level as well because of efficiency differences in product dissociation versus repositioning. We conclude that telomerase can position and copy a dissociable, external RNA template, contrary to its previous classification as an RT specific for an internal template.
TRE Function Is Sequence-Specific.
To determine whether the TRE stimulation of trans-template RNA use requires a specific sequence of RNA or just a specific length of nucleotides, we tested the activity of the cp64–42 telomerase RNP with a series of trans-template RNA oligonucleotides composed of the wild-type template sequence and different 3′ extensions. Consistent with the results shown in Fig. 2, the shorter TTb was copied less efficiently than the longer TTe (Fig. 3B, lanes 3 and 4). When the nucleotides that distinguish TTe from TTb were present but altered to the complementary sequence, activity remained at the lower level of TTb (TTeCOMP, lane 5). This result indicates that trans-template recognition is directed sequence-specifically by residues within positions 55–63. In contrast, when the nucleotides that distinguish TTb (lane 4) from TTa (lane 6) were present but altered to the complementary sequence, the level of activity was similar to that observed with TTb (TTbCOMP, lane 7). In total, results from TRE sequence substitutions indicate two different types of TRE specificity. First, there is a minimal length requirement of between 9 and 12 nt for substantial use of an oligonucleotide as trans-template RNA. Second, the remainder of the TRE functions in a predominantly sequence-specific manner to promote robust copying of adjacent residues. In the context of the unpermuted RNA, substitutions within this second TRE region inhibit overall activity or repeat-addition processivity, which is consistent with their function in template recognition (see Discussion).
Trans-Template RNAs with Sequence Alterations Direct Appropriate Product Synthesis.
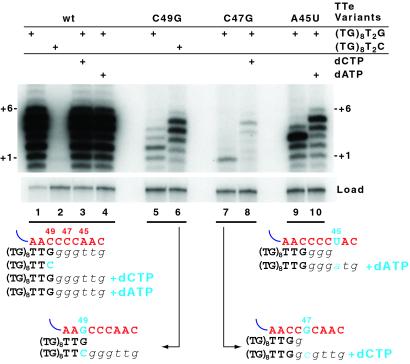
As a final test of the requirements for template recognition and use, we investigated the ability of trans-template RNAs bearing template region substitutions to direct synthesis of altered product sequences. In the unpermuted Tetrahymena telomerase RNA, altered templates can be copied accurately or impose catalytic defects depending on the sequence substitution (20–22). We assayed the activity of RNPs containing cp64–42 RNA with variants of TTe altered in the template region (Fig. 4). TTe C49G RNA is substituted within the region of hybridization to primer (TG)8T2G. When assayed with this standard primer, TTe C49G RNA directed synthesis of only aberrant, short products (lane 5). In contrast, when assayed with the hybridizing primer (TG)8T2C, the C49G template supported strong activity with a normal trans-template product distribution (compare lanes 1 and 6). Conversely, TTe maintained strong activity with (TG)8T2G but not with (TG)8T2C (lanes 1 and 2). For template substitutions 5′ of the region of primer hybridization, synthesis to the template 5′ end should require the presence of all dNTPs cognate to the altered template. TTe C47G RNA assayed with primer (TG)8T2G directed addition of only one dGTP in a standard dGTP/TTP reaction (lane 7) but generated products as long as those produced by unsubstituted TTe in the presence of dCTP (lane 8). Similarly, TTe A45U RNA directed product synthesis resulting from the addition of only three dGTP nucleotides in the absence of dATP (lane 9), but longer products were generated in the presence of dATP (lane 10). The addition of dATP or dCTP to reactions with TTe did not alter the product profile (lanes 3 and 4).
Figure 4.
Sequence specificity of trans-template function. Immunopurified RNP containing TERT and cp64–42 RNA was assayed with the indicated template-substituted TTe-variant RNA and nucleotides. First-repeat addition product lengths are indicated. A loading control was added to each sample before precipitation to verify sample recovery. Schematics of the expected reaction are shown below the gel. Wild-type (wt) sequence template residues are red, primer and product residues are black, and all altered residues are cyan.
These results demonstrate convincingly that telomerase can copy an altered template sequence in the trans-template context, as was observed previously in unpermuted RNAs (23). However, trans-template RNAs with substitutions that involved the majority of templating residues did not support detectable telomerase activity [data not shown; the templates tested were 5′-CUAUAUAUA-3′ (AU4 from ref. 15) and 5′-GUACGCGUA-3′]. The failure to copy these templates may derive in part from a preference of the telomerase active site for dGTP binding (16). In sum, these template-substitution studies demonstrate that telomerase RNP composed of TERT and a template-less telomerase RNA body can recognize the proper template region of a trans-template RNA, suitably align a primer 3′ end, and copy it with reasonable fidelity.
Discussion
The TRE Specifies Adjacent Sequence as Template.
Our analysis of telomerase RNAs with disrupted backbones, along with previous mutagenesis of the telomerase RNA, reveals the presence of a TRE that promotes the copying of nucleotides 43–51. When the template is separated from the remainder of the telomerase RNA on one (Fig. 1) or both (Fig. 2) sides, its use is stimulated dramatically by local RNA backbone connectivity with the TRE. In the unpermuted telomerase RNA, residues of the TRE stimulate overall activity and repeat-addition processivity; the substitution UCU55–57AGA or AG58–59UC inhibits repeat-addition processivity, and substitution C62G strongly inhibits nucleotide addition activity (17, 18).
The defects induced by TRE alteration or disconnection from the template are explained most readily by a loss of template “default” positioning. Ciliate telomerases can extend nontelomeric substrates, both in vitro and in vivo, but always do so from a specific position at the 3′ end of the template (24–27). Likewise, after synthesis to the 5′ end of the template and product dissociation, the RNP returns the 3′ end of the template to the default position, allowing for repeat-addition processivity (28). Mutation of the TRE in an unpermuted RNA can abolish repeat-addition processivity while retaining template-copying activity. This defect is consistent with loss of default positioning, as in this context the template still can be brought to the active site by linkage to the 5′ TERT-binding site and primer-template hybridization but may not be positioned precisely enough for multiple rounds of repeat addition. In the trans-template context, separation of the TRE from the template abolishes all template copying. Because the trans-template RNA is not restrained to the active site via an RNA backbone linkage to the TERT binding site, TRE-mediated default positioning becomes essential for even initial template positioning.
Telomerase Is Specialized to Use an Internal Template.
Although telomerase can reverse transcribe an exchangeable oligonucleotide template, the physiological function of telomerase is restricted to copying its tightly associated internal template. Backbone linkage of the template with motifs other than the TRE also influences RNP catalytic function. Covalent linkage of the template 5′ end and the adjacent high-affinity TERT-binding site/template-boundary element (13, 18, 19) promotes copying to the end of the template and no further, increasing the frequency of generating a precise telomeric repeat (and allowing repeat-addition processivity). This function is evident in the shift in predominant product intensity to 1 nt short of the normal template 5′ boundary with all the trans-template RNAs, as well as with the cp RNAs containing backbone discontinuities 5′ of the template. Backbone linkage of the 3′ end of the TRE with the rest of the telomerase RNA stimulates overall template use and repeat-addition processivity, suggesting that this linkage may facilitate TRE-directed template positioning. These 5′ and 3′ linkages of the template also would increase effective template concentration and prevent copying of spurious templates in vivo. Despite not being required for primer extension, cis linkage of the template TRE to the remainder of the telomerase RNA is essential to proper enzyme function in the cell.
Telomerase Is an RNP Enzyme.
Several previous investigations have identified nontemplate telomerase RNA residues that affect RNP function. Species-specific telomerase RNA motifs important for cellular assembly of the telomerase RNP have been identified (29–32). As described above, mutagenesis studies have revealed important roles for template-adjacent RNA elements in specifying features of cis template use including boundary definition (12, 13, 19). In addition, RNA substitutions distant from the template in primary sequence have been shown to affect catalytic properties of the enzyme (17, 32–35). However, in these studies the substituted RNA residues were still linked in cis with the template, potentially altering template position within the RNP and thus affecting template use indirectly. One example of this phenomenon is the C62G substitution in the TRE, which has a more significant impact in the unpermuted RNA (17) than its deletion or substitution does in the trans-template context (Figs. 2 and 3).
We show here that TERT RT activity strictly depends on its association with nontemplate regions of telomerase RNA. The inactivity of TERT on a template-primer duplex that contains the TRE and is competent as a viral RT substrate (5) reflects not the previously supposed requirement for an internal template but rather a requirement for the nontemplate residues of telomerase RNA. In other words, telomerase is an obligate RNP enzyme even when acting on an exchangeable RNA template. What is the molecular basis of the body RNA requirement? One possibility is that the body RNA executes a template-related function such as forming a template-binding site on the RNP. More likely, the body RNA might contribute to telomerase RNP function by positioning the primer substrate, allosterically activating TERT and/or directly establishing an essential feature of the active site. It will be interesting to investigate the roles of the body RNA in greater detail and determine whether they are shared by other RNP RTs (36, 37).
Acknowledgments
We thank members of the Collins lab for discussion and comments on the manuscript. This work was supported by a National Science Foundation Predoctoral Fellowship (to M.C.M.) and National Institutes of Health Grant GM54198 (to K.C.).
Abbreviations
- RT
reverse transcriptase
- RNP
ribonucleoprotein
- TERT
telomerase RT protein
- cp
circularly permuted
- TRE
template-recognition element
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Blackburn E H. Nat Struct Biol. 2000;7:847–850. doi: 10.1038/79594. [DOI] [PubMed] [Google Scholar]
- 2.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichsteiner S, Kim N W, Trager J B, et al. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 3.Beattie T L, Zhou W, Robinson M O, Harrington L. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg R A, Chin L, Femino A, Lee K-H, Gottlieb G J, Singer R H, Greider C W, DePinho R A. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 5.Collins K, Gandhi L. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero D P, Blackburn E H. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 7.Chen J-L, Blasco M A, Greider C W. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 8.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 9.Bryan T M, Sperger J M, Chapman K B, Cech T R. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller M C, Liu J K, Collins K. EMBO J. 2000;19:4412–4422. doi: 10.1093/emboj/19.16.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia J, Peng Y, Mian I S, Lue N F. Mol Cell Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzfati Y, Fulton T B, Roy J, Blackburn E H. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- 13.Lai C K, Miller M C, Collins K. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaug A J, Cech T R. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]
- 15.Ware T L, Wang H, Blackburn E H. EMBO J. 2000;19:3119–3131. doi: 10.1093/emboj/19.12.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins K. Annu Rev Biochem. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- 17.Licht J D, Collins K. Genes Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai C K, Mitchell J R, Collins K. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Autexier C, Greider C W. Genes Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- 20.Gilley D, Lee M S, Blackburn E H. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 21.Gilley D, Blackburn E H. Mol Cell Biol. 1996;16:66–75. doi: 10.1128/mcb.16.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy C D, Schultz C S, Collins K. J Biol Chem. 2001;276:4863–4871. doi: 10.1074/jbc.M005158200. [DOI] [PubMed] [Google Scholar]
- 23.Yu G, Bradley J D, Attardi L D, Blackburn E H. Nature (London) 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 24.Yu G, Blackburn E H. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
- 25.Melek M, Greene E C, Shippen D E. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Blackburn E H. EMBO J. 1997;16:866–879. doi: 10.1093/emboj/16.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Gilley D, Blackburn E H. EMBO J. 1998;17:1152–1160. doi: 10.1093/emboj/17.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greider C W. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell J R, Cheng J, Collins K. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto A G, Zaug A J, Sobel S G, Wolin S L, Cech T R. Nature (London) 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 31.Gilley D, Blackburn E H. Proc Natl Acad Sci USA. 1999;96:6621–6625. doi: 10.1073/pnas.96.12.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell J R, Collins K. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharyya A, Blackburn E H. Proc Natl Acad Sci USA. 1997;94:2823–2827. doi: 10.1073/pnas.94.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy J, Fulton T B, Blackburn E H. Genes Dev. 1998;12:3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Rivera L, Blasco M A. J Biol Chem. 2001;276:5856–5865. doi: 10.1074/jbc.M008419200. [DOI] [PubMed] [Google Scholar]
- 36.Tavis J E, Ganem D. J Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmerly S, Moran J V, Perlman P S, Lambowitz A M. J Mol Biol. 1999;289:473–490. doi: 10.1006/jmbi.1999.2778. [DOI] [PubMed] [Google Scholar]
- 38.ten Dam E, van Belkum A, Pleij K. Nucleic Acids Res. 1991;19:6951. doi: 10.1093/nar/19.24.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]