ABSTRACT
Influenza A virus (IAV) has become increasingly unpredictable in its seasonal patterns following the COVID-19 pandemic. A key challenge remains identifying optimal moments for public health interventions to inform evidence-based decisions. Real-time PCR was employed to quantify IAV concentrations in wastewater samples from 38 treatment plants in Shenzhen, China (n = 2,764), collected weekly from March 2023 to March 2024. The random forest model was used to estimate IAV infections based on viral concentrations and physico-chemical parameters. Baseline IAV concentrations were established using mean, geometric, and median values, revealing a seasonal IAV pattern with peaks in winter 2023 and spring 2024. The optimized random forest model (mean absolute error = 2,307, R2 = 0.988) integrated IAV concentration, flow rate, wastewater temperature, chemical oxygen demand, total nitrogen, and phosphorus. The established baseline values of IAV concentration provides crucial evidence for optimal timing (early March and mid-November) of seasonal vaccination and nonpharmaceutical interventions. The baseline defined as the median provided 2–4 weeks early warning before influenza season onset (≥ 100 cases/100,000). The median baseline also allows early detection of abnormal increases in respiratory syncytial virus, supporting timely public health interventions. Wastewater surveillance of IAV, which encompasses the entire population, closely aligns with clinical monitoring trends, underscoring its vital role as a complementary tool in public health. The estimated number of infections and the baseline defined by median offer valuable insights to guide vaccination timing and non-pharmaceutical interventions, ultimately improving community health and disease control efforts.
Highlights
By integrating clinical surveillance and wastewater monitoring data, dynamic baselines were established to detect real-time IAV concentration changes, enabling the early identification of epidemic risks.
IAV baselines in wastewater guide optimal timing for vaccination and nonpharmaceutical interventions, especially in developing counties.
The Random Forest model accurately predicts IAV cases using key physico-chemical parameters like IAV concentration, flow rate, wastewater temperature, chemical oxygen demand, total nitrogen and phosphorus.
KEYWORDS: Baseline, influenza A virus, Random Forest model, wastewater-based epidemiology, influenza-like-infection
Introduction
Respiratory infectious diseases, caused by various viruses including SARS-CoV-2, influenza virus, and respiratory syncytial virus (RSV), represent a significant global health burden, standing among the leading causes of morbidity and mortality worldwide [1]. The severity of this public health challenge is underscored by the World Health Organization (WHO), which in 2019 identified influenza as one of the top 10 global health threats [2]. According to estimates, influenza annually results in 3–5 million cases of severe illness and 290,000–650,000 respiratory disease-related deaths globally [3]. The influenza virus poses particular challenges to public health systems due to its frequent mutations, which enable it to evade seasonal vaccines and complicate population-level health response strategies [4]. In China, this health burden is particularly pronounced, with influenza causing an estimated annual average of 88,100–92,102 excess deaths from influenza-associated respiratory illness, accompanied by a substantial economic impact of approximately 226.38 billion CNY [5,6]. This significant epidemiological and economic impact highlights the critical need for effective outbreak monitoring and control strategies. At the community level, accurate understanding of outbreak dynamics (including timing, geographic spread, and case estimates) is essential for local governments to implement timely and targeted disease control measures, as well as to develop evidence-based policies responsive to disease severity [7].
The COVID-19 pandemic has reshaped the global epidemic rhythm of respiratory viruses, creating unprecedented challenges in disease prediction and management. Although traditional influenza surveillance systems, including sentinel hospital and outbreak monitoring, remain fundamental components of public health infrastructure, their inherent limitations have become more apparent during the pandemic [8]. These constraints primarily stem from reliance on symptomatic cases, variations in healthcare-seeking behavior, and limited population coverage [9], potentially underestimating community transmission, particularly among asymptomatic individuals and during early infection stages [10]. In response to these gaps, wastewater surveillance has emerged as a valuable complementary tool. Recent studies have shown that Influenza A virus (IAV) RNA in wastewater reliably reflects community-level transmission and can serve as an early warning system. Wastewater data have been used to estimate the onset, peak, and intensity of influenza outbreaks across local, regional, and national scales, often aligning well with traditional clinical indicators [7-10]. While earlier research focused on small geographical areas, more recent efforts have demonstrated the scalability of wastewater surveillance and its utility in informing timely public health responses. This evolving landscape highlights the need for integrated surveillance strategies to improve our understanding and management of respiratory virus dynamics.
In recent years, wastewater surveillance has been increasingly implemented in several countries for tracking drug abuse [11] and monitoring the infection levels of pathogens associated with intestinal and respiratory infectious diseases due to its advantages of wide population coverage, relatively low cost, and minimal social disturbance [12-16]. Wastewater-based epidemiology (WBE) is an efficient method for COVID-19 surveillance, offering relatively unbiased community level infection estimates at just 0.7–1% of the cost of the population-wide testing [17,18], and enabling early identification of new viral strains and outbreaks [19,20]. A significant correlation between influenza virus concentration in wastewater and the number of clinical cases has been reported in numerous countries [1,10,13-15,21]. Building on these successes, this study integrates clinical surveillance and wastewater monitoring data to establish dynamic baselines for detecting real-time changes in IAV concentration, facilitating early identification of epidemic risks and guiding the optimal timing for vaccination and non-pharmaceutical interventions, particularly in resource-limited settings.
While the effectiveness of multi-pathogen surveillance in wastewater has been demonstrated in numerous studies, its systematic application in public health practice remains underdeveloped. In this study, a random forest model was applied to estimate the number of infections by integrating viral concentrations and physico-chemical parameters in wastewater. By comparing the baseline values of multiple pathogens in wastewater with clinical surveillance data, this study explores the early warning feature of wastewater monitoring aiming to timely initiate appropriate disease control procedures. This approach lays the groundwork for establishing a systematic framework to utilize wastewater epidemiological data in public health practices. By focusing on IAV as a well-characterized virus with extensive clinical data, the study provides insights into the application of wastewater surveillance for future multi-pathogen monitoring efforts.
Materials and methods
Wastewater sample collection and pre-treatment
Wastewater surveillance was conducted once or twice per week at 38 wastewater treatment plants (WWTPs) across ten districts of Shenzhen, equating to approximately 18 million people (Figure S1) [22]. From March, 2023, to October, 2024, 24 h composite influent samples were obtained with 125 mL wastewater collected hourly at each sampling site. During sample collection, information such as the flow rate at the sampling points, chemical oxygen demand (COD), total nitrogen, total phosphorus, ammonia nitrogen, wastewater temperature, pH, and solids in suspension was recorded simultaneously. These physico-chemical parameters were monitored using automated online systems, with COD, total nitrogen, total phosphorus, and ammonia nitrogen measured every 2 h, whereas wastewater temperature, pH, and solids in suspension recorded in real time at high frequency. The 24-hour averages of these parameters were calculated for further analysis. Composite samples were concentrated using a modified “Polyethylene glycol (PEG) precipitation method” [10], whereby approximately 50 mL aliquots were centrifuged at 2000 × g and 4°C for 2 min. Then 40 mL aliquots of supernatant were moved to 50 mL centrifuge tubes with the addition of PEG (10%, w/v) and NaCl (2%, w/v) and shaken vigorously until PEG dissolved completely. The resulting samples were agitated at 180 rpm for 2 h in a FS-50B Constant Temperature Shaking Incubator (Faithful Instrument, Hebei, China). Samples were centrifuged at 4750 ×g and 4°C for 30 min using a L6-6KR Floor Low speed large capacity Refrigerated centrifuge (Hunan Kecheng Instrument Equipment Co., Ltd). The supernatant was discarded, and the pellet was used for RNA extraction on an automatic nucleic acid extraction platform HBNP-9601A (Hybribio, China) to extract nucleic acids with a final elution volume of 50 μL. Positive and negative controls were set up and processed simultaneously with the samples.
RT-qPCR assay
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) testing for viral RNA in the wastewater samples was conducted using the influenza A, B virus and respiratory syncytial virus (RSV) nucleic acid detection kit (Hybribio, Guangzhou), and the SLAN®-96S PCR instrument (Shanghai Hongshi Medical Technology Co., Ltd) following the reference protocol. Fluorescence intensity was measured in four detection channels: FAM (510 nm) for RSV, VIC (565 nm) for IAV, ROX (620 nm) for IBV, and CY5 (665 nm) for the B2M pseudovirus. The SLAN®-96S instrument supports multi-channel fluorescence detection and is factory-calibrated for standard dye sets, eliminating the need for manual spectral calibration by the user. A 25μL mixture was prepared for each well, consisting of 18.75 μL of PCR Mix, 1.25 μL of Enzyme Mix, and 5 μL of RNA templates. Thermal conditions for RT-qPCR were as follows: 55 °C for 5 min, then at 95 °C for 30 s, followed by 45 cycles of 95 °C for 5 s and 60 °C for 20 s.
Analytical evaluation and inhibition testing
To assess the sensitivity of the assay in real wastewater matrices, raw wastewater samples pre-confirmed to be negative for IAV, IBV, and RSV were spiked with known quantities of inactivated virus at six concentrations (0, 2.5, 5, 10, 100, and 1000 copies/mL). After spiking, samples were subjected to virus concentration using PEG precipitation, followed by RNA extraction. This workflow enabled simultaneous evaluation of virus recovery, nucleic acid extraction, reverse transcription and quantitative PCR amplification. The LOD was determined as the lowest concentration that yielded no more than one negative result in 20 replicates (positive rate ≥ 95%) [22]. Reproducibility was evaluated by testing each concentration in triplicate in three independent assays. The intra- and inter-assay coefficients of variation (CVs) were calculated using the Ct values obtained. The presence of RT-qPCR inhibitors in wastewater nucleic acid samples was evaluated based on protocols previously validated by our team [23]. In that study, infectious bronchitis virus RNA was spiked into wastewater nucleic acid extracts and compared to control reactions in distilled water. Inhibition was considered significant if the Ct value of the wastewater nucleic acid sample increased by more than two units compared to the reference Ct value [24,25].
Quantification of virus concentration in wastewater
The extracted nucleic acids of wastewater samples were tested for influenza RNA using the RT-qPCR method mentioned above. A standard curve was generated for each RT-qPCR run using six 10-fold serial dilutions of plasmid DNA (1.0×105–1.0×100 copies/reaction), which was artificially synthesized by Sangon Biotechnology (Shanghai, China). The samples, standards, positive and negative controls were detected in duplicate.
A one-step Pepper Mild Mottle Virus (PMMoV) RT-qPCR assay was performed as a qualitative internal indicator for the process control [26]. Primers and probes for the PMMoV assay can be found in Table S1. IAV concentration dataset was omitted as outliers when PMMoV virus was undetectable.
Calculation of virus concentration at the city-wide level
In this study, WWTPs were sampled to assess the virus concentration. By combining viral concentrations in wastewater with the flow rates of WWTPs from 10 districts, we determined the flowrate-weighted average viral load across Shenzhen at the city-wide level [22], using the following Eq. (1).
| (1) |
Where, C: flowrate-weighted average concentration of virus (copies/L); Cn: concentration of virus in wastewater at WWTP n (copies/L); Qn: the daily flow rate of WWTP n (L/day).
Collection of clinical surveillance datasets
Clinical surveillance data were obtained from three main sources: city-wide clinical IAV case numbers from the China Information System for Disease Control and Prevention [27], influenza clusters (defined as 5 or more cases in a single collective unit within a week) from the Shenzhen Communicable Disease Information System, and the detection rates (DR) and the proxy influenza-like illness rates (ILI) from 14 sentinel hospitals monitored by the Shenzhen Center for Disease Control and Prevention.
Statistical analysis
Paired T-test was used to evaluate the seasonal differences in IAV concentration in wastewater. Spearman's rank correlation was used to evaluate the correlation between IAV concentration in wastewater surveillance and influenza activity in clinical surveillance. The difference was considered significant when the P value was less than 0.05. To identify the onset and offset of IAV epidemics through wastewater surveillance, we calculated wastewater baseline values using three different approaches: (1) calculating the annual mean over of the period of analysis, (2) calculating the annual geometric mean over of the period of analysis [7], (3) calculating the annual median over of the period of analysis.
Predictive models were developed using the Random Forest algorithm, which is known for its intrinsic explainability, robustness to outliers, stability with new data, and proficiency in handling non-linear correlations [28-33]. The city-wide reported influenza case numbers from the China Information System for Disease Control and Prevention were used in the Random Forest model. Other data sources, such as influenza cluster events and sentinel surveillance data, were not included in the modeling to avoid potential duplication. In the case of IAV, the merged dataset was randomly split into 60% training (n = 32), 20% testing (n = 10), and 20% validation (n = 11) data subsets. Here, n refers to the number of epidemiological weeks included in each subset. Three different Random Forest models were run for IAV depending on data set selection: (A) RF1: only including IAV concentrations in 38 WWTPs in Shenzhen. (B) RF2: This model incorporates factors identified in the literature as having a significant influence on Random Forest predictions, including IAV concentrations, flow rate, wastewater temperature, COD, total nitrogen, and phosphorus. These variables were selected based on their established correlations with wastewater characteristics and viral persistence, making them critical for improving model accuracy and predictive performance [30]. (C) RF3: This model utilizes all available data, incorporating both IAV concentrations and a comprehensive set of physico-chemical parameters (IAV, flow rate, wastewater temperature, COD, total nitrogen, phosphorus, ammonia nitrogen, pH, and suspended solids). The inclusion of these parameters is designed to capture the full spectrum of factors that influence viral behavior in wastewater, thus enhancing the model's robustness and ability to provide accurate predictions. All the statistical analysis was conducted using Stata 17.0 (StataCorp).
Results
Quality assurance and control (QA/QC) of wastewater samples
The RT–;qPCR assay exhibited high performance characteristics for quantifying IAV, with an amplification efficiency of 93.11–97.44%. Additionally, the correlation coefficient (R2) was 0.9984–0.9993, indicating a strong linear relationship between the amount of target genes and the resulting fluorescence signal. The standard curves used in this study meet with Minimum Information for Publication of Quantitative Real-Time PCR Experiment (MIQE) guidelines [34] (Supporting Information Table S2). The wastewater samples spiked with inactivated virus (IAV, IBV, and RSV) gave reproducible readouts in 20 replicates when the concentration was equal to or higher than 10 copies/mL; therefore, the LOD under the full analytical workflow (including concentration, RNA extraction, and RT-qPCR) was determined to be 10 copies/mL. For reproducibility, both intra- and inter-assay CVs of Ct values across all targets (IAV, IBV, RSV) and concentrations were below 5%, demonstrating robust performance of the assay in the complex wastewater matrix. Full results, including Ct values, SD, and CV% values, are provided in Supplementary Table S3.
The elevated levels of PMMoV in all samples indicate that there were no significant extraction failures (median = 1.24 × 105 copies/mL, min = 6.79 × 104 copies/mL, max = 2.31 × 105 copies/mL, IQR = 9.84 × 104–1.60 × 105 copies/mL). As demonstrated in our previous study [23], RT-qPCR inhibition was not detected in any of the wastewater samples tested (0/10) using the infectious bronchitis virus assay, validating the suitability of the extraction method applied here. Moreover, he specificity of the assay was further supported by our previous city-wide wastewater surveillance project, in which wastewater samples were simultaneously tested for SARS-CoV-2, norovirus, rotavirus, and enteroviruses [22,23,35].
Reports and detection of IAV in Shenzhen city
From March 3, 2023, to March 3, 2024, a total of 604,257 clinical influenza A cases and 1,086 influenza-associated clusters were reported in Shenzhen, China. Of these, 47.5% of influenza-associated clusters (516 of 1,086) occurred in the spring. Additionally, total of 14,082 respiratory specimens from 14 sentinel hospitals were collected, of which 4,021 tested positive for IAV. Notably, 34.9% of the positive cases (1,403 of 4,021) were occurred in the spring.
Meanwhile, 2,764 samples of untreated wastewater were collected from 38 WWTPs in Shenzhen city, and the detection rate of IAV was 81.2% (2,245 of 2,764) by RT-qPCR. The concentration of IAV in Shenzhen's wastewater ranged from 7.96 × 102 copies/L to 3.12 × 105 copies/L, reaching two peaks, respectively, in mid-to-late April and early December, 2023, and declining to lower levels in early June and mid-September, 2023 (Figure 1(a)). Significant seasonal patterns in IAV concentration were observed, with the highest levels recorded in spring (March, April, and May), followed by winter (December, January, and February), summer (June, July, and August) and autumn (September, October, and November) (Figure 1(b)). Seasonal patterns of IAV concentrations in wastewater aligned with clinical data, where 47.5% of influenza-associated clusters occurred in the spring. From a spatial distribution perspective, the strongest correlation in IAV concentrations in wastewater was observed between Baoan and Longhua District (r = 0.78, P < 0.001). Notably, these two districts were geographically adjacent (Figure 1(c) and (d)). The flowrate-weighted average IAV concentration in wastewater in Shenzhen significantly correlated with both clinical detection rates of IAV in respiratory specimens (Spearman's r = 0.54, 95% CI: 0.31–0.78, degrees of freedom = 51, P < 0.001) and reported clinical influenza A cases (Spearman's r = 0.56, 95% CI: 0.34–0.78, degrees of freedom = 51, P < 0.001) (Figure 1(e) and (f)).
Figure 1.
Influenza A virus (IAV) concentration in wastewater and spatial distribution of clinical influenza clusters in Shenzhen from March 3, 2023 to March 3, 2024. (a) IAV concentration in wastewater in ten districts of Shenzhen. Missing lines indicate that data was not available for calculation. (b) Seasonal patterns of IAV concentration in wastewater at a city-wide level in Shenzhen. (c) Spearman correlation of IAV concentration in wastewater in ten districts of Shenzhen. (d) Spatial distribution of clinical influenza clusters in ten districts of Shenzhen. (e) Correlation between IAV concentration in wastewater and detection rates of IAV in respiratory specimens. (f) Correlation between IAV concentration in wastewater and clinical influenza A cases. The vertical black dotted lines represent the dividing lines between spring, summer, autumn and winter.
Mathematical approximation for IAV cases prediction based on IAV quantification data in wastewater
The Random Forest model with the lowest mean absolute error (2,307 clinical cases) and the highest validation R2 value of 0.988, incorporated a comprehensive set of predictors: IAV concentrations, flow rate, wastewater temperature, COD, total nitrogen, and phosphorus. This model demonstrated superior performance compared to the other two models evaluated. In contrast, the model that relied solely on IAV concentrations showed a higher validation MAE of 3,181, despite achieving an almost perfect validation R2 of 0.999. This suggests that while the model using only IAV concentrations had a very high R2, indicating a near-perfect fit to the validation data, its MAE was higher, reflecting less accurate predictions of actual clinical cases. Furthermore, the third model, which included additional predictors beyond those in RF2, had a validation MAE of 2,472 and an R2 of 0.990. Although it performed well, it did not surpass the performance of RF2 in terms of validation MAE and R2. Overall, RF2′s integration of multiple relevant environmental variables resulted in the most balanced and accurate predictions across training, testing, and validation phases, highlighting the importance of including a diverse set of predictors for improved model performance in estimating IAV concentrations (Figure 2 and Table 1).
Figure 2.
Random Forest's predictions on training and test data for IAV. (a) RF1: Model using only IAV concentrations from 38 WWTPs in Shenzhen. (b) RF2: Model incorporating IAV concentrations and key physico-chemical parameters identified in the literature (flow rate, temperature, COD, total nitrogen, and phosphorus). (c) RF3: Comprehensive model using all available parameters (IAV concentrations, flow rate, temperature, COD, total nitrogen, phosphorus, ammonia nitrogen, pH, and suspended solids).
Table. 1.
Mean absolute error and its percentage over the mean of clinical cases.
| Training | Test | Validation | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MAE | Training %OM | R square | MAE | Test %OM | R square | MAE | Validation %OM | R square | |
| RF1 | 2,793 | 0.263 | 0.872 | 4,053 | 0.308 | 0.657 | 3,181 | 0.259 | 0.999 |
| RF2 | 2,656 | 0.250 | 0.916 | 3,763 | 0.286 | 0.502 | 2,307 | 0.188 | 0.988 |
| RF3 | 2,771 | 0.261 | 0.897 | 3,672 | 0.279 | 0.498 | 2,472 | 0.201 | 0.990 |
Notes: RF1: Model using only IAV concentrations from 38 WWTPs in Shenzhen. RF2: Model incorporating IAV concentrations and key physico-chemical parameters identified in the literature (flow rate, temperature, COD, total nitrogen, and phosphorus). RF3: Comprehensive model using all available parameters (IAV concentrations, flow rate, temperature, COD, total nitrogen, phosphorus, ammonia nitrogen, pH, and suspended solids).
IAV concentration baseline values in wastewater
The onset, offset, and duration of the 2023–2024 influenza A season determined using the ILI baseline and wastewater baseline are compared in Figure 3(a) and Table 2. For a wastewater baseline that aligns with the expected arise in ILI, the geometric mean is the most effective measure, as compared with the mean (5.57 × 103 copies/L). The lowest IAV concentration baseline values in wastewater in Shenzhen was that defined as the median (4.50 × 103 copies/L) whilst the highest baseline was defined as the mean (1.45 × 104 copies/L).
Figure 3.
(a) Comparison of IAV concentration baseline values (y-axis) and corresponding influenza season onset, offset, and duration for 2023−2024 influenza season (x-axis). The green shaded area indicates the period when ILI duration and influenza incidence (>100 cases per 100,000 population) overlapped. (b) Geographical distribution of IAV onset dates in ten districts of Shenzhen. (c) Cumulative fraction of district in ten districts of Shenzhen. (d) Time trend of clinical influenza clusters in ten districts of Shenzhen.
Table. 2.
The onset, offset, and duration of multi-pathogen based on calculation using annual mean, geometric mean, or median of virus concentration in wastewater.
| Baseline | Baseline value of concentration in wastewater | Onset/Offset (duration in days) | Peak date (Maximum concentration in wastewater) | |
|---|---|---|---|---|
| Clinical surveillance | Influenza cases > 50 per 100,000 | – | 3/13/2023–5/7/2023 (55) | – |
| Influenza cases > 100 per 100,000 | – | 3/20/2023–4/30/2023 (41) | – | |
| ILI duration (IAV) | – | 3/20/2023–7/9/2023 (111) | – | |
| ILI duration (IBV) | – | 10/30/2023–6/30/2024 (244) | – | |
| ILI duration (RSV) | – | 1/22/2024–7/24/2024 (184) | – | |
| IAV concentration in wastewater | IAV mean | 1.45 × 104 copies/L | 3/27/2023–4/30/2023 (35) | 4/21/2023 (1.39 × 105 copies/L) |
| IAV geometric mean | 5.57 × 103 copies/L | 3/27/2023–5/14/2023 (49) | ||
| IAV median | 4.50 × 103 copies/L | 3/6/2023–5/14/2023 (69) | ||
| IBV concentration in wastewater | IBV mean | 1.44 × 103 copies/L | 1/1/2024–4/21/2024 (111) | 2/25/2024 (8.34 × 103 copies/L) |
| IBV geometric mean | 5.87 × 102 copies/L | 11/27/2023–6/9/2024 (195) | ||
| IBV median | 6.14 × 102 copies/L | 11/27/2023–6/9/2024 (195) | ||
| RSV concentration in wastewater | RSV mean | 6.62 × 102 copies/L | 1/28/2024–5/19/2024 (112) | 2/25/2024 (2.03 × 103 copies/L) |
| RSV geometric mean | 4.85 × 102 copies/L | 1/28/2024–5/19/2024 (112) | ||
| RSV median | 4.72 × 102 copies/L | 1/28/2024–5/19/2024 (112) |
Notes: Onset/Offset: day when measure was above/below the baseline value and remained there for at least two additional weeks. Duration in days: the number of days between onset and offset. ILI: influenza-like illness rates, that is the total ILI consultation rate from sentinel hospital. ILI baseline values were calculated from the weekly averages of ILI.
When comparing onset determined using ILI and wastewater data, the date of onset differed by less than 7 days. In comparison, the onset determined by the annual mean was the latest among all approaches. Results showed that, compared to the onset of the influenza season defined as more than 100 influenza cases per 100,000 population in Shenzhen, the baseline defined as the median detected the onset 2–4 weeks earlier (exceeded the baseline value for two consecutive weeks). Specifically, on March 13, 2023, the clinical monitoring of influenza cases per 100,000 people began to exceed 50 for two consecutive weeks. On March 20, 2023, the clinical monitoring of influenza cases per 100,000 people started to exceed 100 for two consecutive weeks. However, as early as March 6, 2023, the concentration of influenza A virus in wastewater had already exceeded the baseline value defined by the median for two consecutive weeks. The baseline concentrations of IAV in wastewater for each district are shown in Figure S2. We found that onset dates vary in each district, as shown in Figure 3(b). Overall, exceeding the baseline concentration level may indicate a risk of outbreak. The wastewater in Luohu District, Pingshan District, and Dapeng District exhibited the earliest detection of IAV (March 3, 2023), despite these regions reporting relatively fewer outbreak cases, which may lead to potential oversight by authorities due to insufficient case numbers (Figure 3(c) and (d)).
Baseline validation for multi-pathogen monitoring in wastewater
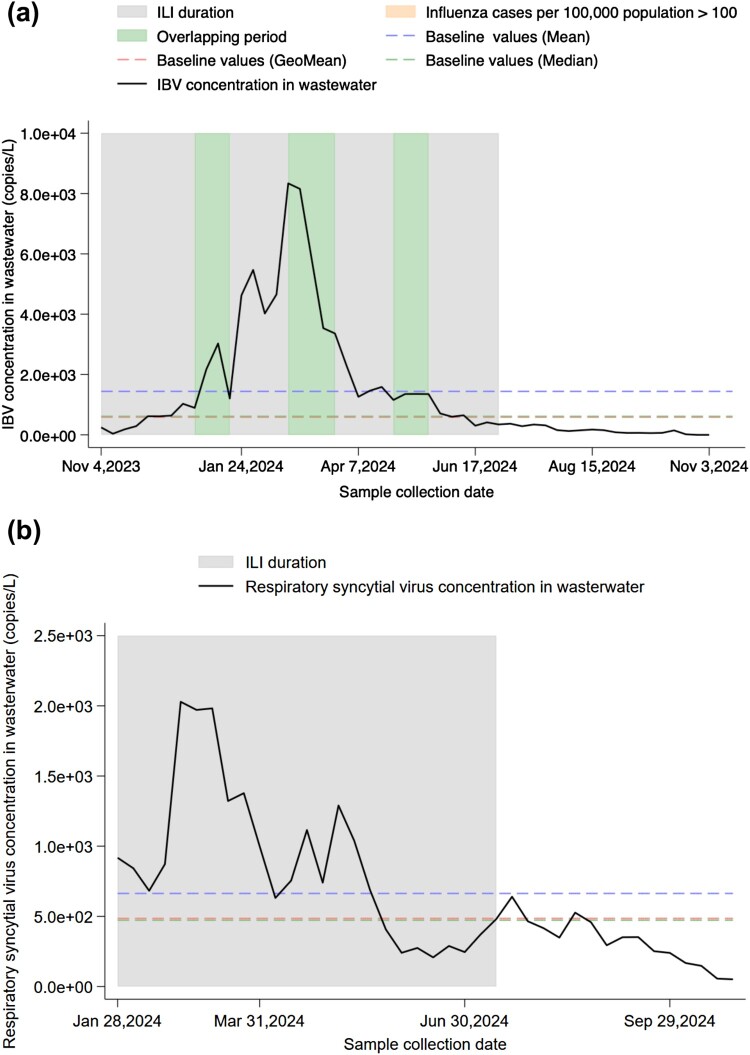
In order to verify the role of the defined concentration baseline for early warning, we established baselines for IBV and RSV in wastewater (Figure 4 and Table 2). Compared to the highest concentration of IAV (1.39 × 105 copies/L), the highest concentration of IBV (8.34 × 103 copies/L) was reached on February 25, 2024. On December 25, 2023, the clinical monitoring of influenza cases per 100,000 people started to exceed 100 for two consecutive weeks. As early as November 27, 2023, the IBV concentration in wastewater had already exceeded the baseline value defined by the median for two consecutive weeks. By comparing the different definitions of baseline concentration levels, we found that the baseline defined by the mean in this study was the highest among all viruses. The average concentration of IBV and RSV was 1.44 × 103 copies/L, and 6.63 × 102 copies/L, respectively. The baseline values defined by the geometric mean and the median were similar (Figure 4(a) and (b)). The baseline values for IBV defined by the geometric mean and the median were 5.87 × 102 copies/L, and 6.14 × 102 copies/L, respectively, while those for RSV were 4.85 × 102 copies/L, and 4.72 × 102 copies/L, respectively.
Figure 4.
Validation of baseline values in early warning for multi-pathogen monitoring in wastewater. (a) IBV concentration baseline values in wastewater in Shenzhen from November, 2023 to October, 2024. The green shaded area indicates the period when ILI duration and influenza incidence (>100 cases per 100,000 population) overlapped. (b) Respiratory syncytial virus concentration baseline values in wastewater in Shenzhen from January, 2024 to October, 2024.
Discussion
This study explores the use of baseline values of influenza A virus (IAV) in wastewater, integrating viral concentrations with physico-chemical parameters, to develop early warning for infection estimation and inform timely public health interventions. Since the outbreak of the COVID-19 pandemic, the transmission patterns of respiratory viruses worldwide have changed, and the epidemic rhythm has become unpredictable. In response, we have integrated clinical surveillance and wastewater monitoring datasets to establish virus concentration baselines using various methods. By dynamically assessing these baseline values, we can capture real-time changes and identify potential epidemic risks in advance. This approach is particularly valuable in low- and middle-income countries, where the lack of sufficient clinical surveillance resources and public health infrastructure. Through dynamic spatiotemporal baselines and multi-source data fusion, we are able to more accurately capture abnormal fluctuations in virus transmission, providing a scientific basis for epidemic warning and control decisions. In this study, the baseline method not only upgrades technical tools but also serves as a strategic necessity in addressing global public health challenges.
By integrating multiple datasets from various surveillance tools, such as wastewater surveillance, clinical case reporting system, sentinel hospital surveillance, and influenza cluster monitoring, policymakers can access more comprehensive information to develop targeted interventions at different times. Our study revealed that the peak of IAV concentration in wastewater in Shenzhen occurred primarily in mid-to-late April, which aligned with influenza clusters. In terms of seasonal distribution, IAV showed a moderate peak in winter and a much more pronounced peak in spring, consistent with reports from Spain and the USA, as recently reported [12,30]. The seasonal pattern of IAV concentration in wastewater aligned with the typical IAV outbreak season, further validating the effectiveness of wastewater surveillance in capturing population-level IAV dynamics. Although densely populated provinces might be expected to report more influenza clusters, differences in healthcare accessibility, early case detection, and local reporting practices may lead to fewer clusters being recorded in these areas. These findings highlight the importance of combining diverse surveillance tools to guide public health decision-making, rather than relying on a single data source, which may provide a limited view of the epidemic landscape.
Moreover, the results of seasonal and regional differences in IAV concentrations in wastewater can provide valuable insights for the development of targeted health interventions and policies. For instance, it suggests bolstering influenza awareness and education efforts during the spring and winter months, alongside the implementation of preventive measures like mask-wearing and vaccination for vulnerable populations [10]. Data collected over multiple IAV seasons can be used to refine the approaches introduced in this study to better capture the variation in IAV concentration in wastewater across seasons. By comparing multiple surveillance datasets over one year, our results highlight that wastewater surveillance provides accurate, near real-time information and effectively captures IAV distribution in the population. In this study, IAV concentrations in wastewater were significantly correlated with clinical detection rates and reported clinical cases number of IAV. This finding is consistent with previous studies showing that IAV wastewater surveillance effectively tracked IAV infection trends in the sampled communities in Australia, the United States, Canada, Spain [10,13-15,21,36]. While existing literature primarily focuses on the early detection capabilities of wastewater surveillance and clinical signals within specific timeframes [10,14,15], which may be arbitrary due to variations in sample size and detection timing. Our findings emphasize the importance of integrating WBE into existing influenza surveillance frameworks to improve the timeliness and efficiency of influenza control measures. Governments can utilize wastewater data to anticipate healthcare burdens, optimize resource allocation, and implement timely public health interventions, particularly in areas where traditional surveillance is delayed or less accessible.
The Random Forest algorithm can be used to estimate the number of clinical cases [30,31], and the performance of the model improves when combined with the physicochemical parameters of the wastewater (Table S4). The results of this study show that water inflow rate, wastewater temperature, COD, total nitrogen and phosphorus provide relatively robust results for the model, which is consistent with findings from model studies conducted in Spain [30]. These environmental variables influence viral detection and persistence in wastewater, affecting the accuracy of epidemiological inferences derived from wastewater surveillance. For instance, wastewater temperature and organic matter content can impact viral stability, while flow rate fluctuations may dilute or concentrate viral signals, thereby influencing model predictions. This integrative approach provides a more comprehensive representation of complex real-world conditions, enhancing the model's predictive accuracy. In addition, internal normalization markers may help improve model robustness. This study used PMMoV as a qualitative indicator to assess sample quality and exclude outliers. While originally developed under Western dietary contexts, PMMoV has also been detected in Chinese wastewater, suggesting potential applicability [37,38]. Future studies should evaluate suitable markers (e.g. crAssphage, PMMoV, Bacteroides rRNA, human 18S rRNA) for quantitative normalization to improve model performance [26,39]. However, some environmental and operational confounders remain unaddressed. While water inflow rate was included in the model, it may not fully capture dilution effects from rainfall [40,41]. For instance, the abrupt decline in viral concentrations observed in mid-September 2023 coincided with Typhoon Haikui, which brought record-breaking rainfall to Shenzhen and likely led to substantial dilution and potential overflow in the sewage system. Without specific precipitation data, the impact of rainfall on viral signal variability remains uncertain. In addition, more complex hydraulic dynamics – such as transport time, sedimentation, and resuspension – were not explicitly modeled, though they may influence viral decay and dispersion [42-44]. Limited sampling frequency may also reduce temporal resolution and lead to missed short-term fluctuations in viral shedding. Future research should integrate rainfall data, detailed sewer system hydraulics, and higher-frequency sampling to improve surveillance reliability.
Quantitatively, the RF model exhibits a lower MAE across training, testing, and validation datasets (e.g. an MAE of 2,307 in the validation set), as well as higher R2 values (e.g. an R2 of 0.988 in the validation set), indicating strong fitting capability and predictive performance across all datasets. This demonstrates the value of wastewater surveillance and supports its potential application for monitoring additional viruses. Future studies should include a sensitivity analysis to quantify the contribution of each environmental variable to model performance, which would help refine the selection of key predictive features. Additionally, incorporating long-term wastewater data across multiple seasons would improve the model's robustness and generalizability. Improving viral concentration prediction models will contribute to the development of a comprehensive epidemiological surveillance system in Shenzhen and support population health decision-making. It is worth noting that wastewater samples are inherently complex. Although prior validation showed no detectable inhibition under our experimental conditions [23], the presence of residual inhibitors or contaminants in wastewater may still pose challenges. These potential matrix effects should be considered when interpreting the results.
The established baseline values for IAV concentrations in wastewater indicate crucial timings for population-levels vaccination and nonpharmaceutical interventions, likely in early March and mid-November. Compared with the baseline values of IAV concentration in wastewater in some states of the United States, onset times were also in mid-November [7]. This study explored the effectiveness of using different baseline indicators to assess the early warning potential of IAV concentrations in wastewater exceeding baseline levels for influenza outbreaks. Compared to the ILI-estimated onset, during the first wave of influenza epidemic, the onset using the baseline corresponding to median was up to 14 days earlier than ILI-estimated onset. Notably, this study found that the distribution of IAV concentrations in wastewater does not conform to a normal or log-normal distribution. Therefore, a baseline was defined based on the median value, as it is less sensitive to outliers [45]. The baseline defined by the median exceeded in wastewater IAV concentrations 2–4 weeks earlier compared to the influenza season defined as more than 100 influenza cases per 100,000 population in Shenzhen, suggesting that the median baseline offers a more advanced early warning capability. Moreover, by understanding the baseline levels of IAV concentration in wastewater across different districts, the onset time for each district can be determined, enabling targeted public health interventions.
Comparing the results of wastewater monitoring and clinical surveillance, the peak timing of the viruses monitored in this study occurred earlier than clinical surveillance or overlapped with the duration of clinical surveillance (gray-shaded areas in Figure 4). This suggests that wastewater surveillance can provide an earlier and more comprehensive signal of viral activity, particularly in regions like Luohu, Pingshan, and Dapeng, where early date of IAV onset in wastewater was observed despite fewer outbreak cases. This suggests that we may miss early virus transmission in low – and middle-income areas with low population density or limited access to health care. Wastewater monitoring, therefore, serves as a critical early warning tool, enabling timely interventions in middle- and low-income regions where healthcare resources are often constrained. Although wastewater surveillance offers a cost-effective alternative to clinical surveillance [17,18], scaling and implementation still face practical challenges. Compared to case-based clinical reporting, wastewater monitoring allows for population-level surveillance without individual testing, reducing the burden on healthcare infrastructure [10]. However, real-world implementation may be limited by gaps in laboratory capacity, analytical expertise, and standardized sampling protocols. Additional barriers include the integration of wastewater data into existing public health systems, and the lack of established policy frameworks to trigger timely interventions [46]. To ensure sustainability, future efforts should invest in workforce training, data infrastructure, and cross-sector coordination [47].
To further validate the effectiveness of early warning based on baseline concentration levels in wastewater, we compared the baseline levels of IBV and RSV. This study found that defining the baseline using the median may identify the optimal timing for public health interventions. When viral concentrations exceed the median for two consecutive weeks, authorities can implement targeted measures, such as online health education to guide public precautions, vaccination campaigns, or non-pharmaceutical interventions like mask-wearing. While using the median as a baseline value provides robustness against outliers and offers earlier warnings, it may also increase the risk of false-positive signals (i.e. baseline exceedance not followed by clinical outbreaks), especially in low-incidence regions or during off-peak seasons. Therefore, public health agencies need to weigh the trade-off between sensitivity and specificity based on available resources and response priorities. In high-risk areas or during vulnerable periods (e.g. early March and mid-November as observed in this study), higher sensitivity may be preferred for early preparedness. Conversely, in low-incidence periods, a more specific threshold might be desirable to avoid unnecessary alerts or public fatigue. As such, baseline definitions should be adapted and validated according to local epidemiological and operational contexts. This study provides preliminary evidence for the utility of the median threshold and highlights the need for further prospective validation studies to determine the most effective and actionable thresholds. Future research should explore additional baseline methods, such as utilizing multiple viruses in wastewater across different regions, to establish thresholds for high, medium, and low infection risk. This would enhance the precision of early warning systems and inform health policy decisions, ensuring a more proactive and equitable public health response.
Taken together, our study demonstrates the feasibility and effectiveness of using wastewater surveillance to monitor influenza activity and to estimate infection dynamics in the population. These findings support the integration of WBE into existing influenza surveillance frameworks for early warning and timely public health response. Looking ahead, the growing body of international WBE research provides a valuable context for situating our findings. For example, collaborative interlaboratory studies during the COVID-19 pandemic helped to standardize methods for quantifying viral RNA in wastewater and validating RT-qPCR assays, offering best practices that remain relevant today [48]. Global genomic surveillance based on wastewater samples has also proven effective in tracking viral variants [49], while innovative approaches such as aircraft-based wastewater monitoring highlight the scalability and adaptability of WBE across regions [50]. Incorporating these insights into future WBE research could further enhance the precision, timeliness, and equity of public health responses, particularly in low-resource settings. Our study adds to this evolving field by illustrating the value of IAV wastewater surveillance as a real-time epidemiological tool with significant public health utility.
Our study has some limitations. Firstly, certain quantitative values related to influenza virus monitoring based on wastewater require further investigation and determination. Key parameters, such as detoxification amount, virus decay rate, and recovery rate, are anticipated to enhance the model's accuracy in predicting human infections. Among these, the viral shedding rates plays a particularly important role, as it may vary considerably between individuals depending on factors such as age and symptomatic status. Incorporating clinical data to establish stratified shedding models or correction factors could further enhance prediction robustness. In addition, future studies may employ subtype-specific assays or sequencing-based approaches to improve the accuracy of viral source attribution, to exclude interference from non-human viral sources, and to enhance the epidemiological interpretation of surveillance data. Moreover, the decision-making process of each tree in a Random Forest is complex, making it challenging to ascertain each variable's specific contribution to the final prediction. Future research could explore various prediction models and algorithms to improve both accuracy and interpretability. This may include deep learning models or hybrid approaches to better understand the relationships between variables. Additionally, by investigating and comparing various methodologies for establishing baselines and thresholds, researchers can enhance the accuracy of influenza risk predictions and develop more effective health management strategies, such as targeted vaccination programs and timely public health interventions. However, it is worth noting that wastewater surveillance does not include individuals not connected to municipal sewer systems, such as those who are unhoused or use septic tanks or well water, which may lead to underrepresentation of certain populations. Future research should aim to address this gap to ensure more inclusive and representative surveillance coverage.
Conclusion
The concentration of IAV in wastewater from Shenzhen, shows a significant correlation with clinical surveillance proxies of influenza activity, illustrating its importance in public health monitoring. Additionally, the application of a Random Forest algorithm to analyze IAV concentrations and physico-chemical parameters in wastewater data effectively reveals realistic clinical trends of IAV cases within the community. For the IAV and IBV, the baseline defined as the median was exceeded 2–4 weeks earlier than the influenza season onset (≥100 cases/100,000) in Shenzhen, enabling earlier warning. Establishing baseline concentration levels in wastewater, such as the median, provides a practical early warning system for detecting abnormal increases in the concentrations of multiple viruses, enabling timely public health interventions, particularly for those lacking widespread clinical surveillance.
Supplementary Material
Funding Statement
This work was supported by the Shenzhen Science and Technology Program (No. KCXFZ20230731093959008), the Sanming Project of Medicine in Shenzhen Municipality (No. SZSM202311015), the Shenzhen Medical Key Discipline (Public Health Key Specialty) (No. SZXK064), and the Shenzhen Key Laboratory of Major Infectious Disease Surveillance (No. CXB201111240109A).
Author contributions
Xiuyuan Shi: Methodology, Formal analysis, Writing – original draft, Writing – review & editing. Shisong Fang: Resources, Methodology, Data curation. Chen Du: Methodology, Writing – review & editing, Data curation. Guixian Luo: Data curation, Formal analysis. Yanpeng Cheng: Resources, Methodology. Zhen Zhang: Resources, Methodology. Qiuying Lv: Methodology. Xin Wang: Methodology. Zhigao Chen: Methodology. Bincai Wei: Writing – review & editing. Ziqi Wu: Writing – review & editing. Bingchan Guo: Methodology. Panpan Yang: Methodology. Miaomiao Luo: Investigation. Weihua Wu: Investigation. Liping Zhou: Writing-review & editing. Ting Huang: Writing-review & editing. Xuan Zou: Writing-review & editing. Xiaolu Shi: Writing-review & editing. Songzhe Fu: Writing-review & editing. Zhanwei Du: Writing-review & editing. Xinxin Han: Conceptualization, Supervision, Writing-review & editing. Yinghui Li: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing-review & editing. Qinghua Hu: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing-review & editing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethical approval
The research protocol was reviewed and approved by the ethics committees of the Shenzhen Center for Disease Prevention and Control, Shenzhen CDC (SZCDC-IRB2024051).
Data availability statement
Data will be made available on request.
Supplemental Material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/22221751.2025.2552724.
References
- 1.Boehm AB, Hughes B, Duong D, et al. Wastewater concentrations of human influenza, metapneumovirus, parainfluenza, respiratory syncytial virus, rhinovirus, and seasonal coronavirus nucleic-acids during the COVID-19 pandemic: a surveillance study. Lancet Microbe. 2023;4(5):e340–e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization W . Ten threats to global health in 2019; 2022.
- 3.Iuliano AD, Roguski KM, Chang HH, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391(10127):1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han AX, de Jong SPJ, Russell CA.. Co-evolution of immunity and seasonal influenza viruses. Nat Rev Microbiol. 2023;21(12):805–817. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Liu Y, Wu P, et al. Influenza-associated excess respiratory mortality in China, 2010–15: a population-based study. Lancet Public Health. 2019;4(9):e473–e481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong H, Shen X, Yan H, et al. Estimating the disease burden of seasonal influenza in China, 2006-2019. Zhonghua Yi Xue Za Zhi. 2021;101(8):560–567. [DOI] [PubMed] [Google Scholar]
- 7.Schoen ME, Bidwell AL, Wolfe MK, et al. United States influenza 2022–2023 season characteristics as inferred from wastewater solids, influenza hospitalization, and syndromic data. Environ Sci Technol. 2023;57(49):20542–20550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehm AB, Wolfe MK, White BJ, et al. More than a tripledemic: influenza A virus, respiratory syncytial virus, SARS-CoV-2, and human metapneumovirus in wastewater during winter 2022–2023. Environ Sci Technol Lett. 2023;10(8):622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polo D, Quintela-Baluja M, Corbishley A, et al. Making waves: wastewater-based epidemiology for COVID-19–approaches and challenges for surveillance and prediction. Water Res. 2020;186:116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Zhao K, Xu X, et al. Development and application of influenza virus wastewater surveillance in Hong Kong. Water Res. 2023;245:120594. [DOI] [PubMed] [Google Scholar]
- 11.Löve ASC, Ásgrímsson V, Ólafsdóttir K.. Illicit drug use in Reykjavik by wastewater-based epidemiology. Sci Total Environ. 2022;803:149795. [DOI] [PubMed] [Google Scholar]
- 12.Wolfe MK, Duong D, Bakker KM, et al. Wastewater-based detection of two influenza outbreaks. Environ Sci Technol Lett. 2022;9(8):687–692. [Google Scholar]
- 13.Wolken M, Sun T, McCall C, et al. Wastewater surveillance of SARS-CoV-2 and influenza in preK-12 schools shows school, community, and citywide infections. Water Res. 2023;231:119648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercier E, D'Aoust PM, Thakali O, et al. Municipal and neighbourhood level wastewater surveillance and subtyping of an influenza virus outbreak. Sci Rep. 2022;12(1):15777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed W, Bivins A, Stephens M, et al. Occurrence of multiple respiratory viruses in wastewater in Queensland, Australia: potential for community disease surveillance. Sci Total Environ. 2023;864:161023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chai X, Liu S, Liu C, et al. Surveillance of SARS-CoV-2 in wastewater by quantitative PCR and digital PCR: a case study in Shijiazhuang city, Hebei province, China. Emerg Microbes Infect. 2024;13(1):2324502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Liu H, Gao L, et al. Wastewater-based epidemiology predicts COVID-19-induced weekly new hospital admissions in over 150 USA counties. Nat Commun. 2023;14(1):4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang G, Wu J, Weidhaas J, et al. Artificial neural network-based estimation of COVID-19 case numbers and effective reproduction rate using wastewater-based epidemiology. Water Res. 2022;218:118451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morvan M, Jacomo AL, Souque C, et al. An analysis of 45 large-scale wastewater sites in England to estimate SARS-CoV-2 community prevalence. Nat Commun. 2022;13(1):4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng Y, Zheng X, Xu X, et al. Use of sewage surveillance for COVID-19: a large-scale evidence-based program in Hong Kong. Environ Health Perspect. 2022;130(5):057008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toribio-Avedillo D, Gomez-Gomez C, Sala-Comorera L, et al. Monitoring influenza and respiratory syncytial virus in wastewater. Beyond COVID-19. Sci Total Environ. 2023;892:164495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du C, Peng Y, Lyu Z, et al. Early detection of the emerging SARS-CoV-2 BA. 2.86 lineage through wastewater surveillance using a mediator probe PCR assay—Shenzhen City, Guangdong Province, China, 2023. China CDC Weekly. 2024;6(15):332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Du C, Lv Z, et al. Rapid and extensive SARS-CoV-2 Omicron variant infection wave revealed by wastewater surveillance in Shenzhen following the lifting of a strict COVID-19 strategy. Sci Total Environ. 2024;949:175235. [DOI] [PubMed] [Google Scholar]
- 24.Fu S, He F, Wang R, et al. Development of quantitative wastewater surveillance models facilitated the precise epidemic management of COVID-19. Sci Total Environ. 2023;857:159357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staley C, Gordon KV, Schoen ME, et al. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl Environ Microbiol. 2012;78(20):7317–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenwald HD, Kennedy LC, Hinkle A, et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Res. 2021;12:100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Wang Y, Yang G, et al. China information system for disease control and prevention (CISDCP). HIT Brief Book. 2013;101–108. [Google Scholar]
- 28.Marin-Ramirez A, Mahoney T, Smith T, et al. Predicting wastewater treatment plant influent in mixed, separate, and combined sewers using nearby surface water discharge for better wastewater-based epidemiology sampling design. Sci Total Environ. 2024;906:167375. [DOI] [PubMed] [Google Scholar]
- 29.Singh KS, Paul D, Gupta A, et al. Indian sewage microbiome has unique community characteristics and potential for population-level disease predictions. Sci Total Environ. 2023;858:160178. [DOI] [PubMed] [Google Scholar]
- 30.Girón-Guzmán I, Cuevas-Ferrando E, Barranquero R, et al. Urban wastewater-based epidemiology for multi-viral pathogen surveillance in the Valencian region, Spain. Water Res. 2024;255:121463. [DOI] [PubMed] [Google Scholar]
- 31.Senaratna KYK, Bhatia S, Giek GS, et al. Estimating COVID-19 cases on a university campus based on Wastewater Surveillance using machine learning regression models. Sci Total Environ. 2024;906:167709. [DOI] [PubMed] [Google Scholar]
- 32.Koureas M, Amoutzias GD, Vontas A, et al. Wastewater monitoring as a supplementary surveillance tool for capturing SARS-COV-2 community spread. A case study in two Greek municipalities. Environ Res. 2021;200:111749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liaw A, Wiener M.. Classification and regression by random forest. 2002;2(3):18–22. [Google Scholar]
- 34.Bivins A, Kaya D, Bibby K, et al. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021;203:117516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue Z, Shi X, Zhang H, et al. The viral trends and genotype diversity of norovirus in the wastewater of Shenzhen, China. Sci Total Environ. 2024;950:174884. [DOI] [PubMed] [Google Scholar]
- 36.Vo V, Harrington A, Chang CL, et al. Identification and genome sequencing of an influenza H3N2 variant in wastewater from elementary schools during a surge of influenza A cases in Las Vegas, Nevada. Sci Total Environ. 2023;872:162058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng X, Li S, Deng Y, et al. Quantification of SARS-CoV-2 RNA in wastewater treatment plants mirrors the pandemic trend in Hong Kong. Sci Total Environ. 2022;844:157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Wang JCF.. Pepper mild mottle virus as an indicator of fecal contamination in water: a review on research progress. Chin J Public Health. 2020;36(2):265–269. [Google Scholar]
- 39.Zheng X, Zhao K, Xue B, et al. Tracking diarrhea viruses and mpox virus using the wastewater surveillance network in Hong Kong. Water Res. 2024;255:121513. [DOI] [PubMed] [Google Scholar]
- 40.Guo Y, Li J, O'Brien J, et al. Back-estimation of norovirus infections through wastewater-based epidemiology: a systematic review and parameter sensitivity. Water Res. 2022;219:118610. [DOI] [PubMed] [Google Scholar]
- 41.Weidhaas J, Aanderud ZT, Roper DK, et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci Total Environ. 2021;775:145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zehnder C, Béen F, Vojinovic Z, et al. Machine learning for detecting virus infection hotspots via wastewater-based epidemiology: the case of SARS-CoV-2 RNA. GeoHealth. 2023;7(10):e2023GH000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu S, Zhang Y, Wang R, et al. Longitudinal wastewater surveillance of four key pathogens during an unprecedented large-scale COVID-19 outbreak in China facilitated a novel strategy for addressing public health priorities–a proof of concept study. Water Res. 2023;247:120751. [DOI] [PubMed] [Google Scholar]
- 44.Ahmed W, Bertsch PM, Bibby K, et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ Res. 2020;191:110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vega T, Lozano JE, Meerhoff T, et al. Influenza surveillance in Europe: establishing epidemic thresholds by the moving epidemic method. Influenza Other Respir Viruses. 2013;7(4):546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilaru P, Hill D, Anderson K, et al. Wastewater surveillance for infectious disease: a systematic review. Am J Epidemiol. 2023;192(2):305–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adhikari S, Halden RU.. Opportunities and limits of wastewater-based epidemiology for tracking global health and attainment of UN sustainable development goals. Environ Int. 2022;163:107217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pecson BM, Darby E, Haas CN, et al. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the US. Environ Sci: Water Res Technol. 2021;7(3):504–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pilapil JD, Notarte KI, Yeung KL.. The dominance of co-circulating SARS-CoV-2 variants in wastewater. Int J Hyg Environ Health. 2023;253:114224. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Hosegood I, Powell D, et al. A global aircraft-based wastewater genomic surveillance network for early warning of future pandemics. Lancet Global Health. 2023;11(5):e791–e795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.